Effective Isolation for Lung Carcinoma Cells Based on Immunomagnetic Separation in a Microfluidic Channel
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
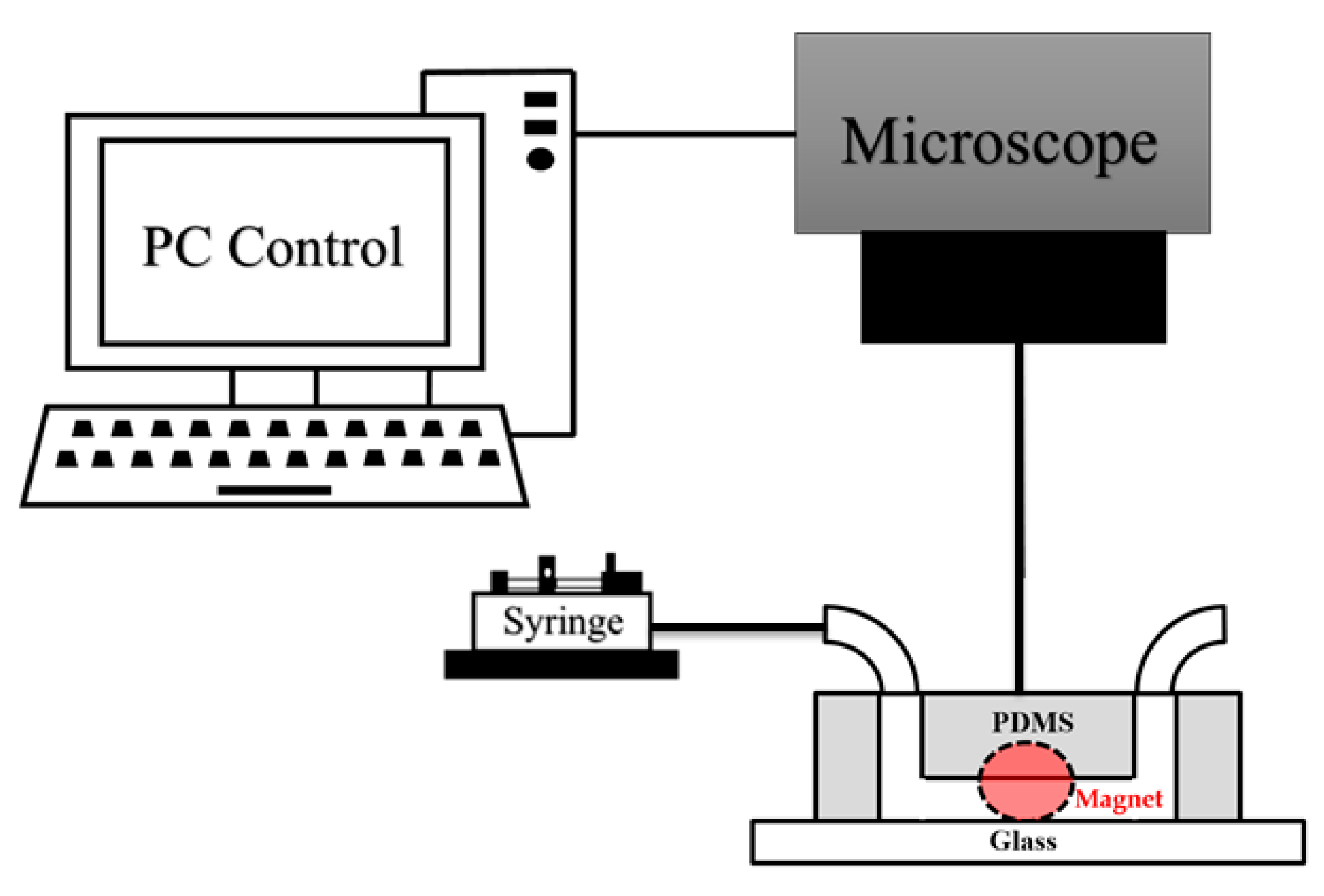
2.2. Microfluidic System Design and Fabrication
2.3. Theory
2.4. Magnetic Bead Conjugation
2.5. Isolation of A549 Cells
3. Results and Discussion
3.1. Characteristics of Functionalized A549 Cells
3.2. Effective Isolation of A549 Cells
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in globocan 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.C.; Lao, X.Q.; Ho, K.-F.; Goggins, W.B.; Shelly, L. Incidence and mortality of lung cancer: Global trends and association with socioeconomic status. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Siegel, R.L.; Jemal, A. Lung cancer statistics. In Lung Cancer and Personalized Medicine. Advances in Experimental Medicine and Biology; Ahmad, A., Gadgeel, S., Eds.; Springer: Cham, Switzerland, 2016; Volume 893, pp. 1–19. [Google Scholar]
- Foster, K.A.; Oster, C.G.; Mayer, M.M.; Avery, M.L.; Audus, K.L. Characterization of the a549 cell line as a type ii pulmonary epithelial cell model for drug metabolism. Exp. Cell Res. 1998, 243, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Luo, H.; Zhou, X.; Zhu, B.; Wang, Y.; Bian, X. Identification of cd90 as a marker for lung cancer stem cells in a549 and h446 cell lines. Oncol. Rep. 2013, 30, 2733–2740. [Google Scholar] [CrossRef] [PubMed]
- Balis, J.U.; Bumgarner, S.D.; Paciga, J.E.; Paterson, J.F.; Shelley, S.A. Synthesis of lung surfactant-associated glycoproteins by a549 cells: Description of an in vitro model for human type ii cell dysfunction. Exp. Lung Res. 1984, 6, 197–213. [Google Scholar] [CrossRef]
- Lieber, M.; Todaro, G.; Smith, B.; Szakal, A.; Nelson-Rees, W. A continuous tumor-cell line from a human lung carcinoma with properties of type ii alveolar epithelial cells. Int. J. Cancer 1976, 17, 62–70. [Google Scholar] [CrossRef]
- Nguyen, N.-V.; Yang, C.-H.; Liu, C.-J.; Kuo, C.-H.; Wu, D.-C.; Jen, C.-P. An aptamer-based capacitive sensing platform for specific detection of lung carcinoma cells in the microfluidic chip. Biosensors 2018, 8, 98. [Google Scholar] [CrossRef]
- Quoc, T.V.; Ngoc, V.N.; Bui, T.T.; Jen, C.-P.; Duc, T.C. High-frequency interdigitated array electrode-based capacitive biosensor for protein detection. BioChip J. 2019, 13, 1–13. [Google Scholar] [CrossRef]
- Iliuk, A.B.; Hu, L.; Tao, W.A. Aptamer in bioanalytical applications. Anal. Chem. 2011, 83, 4440–4452. [Google Scholar] [CrossRef]
- Quinn, J.G.; O’Neill, S.; Doyle, A.; McAtamney, C.; Diamond, D.; MacCraith, B.D.; O’Kennedy, R. Development and application of surface plasmon resonance-based biosensors for the detection of cell–ligand interactions. Anal. Biochem. 2000, 281, 135–143. [Google Scholar] [CrossRef]
