Biosensing Amplification by Hybridization Chain Reaction on Phase-Sensitive Surface Plasmon Resonance
Abstract
1. Introduction
2. Materials and Methods
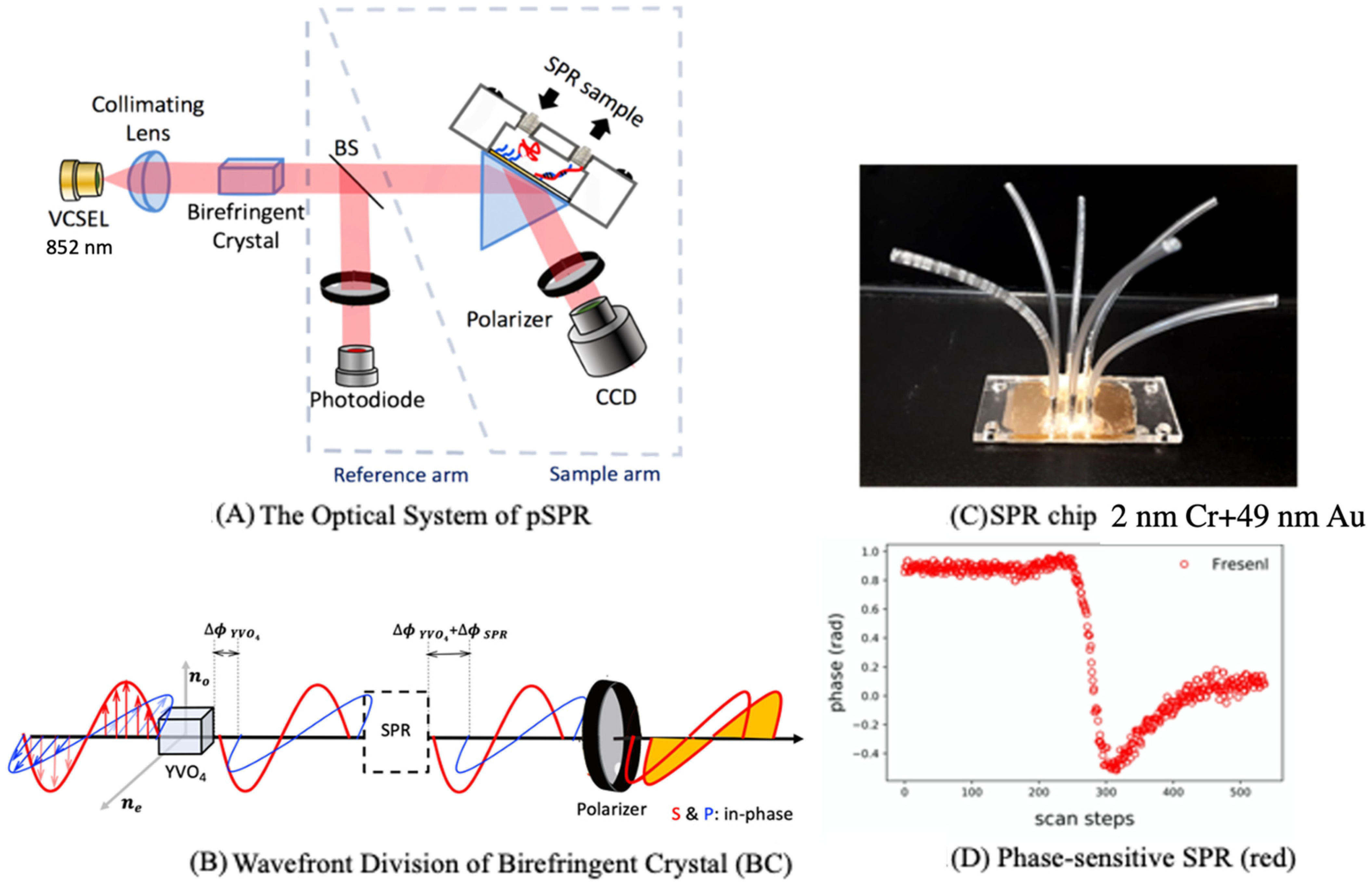
2.1. Phase-Sensitive Surface Plasmon Resonance (pSPR)
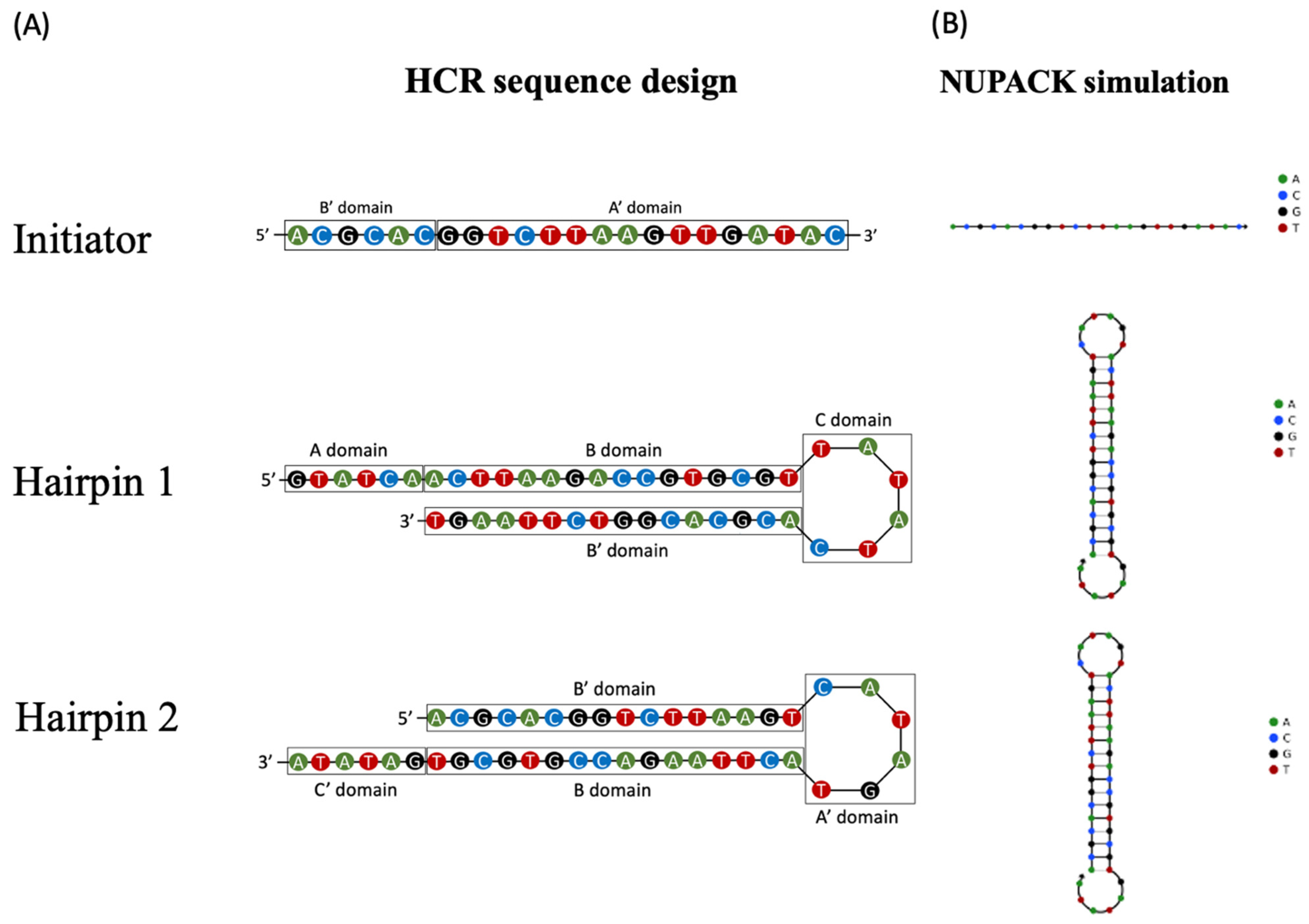
2.2. DNA Aptamers
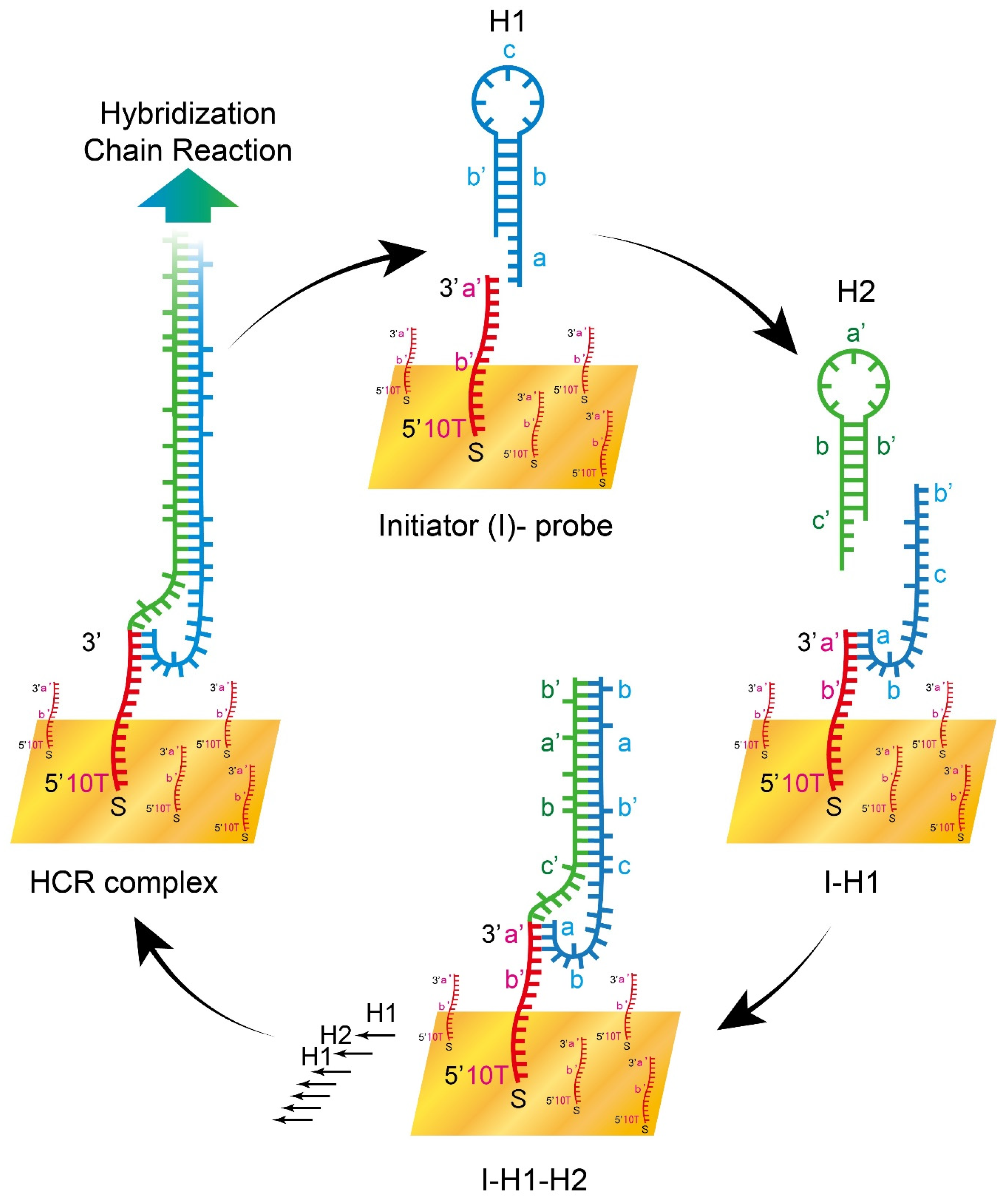
2.3. HCR on pSPR with Pre-Immobilized Initiator Probe
2.4. DNA Gel Electrophoresis
3. Results
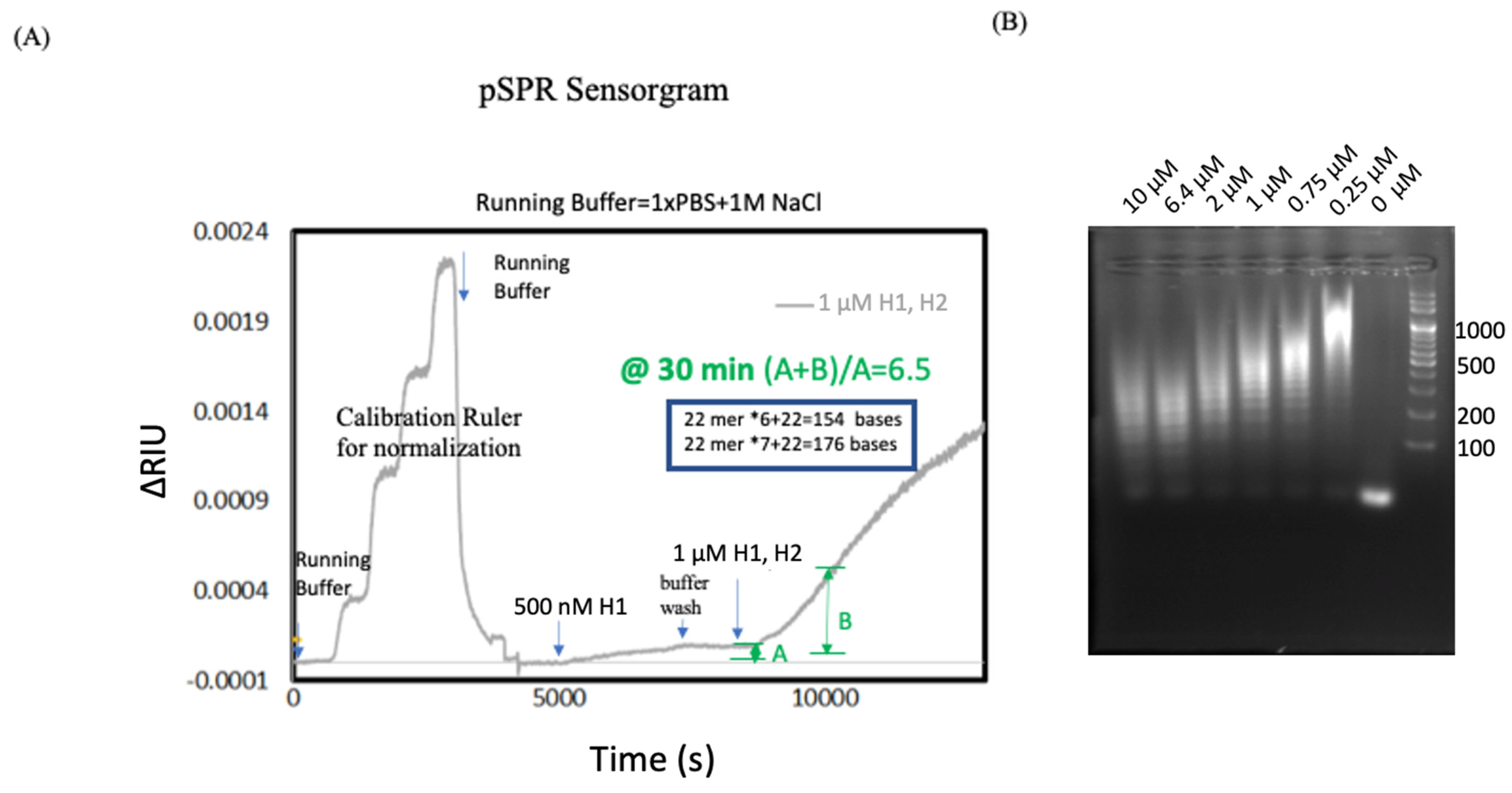
3.1. Validation of HCR on pSPR with Pre-Immobilized Initiator Probe
3.2. Combining pSPR and DNA Gel Electrophoresis
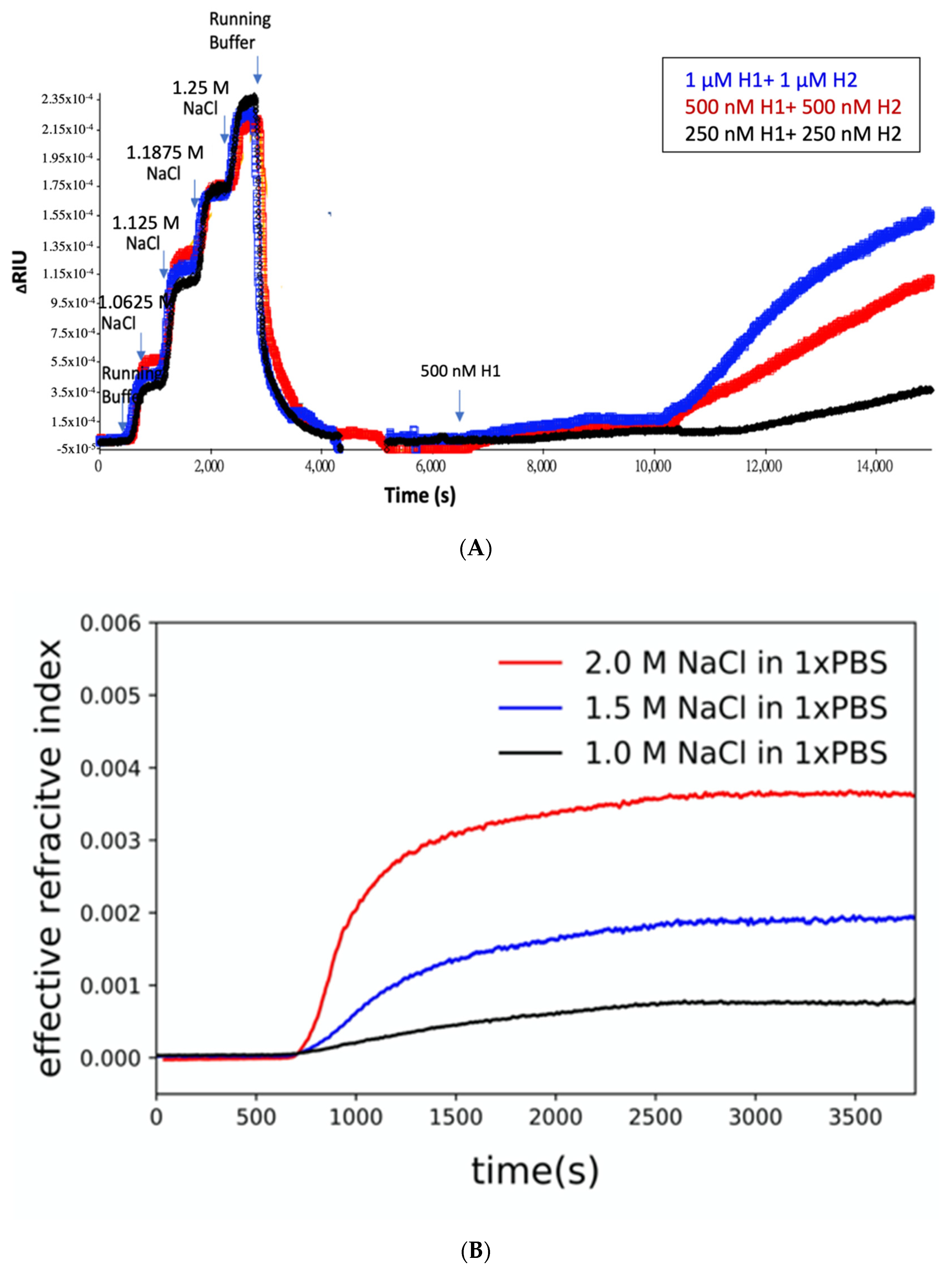
3.3. The Effect of Aptamer and Salt Concentration on HCR Amplification
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Homola, J. Surface plasmon resonance sensors for detection of chemical and biological species. Chem. Rev. 2008, 108, 462–493. [Google Scholar] [CrossRef]
- Wark, A.W.; Lee, H.J.; Corn, R.M. Multiplexed detection methods for profiling microRNA expression in biological samples. Angew. Chem. Int. Ed. 2008, 47, 644–652. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Musick, M.D.; Nicewarner, S.R.; Salinas, F.G.; Benkovic, S.J.; Natan, M.J.; Keating, C.D. Colloidal Au-Enhanced Surface Plasmon Resonance for Ultrasensitive Detection of DNA Hybridization. J. Am. Chem. Soc. 2000, 122, 9071–9077. [Google Scholar] [CrossRef]
