Influence of the Electrolyte Salt Concentration on DNA Detection with Graphene Transistors
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
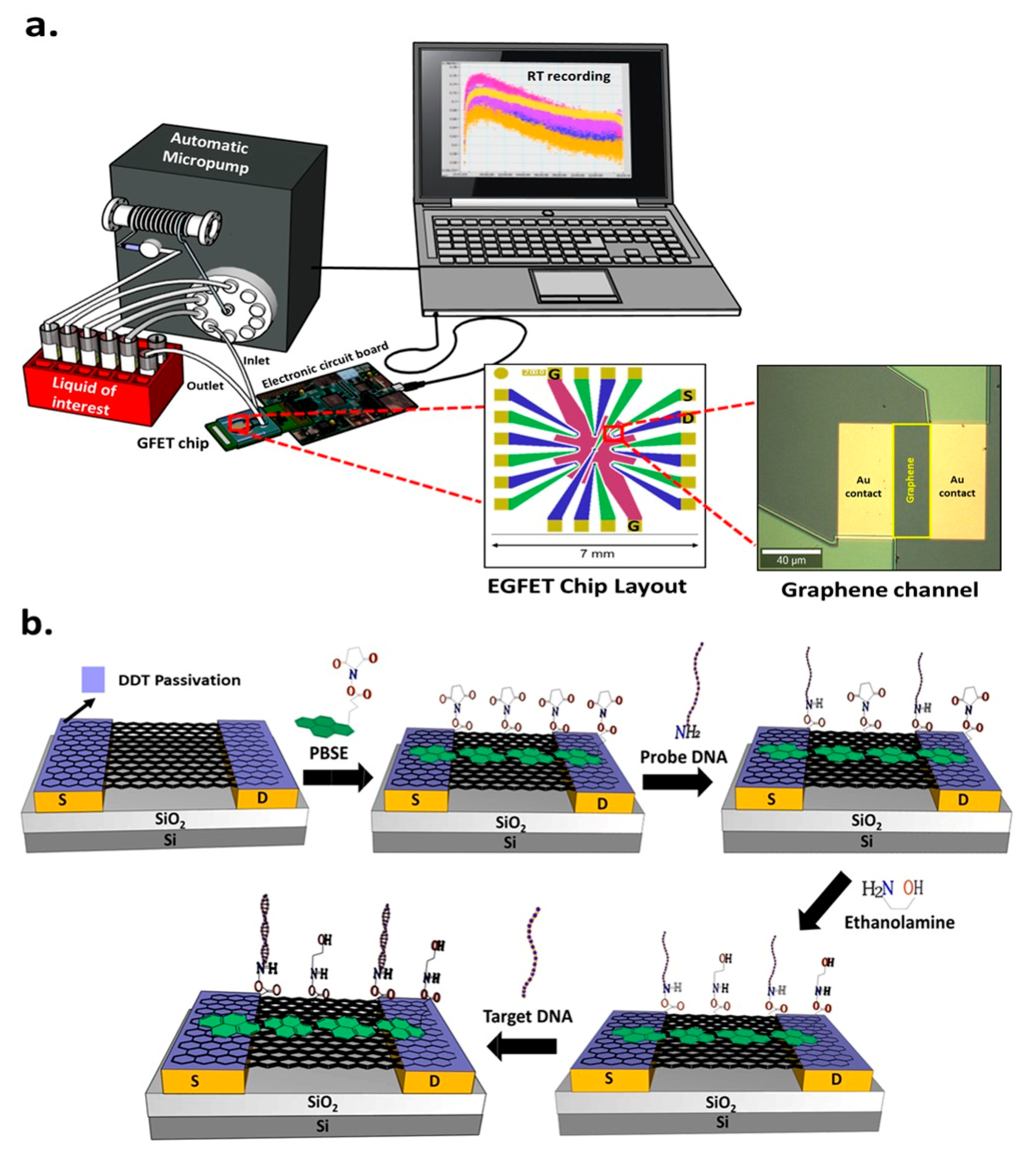
2.2. Apparatus
2.3. Methods
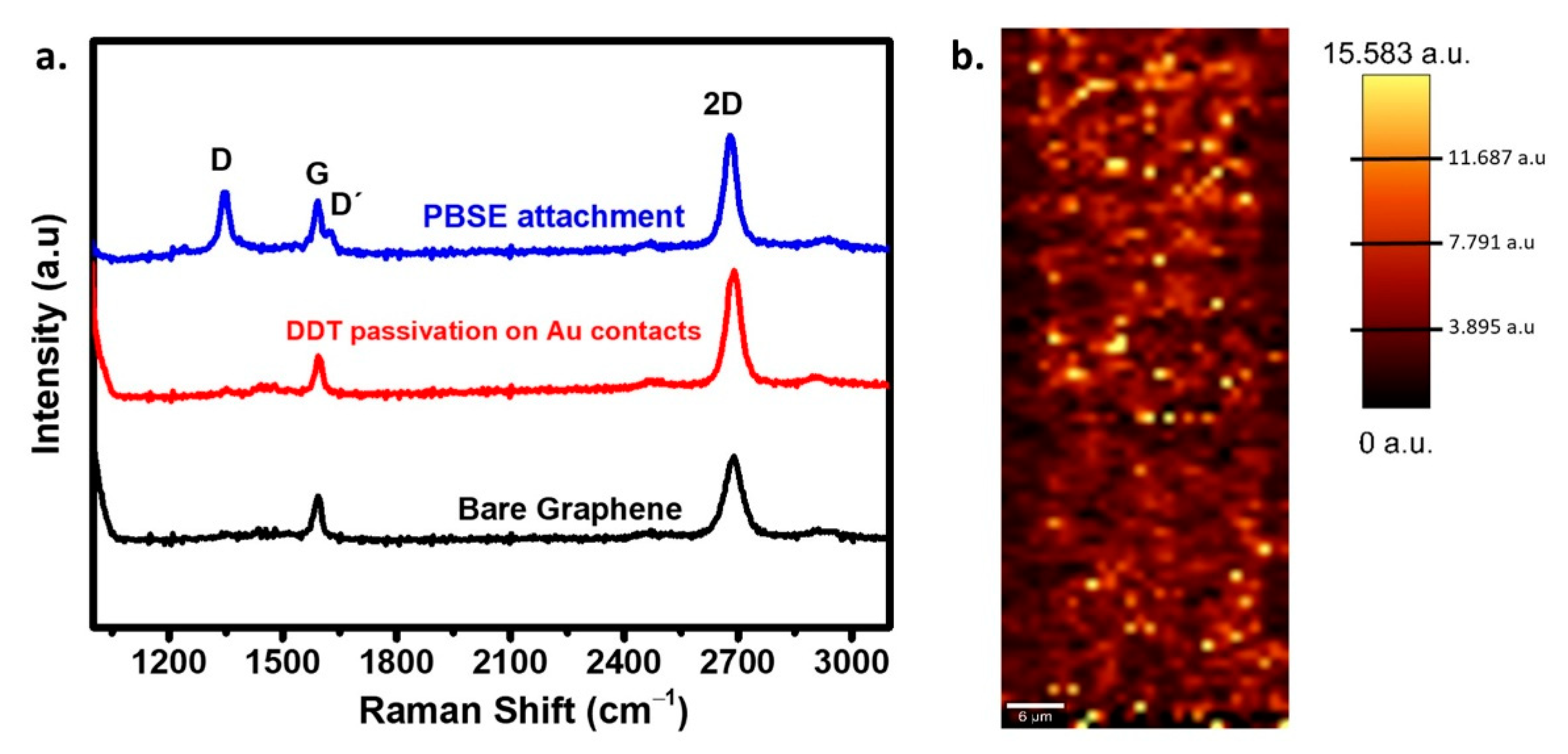
2.3.1. GFET Chip Fabrication and Surface Characterization
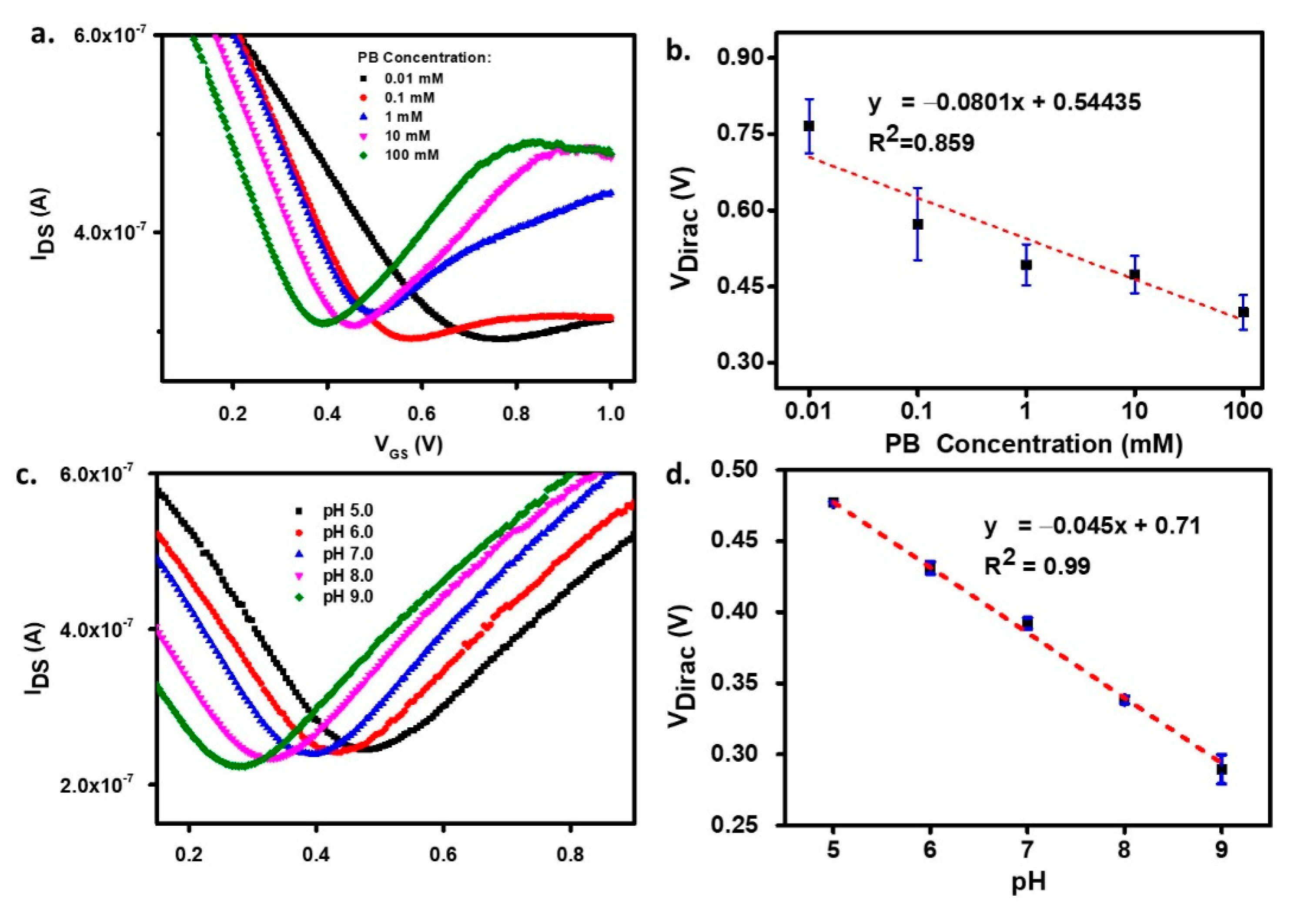
2.3.2. The Study of the Effects of Ionic Strength and pH
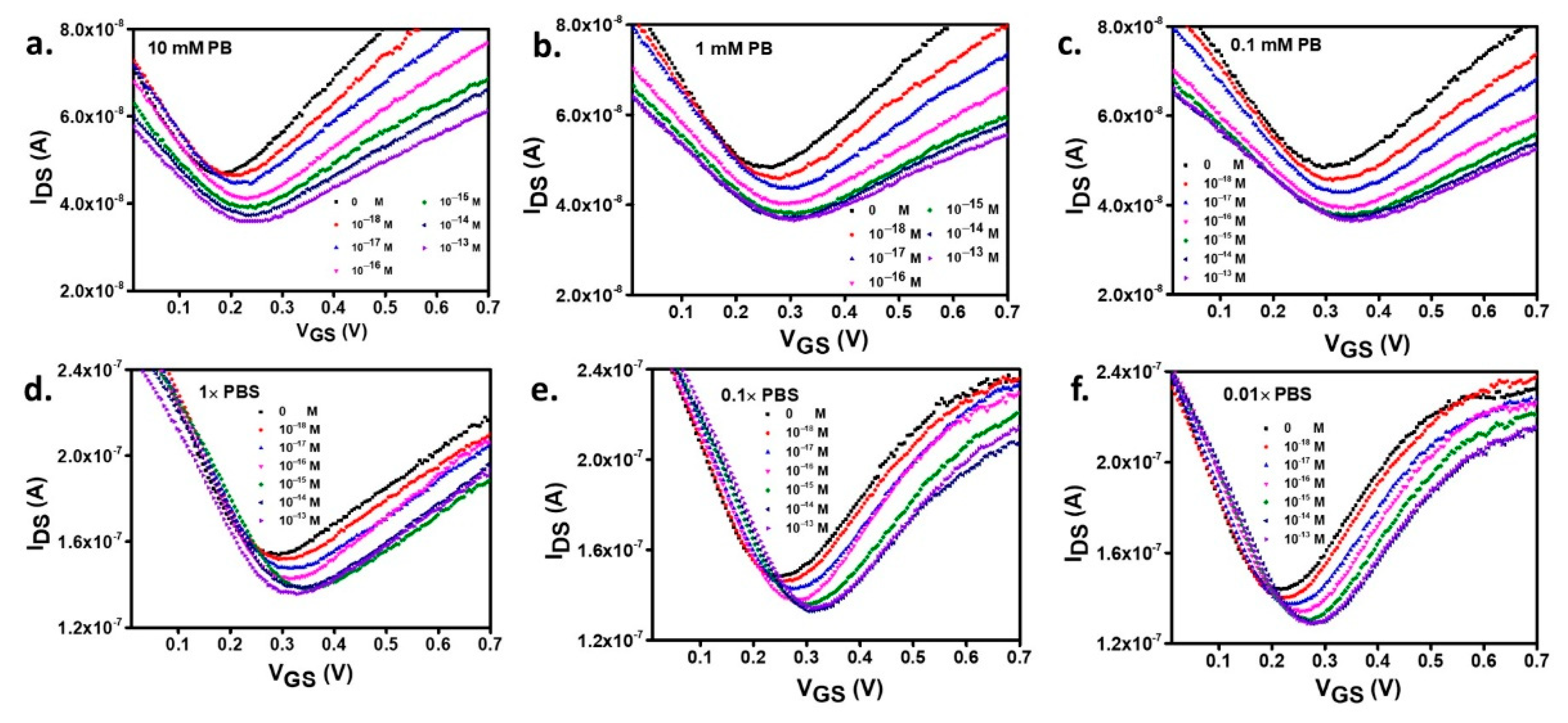
2.3.3. DNA Hybridization Detection with PB and PBS
2.3.4. The Study of Probe DNA Adsorption Kinetic with PB and PBS in the Microfluidic System
3. Results and Discussion
3.1. GFET Characterization and Surface Modification
3.2. The Effect of Ionic Strength and pH on the Graphene Transistor Response
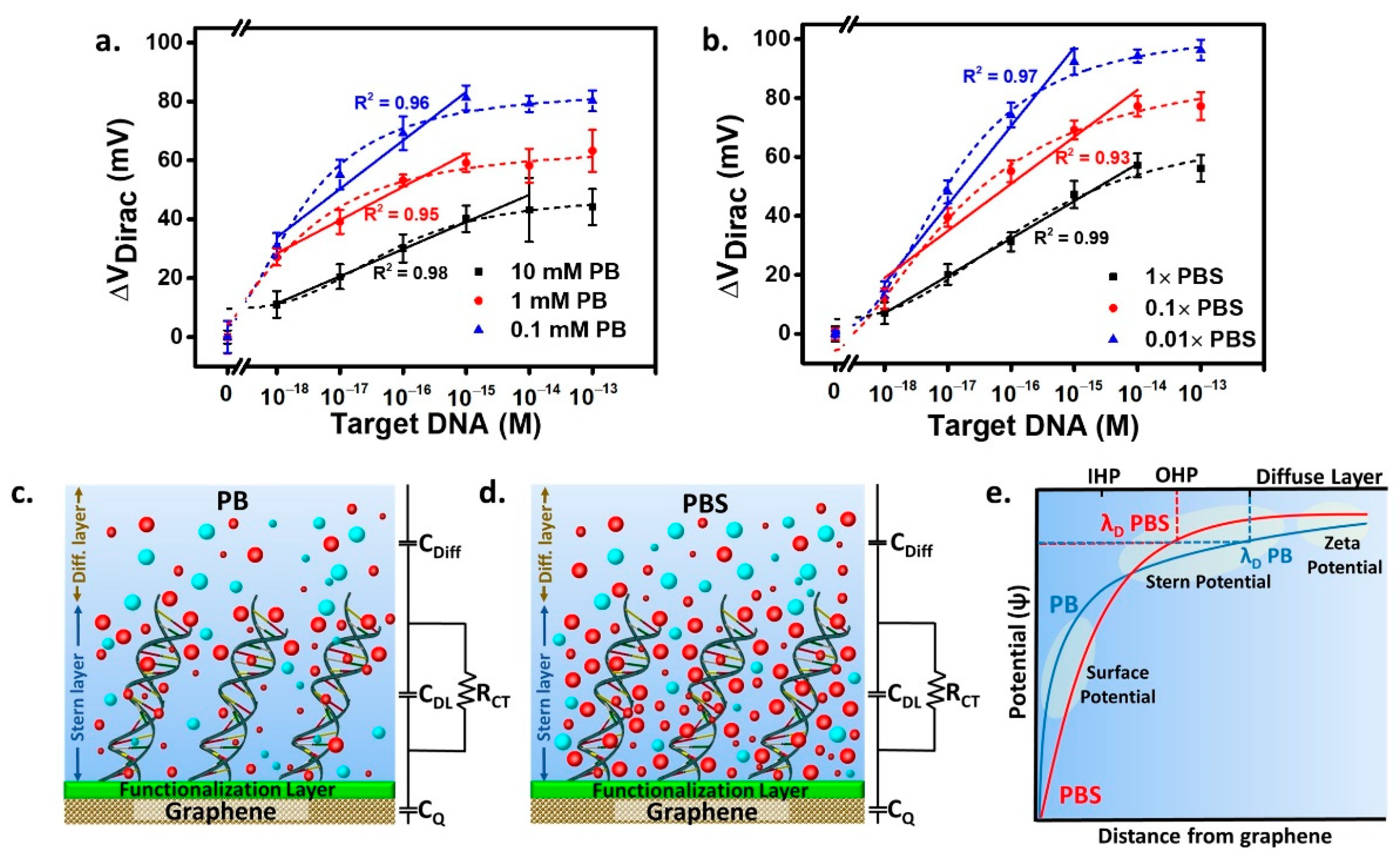
3.3. The Effects of Salts in the Buffer and Buffer Dilution on DNA Hybridization Study
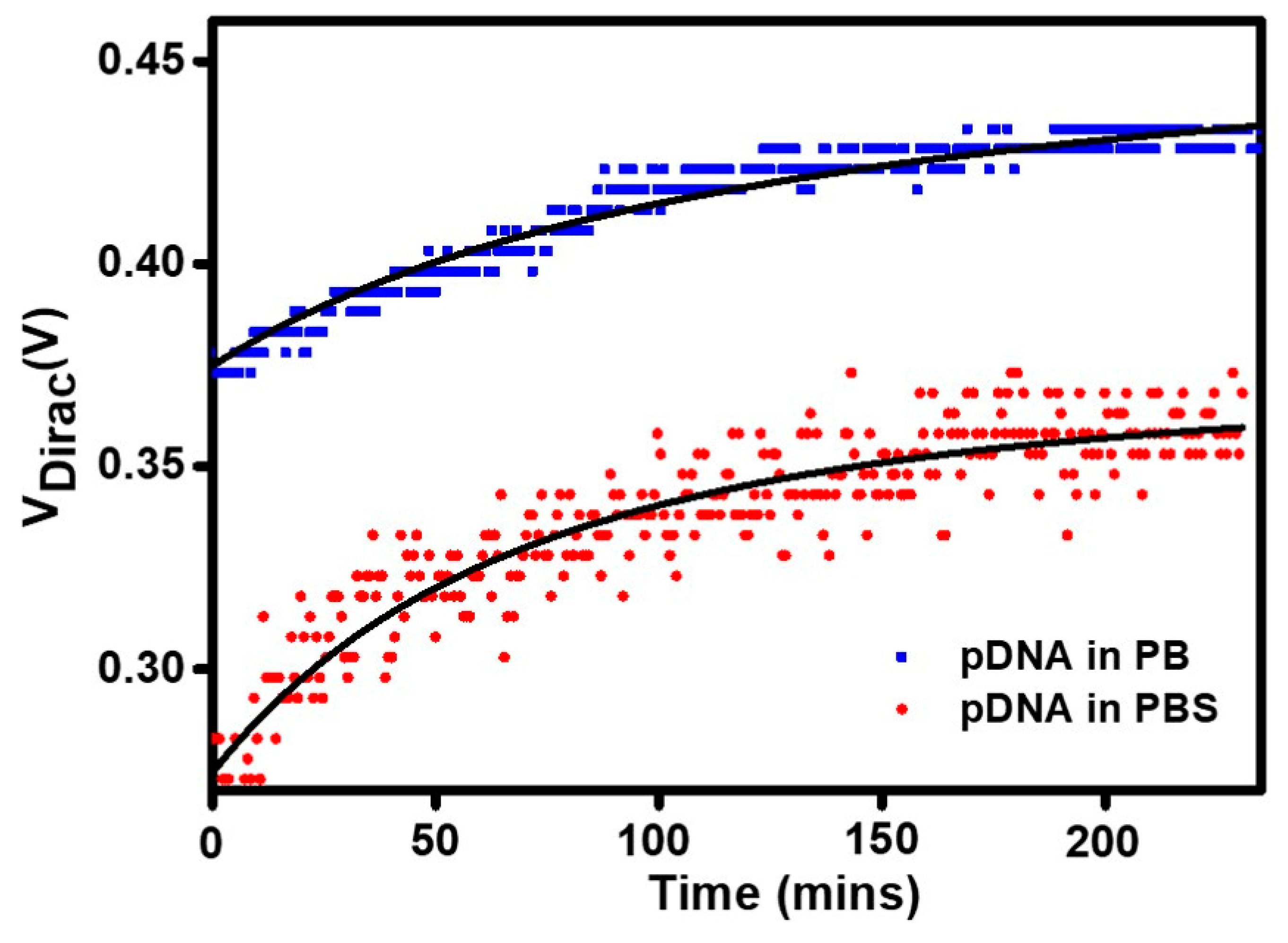