- Ming, Y.; Li, Y.; Xing, H.; Luo, M.; Li, Z.; Chen, J.; Mo, J.; Shi, S. Circulating tumor cells: From theory to nanotechnology-based detection. Front. Pharmacol. 2017, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Gou, Y.; Zhang, S.; Sun, C.; Wang, P.; You, Z.; Yalikun, Y.; Tanaka, Y.; Ren, D. Sheathless inertial focusing chip combining a spiral channel with periodic expansion structures for efficient and stable particle sorting. Anal. Chem. 2019, 92, 1833–1841. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Hassanisaber, H.; Yu, R.; Ma, S.; Verbridge, S.S.; Lu, C. Paramagnetic structures within a microfluidic channel for enhanced immunomagnetic isolation and surface patterning of cells. Sci. Rep. 2016, 6, 29407. [Google Scholar] [CrossRef] [PubMed]
- Park, M.-H.; Reátegui, E.; Li, W.; Tessier, S.N.; Wong, K.H.; Jensen, A.E.; Thapar, V.; Ting, D.; Toner, M.; Stott, S.L. Enhanced isolation and release of circulating tumor cells using nanoparticle binding and ligand exchange in a microfluidic chip. J. Am. Chem. Soc. 2017, 139, 2741–2749. [Google Scholar] [CrossRef]
- Chang, W.C.; Lee, L.P.; Liepmann, D. Adhesion-based capture and separation of cells for microfluidic devices. MRS Online Proc. Libr. Arch. 2002, 729. [Google Scholar] [CrossRef]
- Lin, S.; Zhi, X.; Chen, D.; Xia, F.; Shen, Y.; Niu, J.; Huang, S.; Song, J.; Miao, J.; Cui, D. A flyover style microfluidic chip for highly purified magnetic cell separation. Biosens. Bioelectron. 2019, 129, 175–181. [Google Scholar] [CrossRef]
- Jankovicova, B.; Svobodova, Z.; Hromadkova, L.; Kupcik, R.; Ripova, D.; Bilkova, Z. Benefits of immunomagnetic separation for epitope identification in clinically important protein antigens: A case study using ovalbumin, carbonic anhydrase I and Tau protein. Univers. J. Biomed. Eng. 2015, 3, 1–8. [Google Scholar] [CrossRef]
- Olsvik, O.; Popovic, T.; Skjerve, E.; Cudjoe, K.S.; Hornes, E.; Ugelstad, J.; Uhlen, M. Magnetic separation techniques in diagnostic microbiology. Clin. Microbiol. Rev. 1994, 7, 43–54. [Google Scholar] [CrossRef]
- Yaman, S.; Anil-Inevi, M.; Ozcivici, E.; Tekin, H.C. Magnetic force-based microfluidic techniques for cellular and tissue bioengineering. Front. Bioeng. Biotechnol. 2018, 6, 192. [Google Scholar] [CrossRef]
- Zhang, R.-Q.; Liu, S.-L.; Zhao, W.; Zhang, W.-P.; Yu, X.; Li, Y.; Li, A.-J.; Pang, D.-W.; Zhang, Z.-L. A simple point-of-care microfluidic immunomagnetic fluorescence assay for pathogens. Anal. Chem. 2013, 85, 2645–2651. [Google Scholar] [CrossRef]
- Ruzycka, M.; Cimpan, M.R.; Rios-Mondragon, I.; Grudzinski, I.P. Microfluidics for studying metastatic patterns of lung cancer. J. Nanobiotechnol. 2019, 17, 71. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Phillips, J.A.; Yan, J.; Li, Q.; Fan, Z.H.; Tan, W. Aptamer-based microfluidic device for enrichment, sorting, and detection of multiple cancer cells. Anal. Chem. 2009, 81, 7436–7442. [Google Scholar] [CrossRef] [PubMed]
- Wild, D. The Immunoassay Handbook: Theory and Applications of Ligand Binding, ELISA and Related Techniques; Newnes: Burlington, MA, USA, 2013. [Google Scholar]
- Lakhin, A.; Tarantul, V.; Gening, L. Aptamers: Problems, solutions and prospects. Acta Nat. 2013, 5, 34–43. [Google Scholar] [CrossRef]
- Zhao, Z.; Xu, L.; Shi, X.; Tan, W.; Fang, X.; Shangguan, D. Recognition of subtype non-small cell lung cancer by DNA aptamers selected from living cells. Analyst 2009, 134, 1808–1814. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.-V.; Jen, C.-P. Selective detection of human lung adenocarcinoma cells based on the aptamer-conjugated self-assembled monolayer of gold nanoparticles. Micromachines 2019, 10, 195. [Google Scholar] [CrossRef]
- Sharma, R.; Agrawal, V.V.; Sharma, P.; Varshney, R.; Sinha, R.; Malhotra, B. Aptamer based electrochemical sensor for detection of human lung adenocarcinoma a549 cells. J. Phys. Conf. Ser. 2012, 358, 012001. [Google Scholar] [CrossRef]
- Ni, S.; Yao, H.; Wang, L.; Lu, J.; Jiang, F.; Lu, A.; Zhang, G. Chemical Modifications of Nucleic Acid Aptamers for Therapeutic Purposes. Int. J. Mol. Sci. 2017, 18, 1683. [Google Scholar] [CrossRef]
- Nguyen, N.-V.; Jen, C.-P. Impedance detection integrated with dielectrophoresis enrichment platform for lung circulating tumor cells in a microfluidic channel. Biosens. Bioelectron. 2018, 121, 10–18. [Google Scholar] [CrossRef]
- Hejazian, M.; Li, W.; Nguyen, N.-T. Lab on a chip for continuous-flow magnetic cell separation. Lab Chip 2015, 15, 959–970. [Google Scholar] [CrossRef]
- Kwak, B.; Lee, J.; Lee, D.; Lee, K.; Kwon, O.; Kang, S.; Kim, Y. Selective isolation of magnetic nanoparticle-mediated heterogeneity subpopulation of circulating tumor cells using magnetic gradient based microfluidic system. Biosens. Bioelectron. 2017, 88, 153–158. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vu-Dinh, H.; Feng, H.; Jen, C.-P. Effective Isolation for Lung Carcinoma Cells Based on Immunomagnetic Separation in a Microfluidic Channel. Biosensors 2021, 11, 23. https://doi.org/10.3390/bios11010023
Vu-Dinh H, Feng H, Jen C-P. Effective Isolation for Lung Carcinoma Cells Based on Immunomagnetic Separation in a Microfluidic Channel. Biosensors. 2021; 11(1):23. https://doi.org/10.3390/bios11010023
Chicago/Turabian StyleVu-Dinh, Hien, Hui Feng, and Chun-Ping Jen. 2021. "Effective Isolation for Lung Carcinoma Cells Based on Immunomagnetic Separation in a Microfluidic Channel" Biosensors 11, no. 1: 23. https://doi.org/10.3390/bios11010023
APA StyleVu-Dinh, H., Feng, H., & Jen, C.-P. (2021). Effective Isolation for Lung Carcinoma Cells Based on Immunomagnetic Separation in a Microfluidic Channel. Biosensors, 11(1), 23. https://doi.org/10.3390/bios11010023






