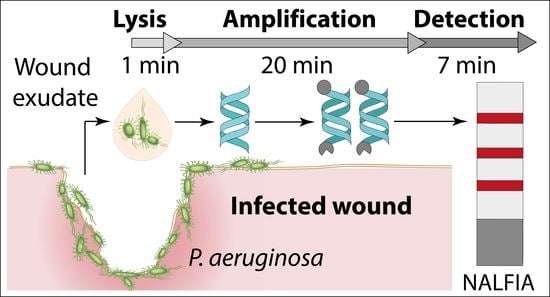
Rapid Detection of Pathogens in Wound Exudate via Nucleic Acid Lateral Flow Immunoassay
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Bacterial Strains and Culture Media
2.3. Spiked Wound Exudate
2.4. Procedure of Bacterial Lysis
2.5. Design of Primers, Probes and Internal Amplification Control
2.6. Synthesis of Antibody-Conjugated Fluorescence Microspheres
2.7. Assembly of the Lateral Flow Dipstick
2.8. RPA-Lateral Flow Assay Procedure
2.9. Detecting P. aeruginosa in Spiked Wound Exudate or Buffer
2.10. Statistical Analysis
3. Results
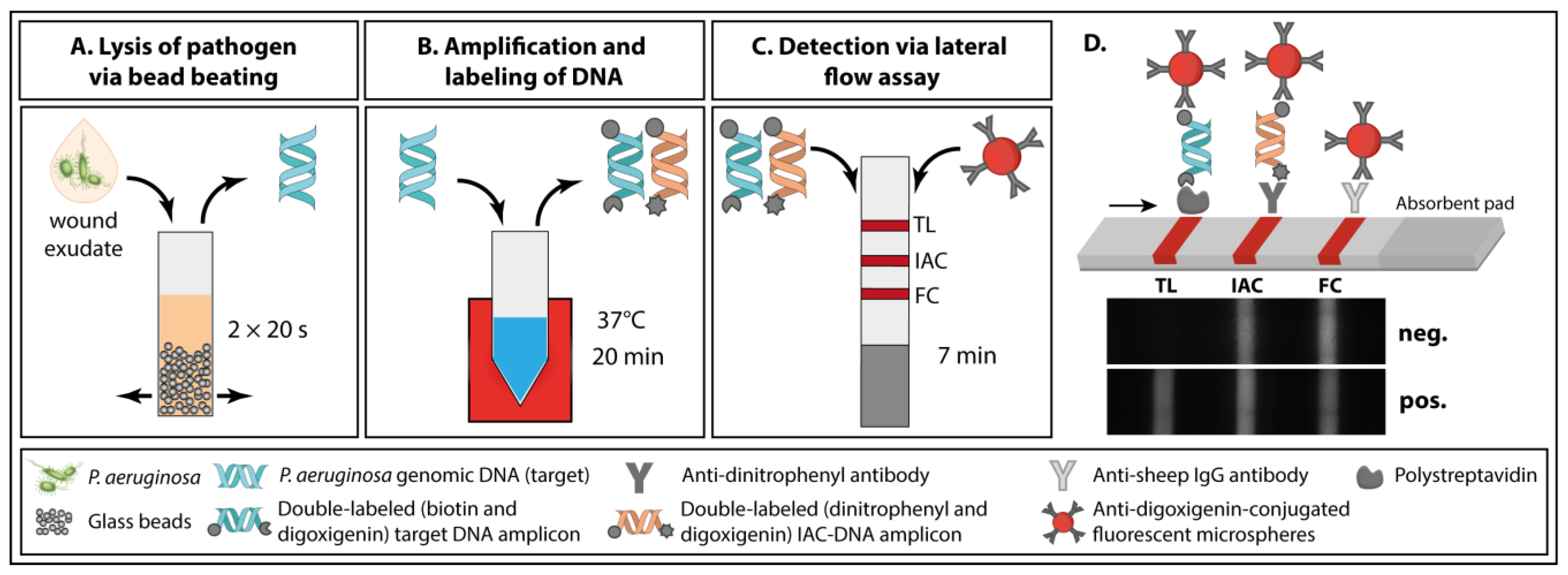
3.1. Principle of the Paper-Based Approach for Pathogen Detection in Wound Exudate
3.2. Analytical Performance of the Paper-Based Approach for Pathogen Detection
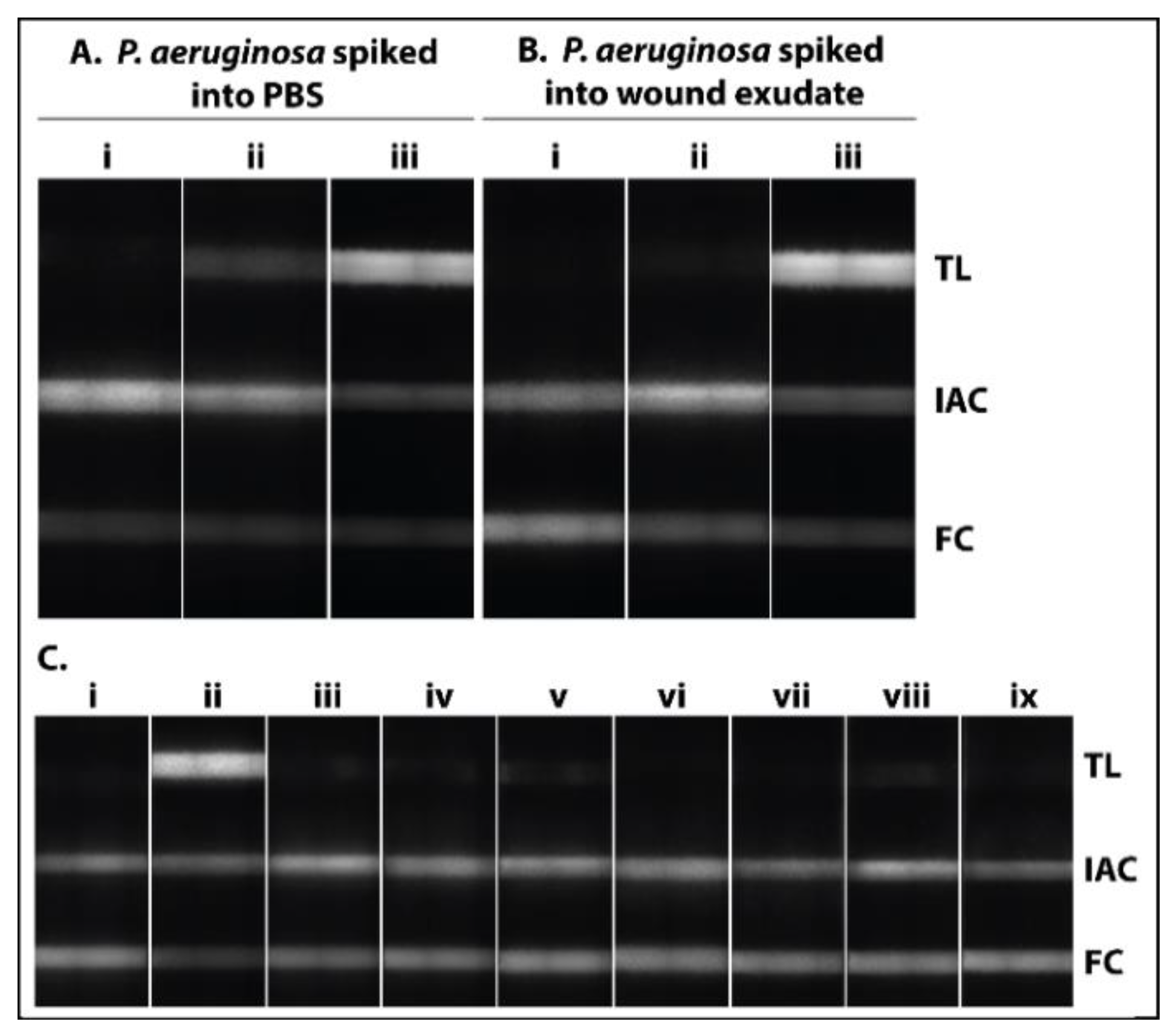
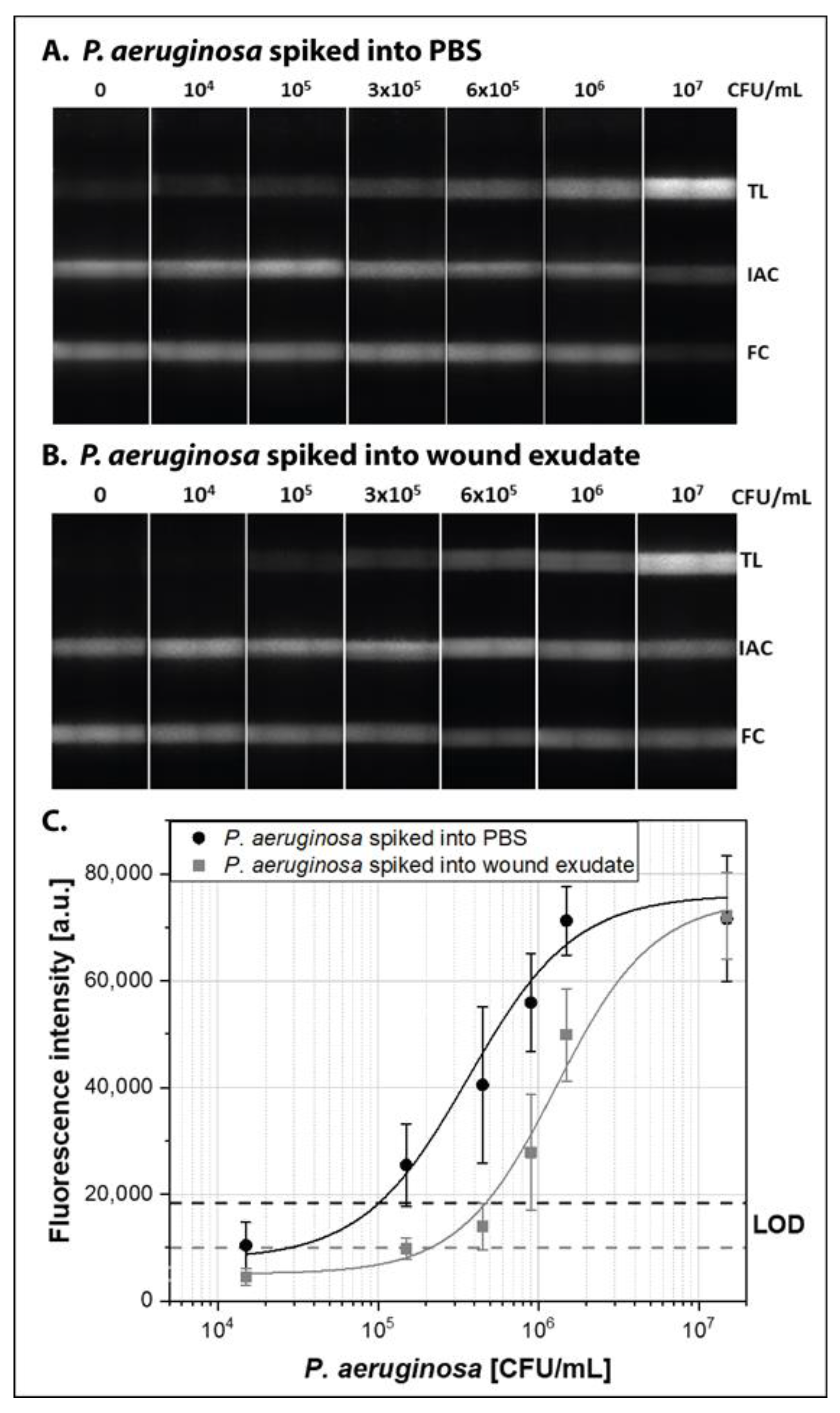
3.3. Demonstration with P. aeruginosa Spiked Human Wound Exudate
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Behnke, M.; Hansen, S.; Leistner, R.; Diaz, L.A.P.; Gropmann, A.; Sohr, D.; Gastmeier, P.; Piening, B. Nosocomial infection and antibiotic use: A second national prevalence study in Germany. Dtsch. Arztebl. Int. 2013, 110, 627–633. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Prävention postoperativer Wundinfektionen: Empfehlung der Kommission für Krankenhaushygiene und Infektionsprävention (KRINKO) beim Robert Koch-Institut. Bundesgesundheitsblatt Gesundh. Gesundh. 2018, 61, 448–473. [CrossRef]
- Anderson, D.J.; Podgorny, K.; Berríos-Torres, S.I.; Bratzler, D.W.; Dellinger, E.P.; Greene, L.; Nyquist, A.-C.; Saiman, L.; Yokoe, D.S.; Maragakis, L.L.; et al. Strategies to prevent surgical site infections in acute care hospitals: 2014 update. Infect. Control Hosp. Epidemiol. 2014, 35, 605–627. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L. Labor und Diagnose: Indikation und Bewertung von Laborbefunden für die medizinische Diagnostik, 8th ed.; Th-Books: Frankfurt/Main, Germany, 2012; ISBN 9783980521581. [Google Scholar]
- Leekha, S.; Terrell, C.L.; Edson, R.S. General principles of antimicrobial therapy. Mayo Clin. Proc. 2011, 86, 156–167. [Google Scholar] [CrossRef]
- Llor, C.; Bjerrum, L. Antimicrobial resistance: Risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. Drug Saf. 2014, 5, 229–241. [Google Scholar] [CrossRef]
- Dargaville, T.R.; Farrugia, B.L.; Broadbent, J.A.; Pace, S.; Upton, Z.; Voelcker, N.H. Sensors and imaging for wound healing: A review. Biosens. Bioelectron. 2013, 41, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Brunauer, A.; Ates, H.C.; Dincer, C.; Früh, S.M. Integrated paper-based sensing devices for diagnostic applications. In Paper Based Sensors; Elsevier: Amsterdam, The Netherlands, 2020; pp. 397–450. ISBN 9780444643452. [Google Scholar]
- Ramsay, S.; Cowan, L.; Davidson, J.M.; Nanney, L.; Schultz, G. Wound samples: Moving towards a standardised method of collection and analysis. Int. Wound J. 2016, 13, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—Occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef]
- Hoorfar, J.; Malorny, B.; Abdulmawjood, A.; Cook, N.; Wagner, M.; Fach, P. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J. Clin. Microbiol. 2004, 42, 1863–1868. [Google Scholar] [CrossRef]
- Mauk, M.G.; Song, J.; Liu, C.; Bau, H.H. Simple Approaches to Minimally-Instrumented, Microfluidic-Based Point-of-Care Nucleic Acid Amplification Tests. Biosensors 2018, 8, 17. [Google Scholar] [CrossRef]
- Niemz, A.; Ferguson, T.M.; Boyle, D.S. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011, 29, 240–250. [Google Scholar] [CrossRef]
- Zhang, L.; Ding, B.; Chen, Q.; Feng, Q.; Lin, L.; Sun, J. Point-of-care-testing of nucleic acids by microfluidics. Trends Anal. Chem. 2017, 94, 106–116. [Google Scholar] [CrossRef]
- Becherer, L.; Hess, J.F.; Frischmann, S.; Bakheit, M.; Nitschko, H.; Stinco, S.; Zitz, F.; Hofer, H.; Porro, G.; Hausladen, F.; et al. Point-of-Care System for HTLV-1 Proviral Load Quantification by Digital Mediator Displacement LAMP. Micromachines 2021, 12, 159. [Google Scholar] [CrossRef]
- Schulz, M.; Calabrese, S.; Hausladen, F.; Wurm, H.; Drossart, D.; Stock, K.; Sobieraj, A.M.; Eichenseher, F.; Loessner, M.J.; Schmelcher, M.; et al. Point-of-care testing system for digital single cell detection of MRSA directly from nasal swabs. Lab Chip 2020, 20, 2549–2561. [Google Scholar] [CrossRef]
- Cesewski, E.; Johnson, B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020, 159, 112214. [Google Scholar] [CrossRef]
- Formisano, N.; Bhalla, N.; Heeran, M.; Reyes Martinez, J.; Sarkar, A.; Laabei, M.; Jolly, P.; Bowen, C.R.; Taylor, J.T.; Flitsch, S.; et al. Inexpensive and fast pathogenic bacteria screening using field-effect transistors. Biosens. Bioelectron. 2016, 85, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Mukherjee, M.D.; Sumana, G.; Gupta, R.K.; Sood, S.; Malhotra, B.D. Biosensors for pathogen detection: A smart approach towards clinical diagnosis. Sens. Actuators B Chem. 2014, 197, 385–404. [Google Scholar] [CrossRef]
- Koczula, K.M.; Gallotta, A. Lateral flow assays. Essays Biochem. 2016, 60, 111–120. [Google Scholar] [CrossRef]
- Dincer, C.; Bruch, R.; Costa-Rama, E.; Fernández-Abedul, M.T.; Merkoçi, A.; Manz, A.; Urban, G.A.; Güder, F. Disposable Sensors in Diagnostics, Food, and Environmental Monitoring. Adv. Mater. 2019, 31, e1806739. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Toley, B.J. Paper-based nucleic acid amplification tests for point-of-care diagnostics. Analyst 2018, 143, 2213–2234. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Nolder, D.; Reboud, J.; Oguike, M.C.; van Schalkwyk, D.A.; Sutherland, C.J.; Cooper, J.M. Paper-Origami-Based Multiplexed Malaria Diagnostics from Whole Blood. Angew. Chem. Int. Ed. Engl. 2016, 55, 15250–15253. [Google Scholar] [CrossRef]
- Choi, J.R.; Hu, J.; Tang, R.; Gong, Y.; Feng, S.; Ren, H.; Wen, T.; Li, X.; Wan Abas, W.A.B.; Pingguan-Murphy, B.; et al. An integrated paper-based sample-to-answer biosensor for nucleic acid testing at the point of care. Lab Chip 2016, 16, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Bender, A.T.; Borysiak, M.D.; Levenson, A.M.; Lillis, L.; Boyle, D.S.; Posner, J.D. Semiquantitative Nucleic Acid Test with Simultaneous Isotachophoretic Extraction and Amplification. Anal. Chem. 2018, 90, 7221–7229. [Google Scholar] [CrossRef] [PubMed]
- Phillips, E.A.; Moehling, T.J.; Ejendal, K.F.K.; Hoilett, O.S.; Byers, K.M.; Basing, L.A.; Jankowski, L.A.; Bennett, J.B.; Lin, L.-K.; Stanciu, L.A.; et al. Microfluidic rapid and autonomous analytical device (microRAAD) to detect HIV from whole blood samples. Lab Chip 2019, 19, 3375–3386. [Google Scholar] [CrossRef]
- Dou, M.; Sanjay, S.T.; Dominguez, D.C.; Liu, P.; Xu, F.; Li, X. Multiplexed instrument-free meningitis diagnosis on a polymer/paper hybrid microfluidic biochip. Biosens. Bioelectron. 2017, 87, 865–873. [Google Scholar] [CrossRef] [PubMed]
- De Silva, R.F.; Longo Cesar Paixão, T.R.; Der Torossian Torres, M.; de Araujo, W.R. Simple and inexpensive electrochemical paper-based analytical device for sensitive detection of Pseudomonas aeruginosa. Sens. Actuators B Chem. 2020, 308, 127669. [Google Scholar] [CrossRef]
- Zhang, Y.; Bai, J.; Ying, J.Y. A stacking flow immunoassay for the detection of dengue-specific immunoglobulins in salivary fluid. Lab Chip 2015, 15, 1465–1471. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Abe, K.; Bennett, S.; Liang, T.; Ladd, P.D.; Yokobe, L.; Anderson, C.E.; Shah, K.; Bishop, J.; Purfield, M.; et al. Disposable Autonomous Device for Swab-to-Result Diagnosis of Influenza. Anal. Chem. 2017, 89, 5776–5783. [Google Scholar] [CrossRef]
- Anderson, C.E.; Buser, J.R.; Fleming, A.M.; Strauch, E.-M.; Ladd, P.D.; Englund, J.; Baker, D.; Yager, P. An integrated device for the rapid and sensitive detection of the influenza hemagglutinin. Lab Chip 2019, 19, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, N.M.; Linnes, J.C.; Fan, A.; Ellenson, C.K.; Pollock, N.R.; Klapperich, C.M. Paper-Based RNA Extraction, in Situ Isothermal Amplification, and Lateral Flow Detection for Low-Cost, Rapid Diagnosis of Influenza A (H1N1) from Clinical Specimens. Anal. Chem. 2015, 87, 7872–7879. [Google Scholar] [CrossRef]
- Rodriguez, N.M.; Wong, W.S.; Liu, L.; Dewar, R.; Klapperich, C.M. A fully integrated paperfluidic molecular diagnostic chip for the extraction, amplification, and detection of nucleic acids from clinical samples. Lab Chip 2016, 16, 753–763. [Google Scholar] [CrossRef]
- Yang, Z.; Xu, G.; Reboud, J.; Ali, S.A.; Kaur, G.; McGiven, J.; Boby, N.; Gupta, P.K.; Chaudhuri, P.; Cooper, J.M. Rapid Veterinary Diagnosis of Bovine Reproductive Infectious Diseases from Semen Using Paper-Origami DNA Microfluidics. ACS Sens. 2018, 3, 403–409. [Google Scholar] [CrossRef]
- Hui, C.Y.; Liu, M.; Li, Y.; Brennan, J.D. A Paper Sensor Printed with Multifunctional Bio/Nano Materials. Angew. Chem. Int. Ed. Engl. 2018, 57, 4549–4553. [Google Scholar] [CrossRef] [PubMed]
- Crannell, Z.; Castellanos-Gonzalez, A.; Nair, G.; Mejia, R.; White, A.C.; Richards-Kortum, R. Multiplexed Recombinase Polymerase Amplification Assay to Detect Intestinal Protozoa. Anal. Chem. 2016, 88, 1610–1616. [Google Scholar] [CrossRef]
- Daher, R.K.; Stewart, G.; Boissinot, M.; Bergeron, M.G. Recombinase Polymerase Amplification for Diagnostic Applications. Clin. Chem. 2016, 62, 947–958. [Google Scholar] [CrossRef]
- Yi, T.-T.; Zhang, H.-Y.; Liang, H.; Gong, G.-Z.; Cai, Y. Betaine-assisted recombinase polymerase assay for rapid hepatitis B virus detection. Biotechnol. Appl. Biochem. 2020. [Google Scholar] [CrossRef]
- Kortli, S.; Jauset-Rubio, M.; Tomaso, H.; Abbas, M.N.; Bashammakh, A.S.; El-Shahawi, M.S.; Alyoubi, A.O.; Ben-Ali, M.; O’Sullivan, C.K. Yersinia pestis detection using biotinylated dNTPs for signal enhancement in lateral flow assays. Anal. Chim. Acta 2020, 1112, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-Z.; Fang, D.-Z.; Chen, F.-F.; Zhao, Q.-F.; Cai, C.-M.; Cheng, M.-G. Utilization of recombinase polymerase amplification method combined with lateral flow dipstick for visual detection of respiratory syncytial virus. Mol. Cell. Probes 2020, 49, 101473. [Google Scholar] [CrossRef]
- Qi, Y.; Shao, Y.; Rao, J.; Shen, W.; Yin, Q.; Li, X.; Chen, H.; Li, J.; Zeng, W.; Zheng, S.; et al. Development of a rapid and visual detection method for Rickettsia rickettsii combining recombinase polymerase assay with lateral flow test. PLoS ONE 2018, 13, e0207811. [Google Scholar] [CrossRef] [PubMed]
- Poulton, K.; Webster, B. Development of a lateral flow recombinase polymerase assay for the diagnosis of Schistosoma mansoni infections. Anal. Biochem. 2018, 546, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Jauset-Rubio, M.; Tomaso, H.; El-Shahawi, M.S.; Bashammakh, A.S.; Al-Youbi, A.O.; O’Sullivan, C.K. Duplex Lateral Flow Assay for the Simultaneous Detection of Yersinia pestis and Francisella tularensis. Anal. Chem. 2018, 90, 12745–12751. [Google Scholar] [CrossRef]
- Natoli, M.E.; Rohrman, B.A.; De Santiago, C.; van Zyl, G.U.; Richards-Kortum, R.R. Paper-based detection of HIV-1 drug resistance using isothermal amplification and an oligonucleotide ligation assay. Anal. Biochem. 2018, 544, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Jia, T.; Yu, Y.; Wang, Y. A recombinase polymerase amplification-based lateral flow strip assay for rapid detection of genogroup II noroviruses in the field. Arch. Virol. 2020, 165, 2767–2776. [Google Scholar] [CrossRef]
- Bai, X.; Ma, X.; Li, M.; Li, X.; Fan, G.; Zhang, R.; Wang, R.; Duan, Q.; Shen, X.; Xie, Y.; et al. Field applicable detection of hepatitis B virus using internal controlled duplex recombinase-aided amplification assay and lateral flow dipstick assay. J. Med. Virol. 2020. [Google Scholar] [CrossRef]
- Lalremruata, A.; Nguyen, T.T.; McCall, M.B.B.; Mombo-Ngoma, G.; Agnandji, S.T.; Adegnika, A.A.; Lell, B.; Ramharter, M.; Hoffman, S.L.; Kremsner, P.G.; et al. Recombinase Polymerase Amplification and Lateral Flow Assay for Ultrasensitive Detection of Low-Density Plasmodium falciparum Infection from Controlled Human Malaria Infection Studies and Naturally Acquired Infections. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef]
- Ma, B.; Fang, J.; Lin, W.; Yu, X.; Sun, C.; Zhang, M. A simple and efficient method for potential point-of-care diagnosis of human papillomavirus genotypes: Combination of isothermal recombinase polymerase amplification with lateral flow dipstick and reverse dot blot. Anal. Bioanal. Chem. 2019, 411, 7451–7460. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Yao, J.; Yuan, S.; Liu, H.; Wei, N.; Zhang, J.; Shan, W. Development of a lateral flow recombinase polymerase amplification assay for rapid and visual detection of Cryptococcus neoformans/C. gattii in cerebral spinal fluid. BMC Infect. Dis. 2019, 19, 108. [Google Scholar] [CrossRef]
- Srisrattakarn, A.; Tippayawat, P.; Chanawong, A.; Tavichakorntrakool, R.; Daduang, J.; Wonglakorn, L.; Sooksongsoontorn, P.; Lulitanond, A. Direct detection of methicillin-resistant in Staphylococcus spp. in positive blood culture by isothermal recombinase polymerase amplification combined with lateral flow dipstick assay. World J. Microbiol. Biotechnol. 2020, 36, 162. [Google Scholar] [CrossRef] [PubMed]
- Molina-Gonzalez, S.J.; Bhattacharyya, T.; AlShehri, H.R.; Poulton, K.; Allen, S.; Miles, M.A.; Arianitwe, M.; Tukahebwa, E.M.; Webster, B.; Russell Stothard, J.; et al. Application of a recombinase polymerase amplification (RPA) assay and pilot field testing for Giardia duodenalis at Lake Albert, Uganda. Parasit. Vectors 2020, 13, 289. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Luo, Y.; Li, H.; Wu, W.; Ruan, X.; Mu, X. Rapid visual detection of dengue virus by combining reverse transcription recombinase-aided amplification with lateral-flow dipstick assay. Int. J. Infect. Dis. 2020, 95, 406–412. [Google Scholar] [CrossRef]
- Sun, N.; Wang, Y.; Yao, X.; Chen, F.; Gao, D.; Wang, W.; Li, X. Visual signal generation for the detection of influenza viruses by duplex recombinase polymerase amplification with lateral flow dipsticks. Anal. Bioanal. Chem. 2019, 411, 3591–3602. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Wu, H.; Zhao, K.; Hu, C.; Ge, Y.; Zhu, X.; Zhang, X.; Zhou, M.; Zhu, F.; Cui, L. Rapid detection of human mastadenovirus species B by recombinase polymerase amplification assay. BMC Microbiol. 2019, 19, 8. [Google Scholar] [CrossRef]
- Sun, N.; Wang, W.; Wang, J.; Yao, X.; Chen, F.; Li, X.; Yinglei, Y.; Chen, B. Reverse transcription recombinase polymerase amplification with lateral flow dipsticks for detection of influenza A virus and subtyping of H1 and H3. Mol. Cell. Probes 2018, 42, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Li, X.; Peng, B.; Wu, W.; Wang, X.; Liu, H.; Yuan, L.; Fang, S.; Lu, J. Rapid Detection of Avian Influenza A Virus (H7N9) by Lateral Flow Dipstick Recombinase Polymerase Amplification. Biol. Pharm. Bull. 2018, 41, 1804–1808. [Google Scholar] [CrossRef] [PubMed]
- James, A.S.; Todd, S.; Pollak, N.M.; Marsh, G.A.; Macdonald, J. Ebolavirus diagnosis made simple, comparable and faster than molecular detection methods: Preparing for the future. Virol. J. 2018, 15, 75. [Google Scholar] [CrossRef]
- Lai, M.-Y.; Ooi, C.-H.; Lau, Y.-L. Recombinase Polymerase Amplification Combined with a Lateral Flow Strip for the Detection of Plasmodium knowlesi. Am. J. Trop. Med. Hyg. 2018, 98, 700–703. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Liu, H.; Ye, F.; Xiang, G.; Shan, W.; Xing, W. Rapid and visual detection of Mycobacterium tuberculosis complex using recombinase polymerase amplification combined with lateral flow strips. Mol. Cell. Probes 2017, 36, 43–49. [Google Scholar] [CrossRef]
- Mesaros, N.; Nordmann, P.; Plésiat, P.; Roussel-Delvallez, M.; van Eldere, J.; Glupczynski, Y.; van Laethem, Y.; Jacobs, F.; Lebecque, P.; Malfroot, A.; et al. Pseudomonas aeruginosa: Resistance and therapeutic options at the turn of the new millennium. Clin. Microbiol. Infect. 2007, 13, 560–578. [Google Scholar] [CrossRef]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef] [PubMed]
- Klebes, M.; Ulrich, C.; Kluschke, F.; Patzelt, A.; Vandersee, S.; Richter, H.; Bob, A.; von Hutten, J.; Krediet, J.T.; Kramer, A.; et al. Combined antibacterial effects of tissue-tolerable plasma and a modern conventional liquid antiseptic on chronic wound treatment. J. Biophotonics 2015, 8, 382–391. [Google Scholar] [CrossRef]
- Bui, U.T.; Finlayson, K.; Edwards, H. The diagnosis of infection in chronic leg ulcers: A narrative review on clinical practice. Int. Wound J. 2019, 16, 601–620. [Google Scholar] [CrossRef]
- Baranoski, S. Wound Care Essentials, 2nd ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2008; ISBN 978-1-58255-469-3. [Google Scholar]
- Bowler, P.G.; Duerden, B.I.; Armstrong, D.G. Wound microbiology and associated approaches to wound management. Clin. Microbiol. Rev. 2001, 14, 244–269. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.; Harding, K.G. Bacteria and wound healing. Curr. Opin. Infect. Dis. 2004, 17, 91–96. [Google Scholar] [CrossRef] [PubMed]
- TwistDx Limited. TwistAmp® DNA Amplification Kits—Assay Design Manual. Available online: https://www.twistdx.co.uk/docs/default-source/RPA-assay-design/twistamp-assay-design-manual-v2-5.pdf?sfvrsn=b3be0efc_29 (accessed on 7 June 2020).
- Raja, B.; Goux, H.J.; Marapadaga, A.; Rajagopalan, S.; Kourentzi, K.; Willson, R.C. Development of a panel of recombinase polymerase amplification assays for detection of common bacterial urinary tract infection pathogens. J. Appl. Microbiol. 2017, 123, 544–555. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Armbruster, D.A.; Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008, 29 (Suppl. S1), S49–S52. [Google Scholar]
- De Boer, R.; Peters, R.; Gierveld, S.; Schuurman, T.; Kooistra-Smid, M.; Savelkoul, P. Improved detection of microbial DNA after bead-beating before DNA isolation. J. Microbiol. Methods 2010, 80, 209–211. [Google Scholar] [CrossRef] [PubMed]
- Shehadul Islam, M.; Aryasomayajula, A.; Selvaganapathy, P. A Review on Macroscale and Microscale Cell Lysis Methods. Micromachines 2017, 8, 83. [Google Scholar] [CrossRef]
- Lee, L.G.; Nordman, E.S.; Johnson, M.D.; Oldham, M.F. A low-cost, high-performance system for fluorescence lateral flow assays. Biosensors 2013, 3, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jin, G.; Liu, L.; Kuang, H.; Xiao, J.; Xu, C. A portable fluorescent microsphere-based lateral flow immunosensor for the simultaneous detection of colistin and bacitracin in milk. Analyst 2020. [Google Scholar] [CrossRef] [PubMed]
- O’Farrell, B. Evolution in Lateral Flow–Based Immunoassay Systems. In Lateral Flow Immunoassay; Wong, R., Tse, H., Eds.; Humana Press: Totowa, NJ, USA, 2009; pp. 1–33. ISBN 978-1-58829-908-6. [Google Scholar]
- Walker, F.M.; Hsieh, K. Advances in Directly Amplifying Nucleic Acids from Complex Samples. Biosensors 2019, 9, 117. [Google Scholar] [CrossRef]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase polymerase amplification: Basics, applications and recent advances. Trends Analyt. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.-H.; Zhang, H.; Zhang, Y.; Li, X.-N.; Shen, X.-X.; Qi, J.-J.; Fan, G.-H.; Xiang, X.-Y.; Zhan, Z.-F.; Chen, Z.-W.; et al. Development and evaluation of recombinase-aided amplification assays incorporating competitive internal controls for detection of human adenovirus serotypes 3 and 7. Virol. J. 2019, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Crannell, Z.A.; Rohrman, B.; Richards-Kortum, R. Development of a quantitative recombinase polymerase amplification assay with an internal positive control. J. Vis. Exp. 2015. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.-L.; Wei, S.; Gooneratne, R.; Mutukumira, A.N.; Ma, X.-J.; Tang, S.-Z.; Wu, X.-Y. Development of a recombinase polymerase amplification assay for Vibrio parahaemolyticus detection with an internal amplification control. Can. J. Microbiol. 2018, 64, 223–230. [Google Scholar] [CrossRef]
- Higgins, O.; Clancy, E.; Forrest, M.S.; Piepenburg, O.; Cormican, M.; Boo, T.W.; O’Sullivan, N.; McGuinness, C.; Cafferty, D.; Cunney, R.; et al. Duplex recombinase polymerase amplification assays incorporating competitive internal controls for bacterial meningitis detection. Anal. Biochem. 2018, 546, 10–16. [Google Scholar] [CrossRef]
- Crannell, Z.A.; Rohrman, B.; Richards-Kortum, R. Quantification of HIV-1 DNA using real-time recombinase polymerase amplification. Anal. Chem. 2014, 86, 5615–5619. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.-Q.; Li, G.-X.; Li, X.-N.; Shen, X.-X.; Gao, Y.; Wang, L.; Fan, T.; Duan, Q.-X.; Wang, Y.-K.; Wang, J.; et al. A rapid and sensitive recombinase aided amplification assay incorporating competitive internal control to detect Bordetella pertussis using the DNA obtained by boiling. Int. J. Infect. Dis. 2019, 86, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Li, X.; Zhang, Y.; Shen, X.; Song, G.; Pan, J.; Fan, T.; Wang, R.; Li, L.; Ma, X. Development of a duplex reverse transcription recombinase-aided amplification assay for respiratory syncytial virus incorporating an internal control. Arch. Virol. 2019, 164, 1843–1850. [Google Scholar] [CrossRef] [PubMed]
- Thet, N.T.; Alves, D.R.; Bean, J.E.; Booth, S.; Nzakizwanayo, J.; Young, A.E.R.; Jones, B.V.; Jenkins, A.T.A. Prototype Development of the Intelligent Hydrogel Wound Dressing and Its Efficacy in the Detection of Model Pathogenic Wound Biofilms. ACS Appl. Mater. Interfaces 2016, 8, 14909–14919. [Google Scholar] [CrossRef] [PubMed]
- Mok, E.; Wee, E.; Wang, Y.; Trau, M. Comprehensive evaluation of molecular enhancers of the isothermal exponential amplification reaction. Sci. Rep. 2016, 6, 37837. [Google Scholar] [CrossRef]
- Dincer, C.; Bruch, R.; Kling, A.; Dittrich, P.S.; Urban, G.A. Multiplexed Point-of-Care Testing—xPOCT. Trends Biotechnol. 2017, 35, 728–742. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Macdonald, J. Multiplexed lateral flow biosensors: Technological advances for radically improving point-of-care diagnoses. Biosens. Bioelectron. 2016, 83, 177–192. [Google Scholar] [CrossRef] [PubMed]
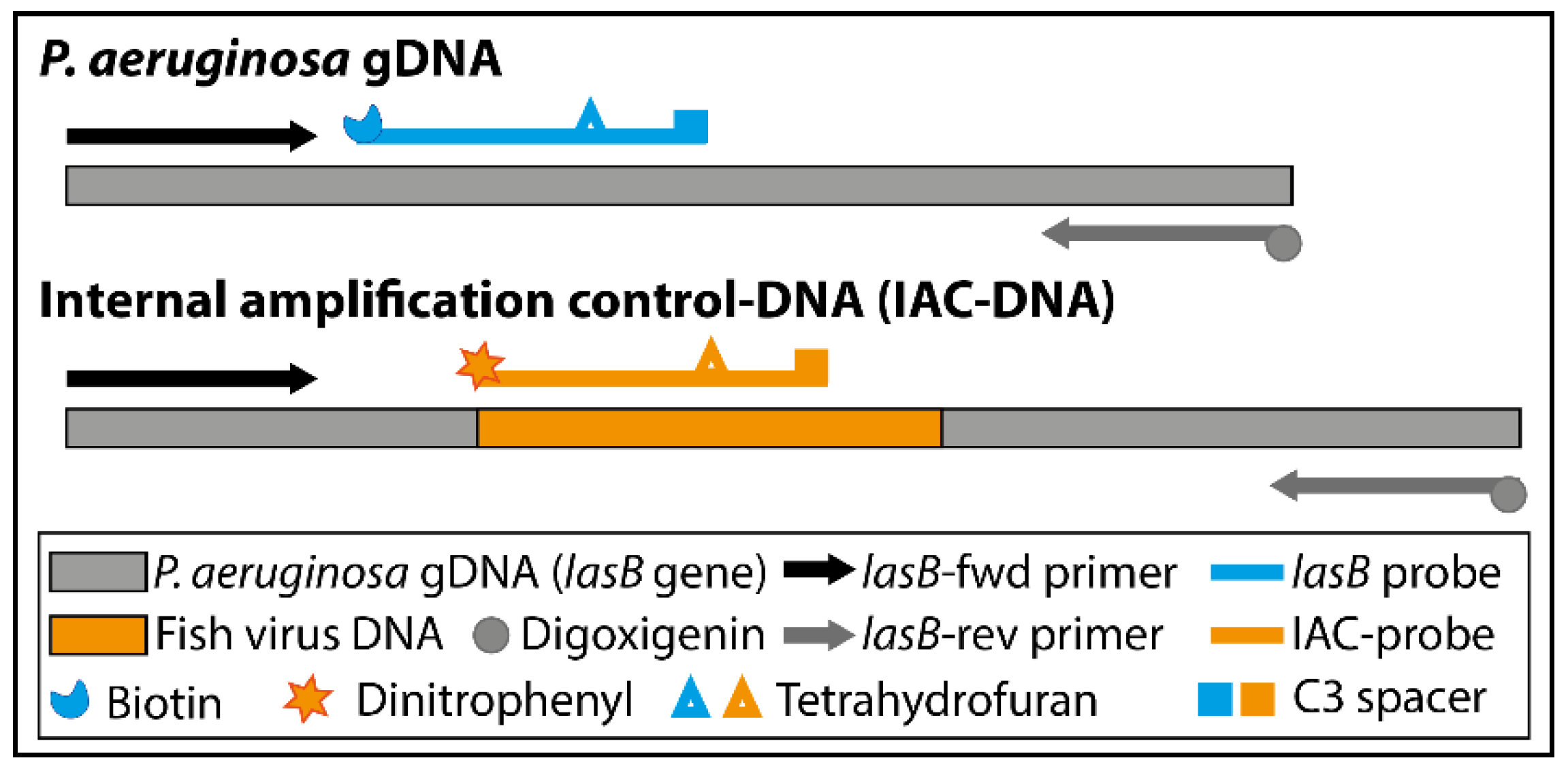
| Name | Sequence (5′-3′) |
|---|---|
| lasB-fwd primer | GAGAATGACAAAGTGGAACTGGTGATCCGCCTG |
| lasB-rev primer | Dig-GCCAGGCCTTCCCACTGATCGAGCACTTCGCCG |
| lasB probe | Biotin-GAACAACATCGCCCAACTGGTCTA CAACGT[H]TCCTACCTGATTCCC-C3 spacer |
| IAC-probe | DNP-CAACTGCAGGGACGATTCCTTTGTCC CGAT[H]CGACCAGCTCAACTC-C3 spacer |
| IAC-DNA | AAGACCGAGAATGACAAAGTGGAACTGGTGATCCGCCTGGGCGATATACACTCATCCCTCCAACTGCAGGGACGATTCCTTTGTCCCGATTCGACCAGCTCAACTCAGGTGTCCTCATGAAGGCGAGGGACTGTCGCGGCCGCATTTCGTCATCGACGCCAAGACCGGCGAAGTGCTCGATCAGTGGGAAGGCCTGGCCCACGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brunauer, A.; Verboket, R.D.; Kainz, D.M.; von Stetten, F.; Früh, S.M. Rapid Detection of Pathogens in Wound Exudate via Nucleic Acid Lateral Flow Immunoassay. Biosensors 2021, 11, 74. https://doi.org/10.3390/bios11030074
Brunauer A, Verboket RD, Kainz DM, von Stetten F, Früh SM. Rapid Detection of Pathogens in Wound Exudate via Nucleic Acid Lateral Flow Immunoassay. Biosensors. 2021; 11(3):74. https://doi.org/10.3390/bios11030074
Chicago/Turabian StyleBrunauer, Anna, René D. Verboket, Daniel M. Kainz, Felix von Stetten, and Susanna M. Früh. 2021. "Rapid Detection of Pathogens in Wound Exudate via Nucleic Acid Lateral Flow Immunoassay" Biosensors 11, no. 3: 74. https://doi.org/10.3390/bios11030074
APA StyleBrunauer, A., Verboket, R. D., Kainz, D. M., von Stetten, F., & Früh, S. M. (2021). Rapid Detection of Pathogens in Wound Exudate via Nucleic Acid Lateral Flow Immunoassay. Biosensors, 11(3), 74. https://doi.org/10.3390/bios11030074