- Zhang, Z.; Cheng, Q.; Feng, P. Selective removal of DNA-labeled nanoparticles from planar substrates by DNA displacement reactions. Angew. Chem. Int. Ed. 2009, 48, 118–122. [Google Scholar] [CrossRef]
- Yi, X.; Hao, Y.; Ning, X.; Wang, J.; Quintero, M.; Li, D.; Zhou, F. Sensitive and continuous screening of inhibitors of β-site amyloid precursor protein cleaving enzyme 1 (BACE1) at single SPR chips. Anal. Chem. 2013, 85, 3660–3666. [Google Scholar] [CrossRef]
- Bai, Y.; Feng, F.; Zhao, L.; Wang, C.; Wang, H.; Tian, M.; Qin, J.; Duan, Y.; He, X. Aptamer/thrombin/aptamer-AuNPs sandwich enhanced surface plasmon resonance sensor for the detection of subnanomolar thrombin. Biosens. Bioelectron. 2013, 47, 265–270. [Google Scholar] [CrossRef]
- Rich, R.L.; Myszka, D.G. Advances in surface plasmon resonance biosensor analysis. Curr. Opin. Biotechnol. 2000, 11, 54–61. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Wang, L.; Wei, Q. A surface plasmon resonance assay coupled with a hybridization chain reaction for amplified detection of DNA and small molecules. Chem. Commun. 2014, 50, 5049–5052. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Wang, P.; Yu, X. Phase-Sensitive Surface Plasmon Resonance Sensors: Recent Progress and Future Prospects. Sensors 2017, 17, 2819. [Google Scholar] [CrossRef]
- Nelson, S.G.; Johnston, K.S.; Yee, S.S. High sensitivity surface plasmon resonace sensor based on phase detection. Sens. Actuators B Chem. 1996, 35, 187–191. [Google Scholar] [CrossRef]
- Kabashin, A.V.; Patskovsky, S.; Grigorenko, A.N. Phase and amplitude sensitivities in surface plasmon resonance bio and chemical sensing. Opt. Express 2009, 17, 21191–21204. [Google Scholar] [CrossRef]
- Spiga, F.M.; Bonyár, A.; Ring, B.; Onofri, M.; Vinelli, A.; Sántha, H.; Guiducci, C.; Zuccheri, G. Hybridization chain reaction performed on a metal surface as a means of signal amplification in SPR and electrochemical biosensors. Biosens. Bioelectron. 2014, 54, 102–108. [Google Scholar] [CrossRef]
- Luan, Q.; Xue, Y.; Yao, X. A simple hairpin DNA sensor for label-free detection of sub-attomole DNA target. Sens. Actuators B Chem. 2010, 147, 561–565. [Google Scholar] [CrossRef]
- Gorodkiewicz, E.; Lukaszewski, Z. Recent Progress in Surface Plasmon Resonance Biosensors (2016 to Mid-2018). Biosensors 2018, 8, 132. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface plasmon resonance: A versatile technique for biosensor applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef] [PubMed]
- Šípová, H.; Homola, J. Surface plasmon resonance sensing of nucleic acids: A review. Anal. Chim. Acta 2013, 773, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Zadeh, J.N.; Steenberg, C.D.; Bois, J.S.; Wolfe, B.R.; Pierce, M.B.; Khan, A.R.; Dirks, R.M.; Pierce, N.A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 32, 170–173. [Google Scholar] [CrossRef]
- Cai, S.; Yan, J.; Xiong, H.; Liu, Y.; Peng, D.; Liu, Z. Investigations on the interface of nucleic acid aptamers and binding targets. Analyst 2018, 143, 5317–5338. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Gómez, R.; Fernández-Alonso, N.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J. Unravelling the lipocalin 2 interaction with aptamers: May rolling circle amplification improve their functional affinity? Talanta 2019, 197, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Song, W.; Guo, X.; Wang, Z. Ultrasensitive detection of nucleic acids and proteins using quartz crystal microbalance and surface plasmon resonance sensors based on target-triggering multiple signal amplification strategy. Anal. Chim. Acta 2017, 978, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Fathi, F.; Rashidi, M.-R.; Omidi, Y. Ultra-sensitive detection by metal nanoparticles-mediated enhanced SPR biosensors. Talanta 2019, 192, 118–127. [Google Scholar] [CrossRef]
- Li, Q.; Wang, Q.; Yang, X.; Wang, K.; Zhang, H.; Nie, W. High sensitivity surface plasmon resonance biosensor for detection of microRNA and small molecule based on graphene oxide-gold nanoparticles composites. Talanta 2017, 174, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Song, J.; Li, M.X.; Zhang, T.T.; Zhao, W.; Xu, J.J.; Liu, M.; Chen, H.Y. Ultrasensitive MicroRNA Assay via Surface Plasmon Resonance Responses of Au@Ag Nanorods Etching. Anal. Chem. 2017, 89, 10585–10591. [Google Scholar] [CrossRef]
- Wang, C.; Liu, M.; Wang, Z.; Li, S.; Deng, Y.; He, N. Point-of-care diagnostics for infectious diseases: From methods to devices. Nano Today 2021, 37, 101092. [Google Scholar] [CrossRef] [PubMed]
- Diao, W.; Tang, M.; Ding, S.; Li, X.; Cheng, W.; Mo, F.; Yan, X.; Ma, H.; Yan, Y. Highly sensitive surface plasmon resonance biosensor for the detection of HIV-related DNA based on dynamic and structural DNA nanodevices. Biosens. Bioelectron. 2018, 100, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Falkowski, P.; Lukaszewski, Z.; Gorodkiewicz, E. Potential of surface plasmon resonance biosensors in cancer detection. J. Pharm. Biomed. Anal. 2021, 194, 113802. [Google Scholar] [CrossRef]
- Dirks, R.M.; Pierce, N.A. Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. USA 2004, 101, 15275–15278. [Google Scholar] [CrossRef]
- Bi, S.; Yue, S.; Zhang, S. Hybridization chain reaction: A versatile molecular tool for biosensing, bioimaging, and biomedicine. Chem. Soc. Rev. 2017, 46, 4281–4298. [Google Scholar] [CrossRef]
- Wu, T.-H.; Chang, C.-C.; Yang, C.-H.; Lin, W.-Y.; Ee, T.J.; Lin, C.-W. Hybridization Chain Reactions Targeting the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Int. J. Mol. Sci. 2020, 21, 3216. [Google Scholar] [CrossRef]
- Abid, S.A.; Suhail, A.; Al-Kadmy, I.M.S.; Sattar, A.A.; Beshbishy, A.M.; Batiha, G.E.; Hetta, H.F. Biosensors as a future diagnostic approach for COVID-19. Life Sci. 2021, 119117. [Google Scholar] [CrossRef]
- Du, A.; Zheng, R.; Disoma, C.; Li, S.; Chen, Z.; Li, S.; Liu, P.; Zhou, Y.; Shen, Y.; Liu, S.; et al. Epigallocatechin-3-gallate, an active ingredient of Traditional Chinese Medicines, inhibits the 3CLpro activity of SARS-CoV-2. Int. J. Biol. Macromol. 2021, 176, 1–12. [Google Scholar] [CrossRef]
- Yokoyama, K.; Ichiki, A. Nano-size dependence in the adsorption by the SARS-CoV-2 spike protein over gold colloid. Colloids Surf. A Physicochem. Eng. Asp. 2021, 615, 126275. [Google Scholar] [CrossRef]
- Yokoyama, K.; Ichiki, A. Spectroscopic investigation on the affinity of SARS-CoV-2 spike protein to gold nano-particles. Colloid Interface Sci. Commun. 2021, 40, 100356. [Google Scholar] [CrossRef]
- Wu, T.-H.; Chang, C.-C.; Vaillant, J.; Bruyant, A.; Lin, C.-W. DNA biosensor combining single-wavelength colorimetry and a digital lock-in amplifier within a smartphone. Lab Chip 2016, 16, 4527–4533. [Google Scholar] [CrossRef]
- Liang, Y.H.; Chang, C.C.; Chen, C.C.; Chu-Su, Y.; Lin, C.W. Development of an Au/ZnO thin film surface plasmon resonance-based biosensor immunoassay for the detection of carbohydrate antigen 15-3 in human saliva. Clin. Biochem. 2012, 45, 1689–1693. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Rana, S.; Giese, N.; Büchler, M.W.; Zöller, M. Tspan8, CD44v6 and α6β4 are biomarkers of migrating pancreatic cancer-initiating cells. Int. J. Cancer 2013, 133, 416–426. [Google Scholar] [CrossRef]
- Chang, C.C.; Lin, S.; Lee, C.H.; Chuang, T.L.; Hsueh, P.R.; Lai, H.C.; Lin, C.W. Amplified surface plasmon resonance immunosensor for interferon-gamma based on a streptavidin-incorporated aptamer. Biosens. Bioelectron. 2012, 37, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Suebsing, R.; Prombun, P.; Kiatpathomchai, W. Reverse transcription loop-mediated isothermal amplification (RT-LAMP) combined with colorimetric gold nanoparticle (AuNP) probe assay for visual detection of Penaeus vannamei nodavirus (Pv NV). Lett. Appl. Microbiol. 2013, 56, 428–435. [Google Scholar] [CrossRef]
- Podushkina, D.; West, N.W.; Golenberg, E.M. Utilizing multiplex fluor LAMPs to illuminate multiple gene expressions in situ. PLoS ONE 2019, 14, e0223333. [Google Scholar] [CrossRef] [PubMed]
- Gantt, S.; Goldfarb, D.M.; Park, A.; Rawlinson, W.; Boppana, S.B.; Lazzarotto, T.; Mertz, L.M. Performance of the Alethia CMV Assay for Detection of Cytomegalovirus by Use of Neonatal Saliva Swabs. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef] [PubMed]
- Tehrani, B.S.; Mirzajani, E.; Fallahi, S.; Naeini, K.M.; Mahmoudi, M.R.; Kavishahi, M.S.; Eskandari, V.; Zebardast, N. Challenging TaqMan probe-based real-time PCR and loop-mediated isothermal amplification (LAMP): The two sensitive molecular techniques for the detection of toxoplasmosis, a potentially dangerous opportunistic infection in immunocompromised patients. Arch. Microbiol. 2020, 202, 1881–1888. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef] [PubMed]
| Aptamer Sequence (5′→3′) | |
|---|---|
| Initiator | |
| (thiolated-10T + 22 mer) | HS-TTTTTTTTTTACGCACGGTCTTAAGTTGATAC |
| H1 (44 mer) | GTATCAACTTAAGACCGTGCGTTATATCACGCACGGTCTTAAGT |
| H2 (44 mer) | ACGCACGGTCTTAAGTTGATACACTTAAGACCGTGCGTGATATA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.-H.; Wu, T.-H.; Chang, C.-C.; Lo, H.-Y.; Liu, H.-W.; Huang, N.-T.; Lin, C.-W. Biosensing Amplification by Hybridization Chain Reaction on Phase-Sensitive Surface Plasmon Resonance. Biosensors 2021, 11, 75. https://doi.org/10.3390/bios11030075
Yang C-H, Wu T-H, Chang C-C, Lo H-Y, Liu H-W, Huang N-T, Lin C-W. Biosensing Amplification by Hybridization Chain Reaction on Phase-Sensitive Surface Plasmon Resonance. Biosensors. 2021; 11(3):75. https://doi.org/10.3390/bios11030075
Chicago/Turabian StyleYang, Ching-Hsu, Tzu-Heng Wu, Chia-Chen Chang, Hui-Yun Lo, Hui-Wen Liu, Nien-Tsu Huang, and Chii-Wann Lin. 2021. "Biosensing Amplification by Hybridization Chain Reaction on Phase-Sensitive Surface Plasmon Resonance" Biosensors 11, no. 3: 75. https://doi.org/10.3390/bios11030075
APA StyleYang, C.-H., Wu, T.-H., Chang, C.-C., Lo, H.-Y., Liu, H.-W., Huang, N.-T., & Lin, C.-W. (2021). Biosensing Amplification by Hybridization Chain Reaction on Phase-Sensitive Surface Plasmon Resonance. Biosensors, 11(3), 75. https://doi.org/10.3390/bios11030075