3.4. The Effects of Salts in the Buffer on DNA Adsorption Kinetics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Neto, A.H.C.; Guinea, F.; Peres, N.M.R.; Novoselov, K.S.; Geim, A.K. The electronic properties of grapheme. Rev. Mod. Phys. 2009, 81, 109. [Google Scholar] [CrossRef]
- Yang, Y.; Asiri, A.M.; Tang, Z.; Du, D.; Lin, Y. Graphene based materials for biomedical applications. Mater. Today 2013, 16, 365–373. [Google Scholar] [CrossRef]
- Forsyth, R.; Devadoss, A.; Guy, O.J. Graphene Field Effect Transistors for Biomedical Applications: Current Status and Future Prospects. Diagnostics 2017, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Szunerits, S.; Boukherroub, R. Graphene-based biosensors. Interface Focus 2018, 8, 20160132. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.-H.; Sarangadharan, I.; Regmi, A.; Chen, Y.-W.; Hsu, C.-P.; Chang, W.-H.; Lee, G.-Y.; Chyi, J.-I.; Chen, C.-C.; Shiesh, S.-C.; et al. Beyond the Debye length in high ionic strength solution: Direct protein detection with field-effect transistors (FETs) in human serum. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Munoz, R.S.; Gómez-Aleixandre, C. Review of CVD synthesis of graphene. Chem. Vap. Depos. 2013, 19, 297–322. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, L.; Zhou, C. Review of chemical vapor deposition of graphene and related applications. Acc. Chem. Res. 2013, 46, 2329–2339. [Google Scholar] [CrossRef]
- Xu, Y.; Yao, B.; Li, Y.; Ding, Z.; Li, J.; Wang, H.; Zhang, Z.; Zhang, L.; Zhao, H.; Shen, D.Z. Chemical states of gold doped in ZnO films and its effect on electrical and optical properties. J. Alloys Compd. 2014, 585, 479–484. [Google Scholar] [CrossRef]
- Kuila, T.; Bose, S.; Khanra, P.; Mishra, A.K.; Kim, N.H.; Lee, J.H. Recent advances in graphene-based biosensors. Biosens. Bioelectron. 2011, 26, 4637–4648. [Google Scholar] [CrossRef]
- Tsang, D.K.H.; Lieberthal, T.J.; Watts, C.; Dunlop, I.E.; Ramadan, S.; Hernández, A.D.R.; Klein, N. Chemically Functionalised Graphene FET Biosensor for the Label-free Sensing of Exosomes. Sci. Rep. 2019, 9, 2–11. [Google Scholar]
- Hwang, M.T.; Heiranian, M.; Kim, Y.; You, S.; Leem, J.; Taqieddin, A.; Faramarzi, V.; Jing, Y.; Park, I.; Van Der Zande, A.M.; et al. Ultrasensitive detection of nucleic acids using deformed graphene channel field effect biosensors. Nat. Commun. 2020, 11, 1543. [Google Scholar] [CrossRef] [PubMed]
- Campos, R.; Borme, J.; Guerreiro, J.R.; Machado, G.; Cerqueira, M.F.G.; Petrovykh, D.Y.; Alpuim, P. Attomolar label-free detection of dna hybridization with electrolyte-gated graphene field-effect transistors. ACS Sens. 2019, 4, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Green, N.S.; Norton, M.L. Interactions of DNA with graphene and sensing applications of graphene field-effect transistor devices: A review. Anal. Chim. Acta 2015, 853, 127–142. [Google Scholar] [CrossRef] [PubMed]
- Ameri, S.K.; Singh, P.K.; Sonkusale, S.R. Liquid gated three dimensional graphene network transistor. Carbon 2014, 79, 572–577. [Google Scholar] [CrossRef]
- Reiner-Rozman, C.; Larisika, M.; Nowak, C.; Knoll, W. Graphene-based liquid-gated field effect transistor for biosensing: Theory and experiments. Biosens. Bioelectron. 2015, 70, 21–27. [Google Scholar] [CrossRef]
- Lin, Y.-T.; Purwidyantri, A.; Luo, J.-D.; Chiou, C.-C.; Yang, C.-M.; Lo, C.-H.; Hwang, T.-L.; Yen, T.; Lai, C.-S. Programming a nonvolatile memory-like sensor for KRAS gene sensing and signal enhancement. Biosens. Bioelectron. 2016, 79, 63–70. [Google Scholar] [CrossRef]
- Pandey, A.; Gurbuz, Y.; Ozguz, V.; Niazi, J.H.; Qureshi, A. Graphene-interfaced electrical biosensor for label-free and sensitive detection of foodborne pathogenic E. coli O157:H7. Biosens. Bioelectron. 2017, 91, 225–231. [Google Scholar] [CrossRef]
- Hong, G.; Han, Y.; Schutzius, T.M.; Wang, Y.; Pan, Y.; Hu, M.; Jie, J.; Sharma, C.S.; Müller, U.; Poulikakos, D. On the mechanism of hydrophilicity of graphene. Nano Lett. 2016, 16, 4447–4453. [Google Scholar] [CrossRef]
- Cao, G.; Liu, W.; Cao, M.; Li, X.; Zhang, A.; Wang, X.; Chen, B. Improve the transconductance of a graphene field-effect transistor by folding graphene into a wedge. J. Phys. D Appl. Phys. 2016, 49, 275108. [Google Scholar] [CrossRef]
- Purwidyantri, A.; Kamajaya, L.; Chen, C.-H.; Luo, J.-D.; Chiou, C.-C.; Tian, Y.-C.; Lin, C.-Y.; Yang, C.-M.; Lai, C.-S. A colloidal nanopatterning and downscaling of a highly periodic au nanoporous EGFET biosensor. J. Electrochem. Soc. 2018, 165, H3170–H3177. [Google Scholar] [CrossRef]
- Watson, F.; Austin, P. Physiology of human fluid balance. Anaesth. Intensiv. Care Med. 2018, 19, 494–501. [Google Scholar] [CrossRef]
- Stern, E.; Wagner, R.; Sigworth, F.J.; Breaker, R.; Fahmy, T.M.; Reed, M.A. Importance of the debye screening length on nanowire field effect transistor sensors. Nano Lett. 2007, 7, 3405–3409. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Crittenden, S.R. Stern potential and Debye length measurements in dilute ionic solutions with electrostatic force microscopy. Nanotechnology 2013, 24, 435701. [Google Scholar] [CrossRef]
- Lu, M.-P.; Vire, E.; Montès, L. Ionic screening effect on low-frequency drain current fluctuations in liquid-gated nanowire FETs. Nanotechnology 2015, 26, 495501. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Kim, K.; Kwak, H.-T.; Lee, S.; Meyyappan, M.; Baek, C.-K. Buffer solution optimization for accurate fluoride ion detection in tap water. J. Electroanal. Chem. 2020, 858, 113837. [Google Scholar] [CrossRef]
- Yang, H.; Sakata, T. Molecular-charge-contact-based ion-sensitive field-effect transistor sensor in microfluidic system for protein sensing. Sensors 2019, 19, 3393. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Su, Z.; Zhang, S. Raman spectra of bilayer graphene covered with Poly(methyl methacrylate) thin film. AIP Adv. 2012, 2, 032122. [Google Scholar] [CrossRef]
- Ping, J.; Xi, J.; Saven, J.G.; Liu, R.; Johnson, A.T.C. Quantifying the effect of ionic screening with protein-decorated graphene transistors. Biosens. Bioelectron. 2017, 89, 689–692. [Google Scholar] [CrossRef]
- Zubiarrain-Laserna, A.; Kruse, P. Review—Graphene-Based Water Quality Sensors. J. Electrochem. Soc. 2020, 167, 037539. [Google Scholar] [CrossRef]
- Vieira, N.C.S.; Borme, J.; Machado, G.; Cerqueira, M.; Cardoso, S.; Zucolotto, V.; Peres, N.M.R.; Alpuim, P.; Machado, G., Jr. Graphene field-effect transistor array with integrated electrolytic gates scaled to 200 mm. J. Phys. Condens. Matter 2016, 28, 85302. [Google Scholar] [CrossRef]
- Ferreira, A.; Gomes, J.C.V.; Nilsson, J.; Mucciolo, E.R.; Peres, N.M.R.; Neto, A.H.C. Unified description of the dc conductivity of monolayer and bilayer graphene at finite densities based on resonant scatterers. Phys. Rev. B 2011, 83, 165402. [Google Scholar] [CrossRef]
- DeRouchey, J.E.; Parsegian, V.A.; Rau, D.C. Cation Charge Dependence of the Forces Driving DNA Assembly. Biophys. J. 2010, 99, 2608–2615. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Singh, N. Effect of salt concentration on the stability of heterogeneous DNA. Phys. A Stat. Mech. Appl. 2015, 419, 328–334. [Google Scholar] [CrossRef]
- Lloret, N.; Frederiksen, R.S.; Møller, T.C.; Rieben, N.I.; Upadhyay, S.; De Vico, L.; Jensen, J.H.; Nygård, J.; Martinez, K.L. Effects of buffer composition and dilution on nanowire field-effect biosensors. Nanotechnology 2012, 24, 035501. [Google Scholar] [CrossRef] [PubMed]
- Marconcini, P.; Macucci, M. