2LTRZFP Interacts Specifically to HIV-1 DNA without Off-Target Effects as Determined by Biolayer Interferometry
Abstract
1. Introduction
2. Materials and Methods
2.1. Recombinant 2LTRZFP-GFP and HIV-1 Integrase (IN) Enzyme Production and Purification
2.2. Double-Stranded 2LTR DNA Preparation
2.3. Determination of the Binding Activity of 2LTRZFP-GFP towards Target DNA by Enzyme-Linked Immunosorbent Assay (ELISA)
2.4. Binding Kinetic Analysis by Bio-Layer Interferometry
2.5. Statistics
3. Results
3.1. Preparation of Recombinant Proteins
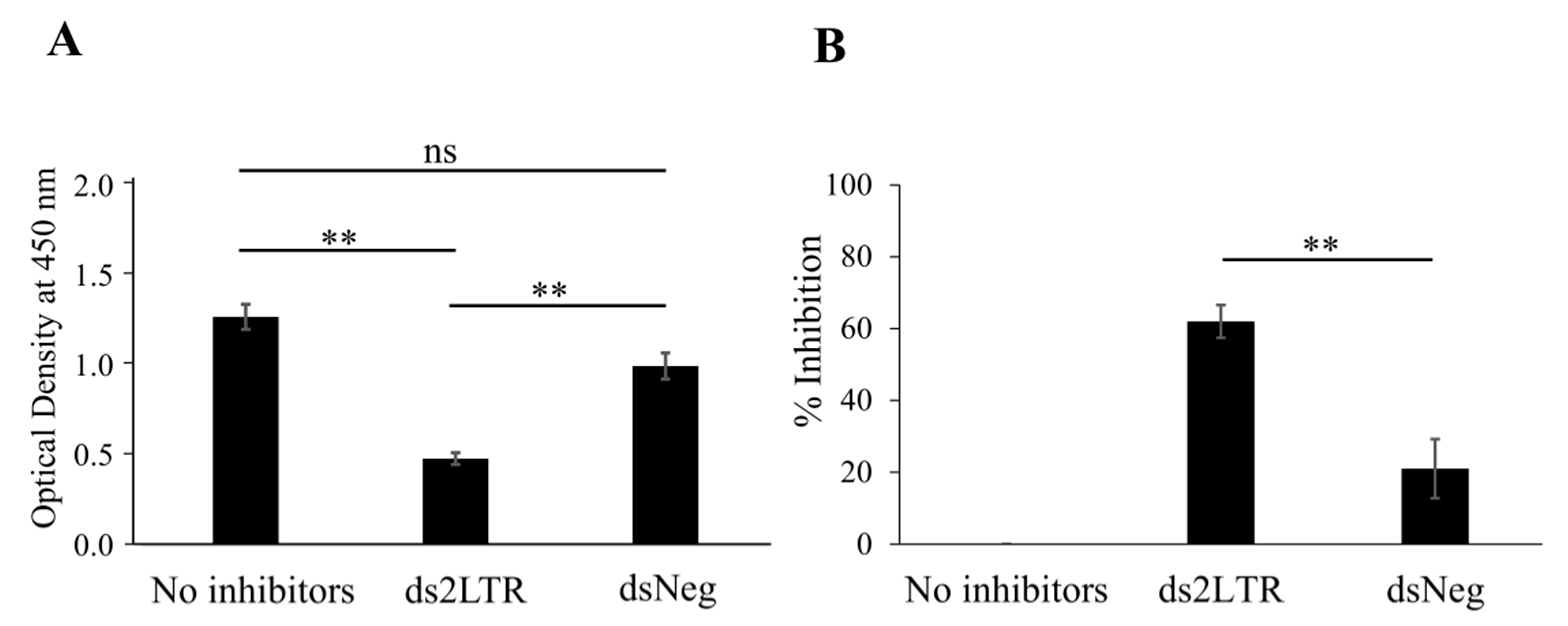
3.2. Determination of 2LTRZFP-GFP Binding to Its Target DNA
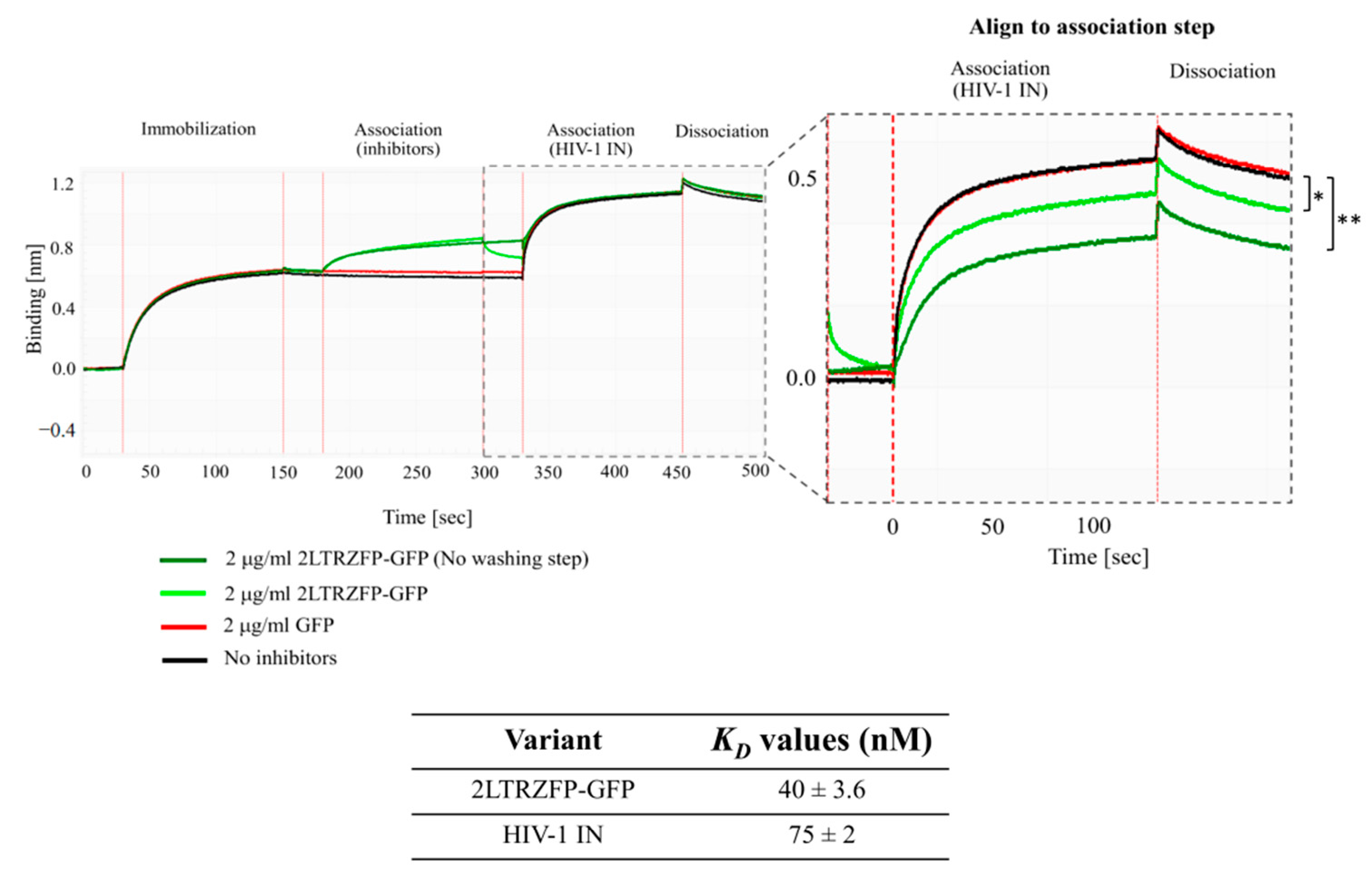
3.3. Binding Kinetics of 2LTRZFP-GFP Using BLItz
3.4. Study of the Inhibitory Effect of 2LTRZFP-GFP on HIV-1 IN Enzyme
3.5. Off-Target Effect of 2LTRZFP-GFP to SupT1 Genomic DNA
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, M.J.; Kim, J.; Li, S.; Zaia, J.; Yee, J.K.; Anderson, J.; Akkina, R.; Rossi, J.J. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol. Ther. 2005, 12, 900–909. [Google Scholar] [CrossRef]
- Mitsuyasu, R.T.; Merigan, T.C.; Carr, A.; Zack, J.A.; Winters, M.A.; Workman, C.; Bloch, M.; Lalezari, J.; Becker, S.; Thornton, L.; et al. Phase 2 gene therapy trial of an anti-HIV ribozyme in autologous CD34+ cells. Nat. Med. 2009, 15, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Binz, H.K.; Stumpp, M.T.; Forrer, P.; Amstutz, P.; Pluckthun, A. Designing repeat proteins: Well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J. Mol. Biol. 2003, 332, 489–503. [Google Scholar] [CrossRef]
- Boersma, Y.L.; Pluckthun, A. DARPins and other repeat protein scaffolds: Advances in engineering and applications. Curr. Opin. Biotechnol. 2011, 22, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Nangola, S.; Urvoas, A.; Valerio-Lepiniec, M.; Khamaikawin, W.; Sakkhachornphop, S.; Hong, S.S.; Boulanger, P.; Minard, P.; Tayapiwatana, C. Antiviral activity of recombinant ankyrin targeted to the capsid domain of HIV-1 Gag polyprotein. Retrovirology 2012, 9, 17. [Google Scholar] [CrossRef] [PubMed]
- Holt, N.; Wang, J.; Kim, K.; Friedman, G.; Wang, X.; Taupin, V.; Crooks, G.M.; Kohn, D.B.; Gregory, P.D.; Holmes, M.C.; et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 2010, 28, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Saydaminova, K.; Ye, X.; Wang, H.; Richter, M.; Ho, M.; Chen, H.; Xu, N.; Kim, J.S.; Papapetrou, E.; Holmes, M.C.; et al. Efficient genome editing in hematopoietic stem cells with helper-dependent Ad5/35 vectors expressing site-specific endonucleases under microRNA regulation. Mol. Ther. Methods Clin. Dev. 2015, 1, 14057. [Google Scholar] [CrossRef] [PubMed]
- Sakkhachornphop, S.; Jiranusornkul, S.; Kodchakorn, K.; Nangola, S.; Sirisanthana, T.; Tayapiwatana, C. Designed zinc finger protein interacting with the HIV-1 integrase recognition sequence at 2-LTR-circle junctions. Protein Sci. 2009, 18, 2219–2230. [Google Scholar] [CrossRef]
- Sakkhachornphop, S.; Barbas, C.F., 3rd; Keawvichit, R.; Wongworapat, K.; Tayapiwatana, C. Zinc finger protein designed to target 2-long terminal repeat junctions interferes with human immunodeficiency virus integration. Hum. Gene Ther. 2012, 23, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Khamaikawin, W.; Saoin, S.; Nangola, S.; Chupradit, K.; Sakkhachornphop, S.; Hadpech, S.; Onlamoon, N.; Ansari, A.A.; Byrareddy, S.N.; Boulanger, P.; et al. Combined Antiviral Therapy Using Designed Molecular Scaffolds Targeting Two Distinct Viral Functions, HIV-1 Genome Integration and Capsid Assembly. Mol. Ther. Nucleic Acids. 2015, 4, e249. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [PubMed]
- Barski, A.; Zhao, K. Genomic location analysis by ChIP-Seq. J. Cell. Biochem. 2009, 107, 11–18. [Google Scholar] [CrossRef]
- Indyk, L.; Fisher, H.F. Theoretical aspects of isothermal titration calorimetry. Methods Enzymol. 1998, 295, 350–364. [Google Scholar] [PubMed]
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface plasmon resonance: A versatile technique for biosensor applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef]
- Rich, R.L.; Myszka, D.G. Why you should be using more SPR biosensor technology. Drug Discov. Today Technol. 2004, 1, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Concepcion, J.; Witte, K.; Wartchow, C.; Choo, S.; Yao, D.; Persson, H.; Wei, J.; Li, P.; Heidecker, B.; Ma, W.; et al. Label-free detection of biomolecular interactions using BioLayer interferometry for kinetic characterization. Comb. Chem. High Throughput Screen. 2009, 12, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Abdiche, Y.; Malashock, D.; Pinkerton, A.; Pons, J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal. Biochem. 2008, 377, 209–217. [Google Scholar] [CrossRef]
- Wilson, J.L.; Scott, I.M.; McMurry, J.L. Optical biosensing: Kinetics of protein A-IGG binding using biolayer interferometry. Biochem. Mol. Biol. Educ. 2010, 38, 400–407. [Google Scholar] [CrossRef]
- Li, D.; Wei, T.; Jin, H.; Rose, A.; Wang, R.; Lin, M.H.; Spann, K.; Harrich, D. Binding of the eukaryotic translation elongation factor 1A with the 5’UTR of HIV-1 genomic RNA is important for reverse transcription. Virol. J. 2015, 12, 118. [Google Scholar] [CrossRef]
- West, G.M.; Tucker, C.L.; Xu, T.; Park, S.K.; Han, X.; Yates, J.R., 3rd; Fitzgerald, M.C. Quantitative proteomics approach for identifying protein-drug interactions in complex mixtures using protein stability measurements. Proc. Natl. Acad. Sci. USA 2010, 107, 9078–9082. [Google Scholar] [CrossRef]
- Lagasse, H.A.; Alexaki, A.; Simhadri, V.L.; Katagiri, N.H.; Jankowski, W.; Sauna, Z.E.; Kimchi-Sarfaty, C. Recent advances in (therapeutic protein) drug development. F1000Research 2017, 6, 113. [Google Scholar] [CrossRef]
- Dimitrov, D.S. Therapeutic proteins. Methods Mol. Biol. 2012, 899, 1–26. [Google Scholar] [PubMed]
- Werkmeister, J.A.; Ramshaw, J.A. Recombinant protein scaffolds for tissue engineering. Biomed. Mater. 2012, 7, 012002. [Google Scholar] [CrossRef]
- Petersen, R.L. Strategies Using Bio-Layer Interferometry Biosensor Technology for Vaccine Research and Development. Biosensors 2017, 7, 49. [Google Scholar] [CrossRef] [PubMed]
- Verzijl, D.; Riedl, T.; Pwhi, P.; Gerritsen, A.F. A novel label-free cell-based assay technology using biolayer interferometry. Biosens. Bioelectron. 2017, 87, 388–395. [Google Scholar] [CrossRef]
- Muller-Esparza, H.; Osorio-Valeriano, M.; Steube, N.; Thanbichler, M.; Randau, L. Bio-Layer Interferometry Analysis of the Target Binding Activity of CRISPR-Cas Effector Complexes. Front. Mol. Biosci. 2020, 7, 98. [Google Scholar] [CrossRef] [PubMed]
- Cradick, T.J.; Fine, E.J.; Antico, C.J.; Bao, G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013, 41, 9584–9592. [Google Scholar] [CrossRef]
- Xie, L.; Xie, L.; Bourne, P.E. Structure-based systems biology for analyzing off-target binding. Curr. Opin. Struct. Biol. 2011, 21, 189–199. [Google Scholar] [CrossRef]
- Kuscu, C.; Arslan, S.; Singh, R.; Thorpe, J.; Adli, M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 2014, 32, 677–683. [Google Scholar] [CrossRef]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef]
- Lin, Y.; Cradick, T.J.; Brown, M.T.; Deshmukh, H.; Ranjan, P.; Sarode, N.; Wile, B.M.; Vertino, P.M.; Stewart, F.J.; Bao, G. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014, 42, 7473–7485. [Google Scholar] [CrossRef] [PubMed]
- Coelho, M.A.; De Braekeleer, E.; Firth, M.; Bista, M.; Lukasiak, S.; Cuomo, M.E.; Taylor, B.J.M. CRISPR GUARD protects off-target sites from Cas9 nuclease activity using short guide RNAs. Nat. Commun. 2020, 11, 4132. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chupradit, K.; Thongkum, W.; Juntit, O.-a.; Sornsuwan, K.; Tayapiwatana, C. 2LTRZFP Interacts Specifically to HIV-1 DNA without Off-Target Effects as Determined by Biolayer Interferometry. Biosensors 2021, 11, 76. https://doi.org/10.3390/bios11030076
Chupradit K, Thongkum W, Juntit O-a, Sornsuwan K, Tayapiwatana C. 2LTRZFP Interacts Specifically to HIV-1 DNA without Off-Target Effects as Determined by Biolayer Interferometry. Biosensors. 2021; 11(3):76. https://doi.org/10.3390/bios11030076
Chicago/Turabian StyleChupradit, Koollawat, Weeraya Thongkum, On-anong Juntit, Kanokporn Sornsuwan, and Chatchai Tayapiwatana. 2021. "2LTRZFP Interacts Specifically to HIV-1 DNA without Off-Target Effects as Determined by Biolayer Interferometry" Biosensors 11, no. 3: 76. https://doi.org/10.3390/bios11030076
APA StyleChupradit, K., Thongkum, W., Juntit, O.-a., Sornsuwan, K., & Tayapiwatana, C. (2021). 2LTRZFP Interacts Specifically to HIV-1 DNA without Off-Target Effects as Determined by Biolayer Interferometry. Biosensors, 11(3), 76. https://doi.org/10.3390/bios11030076



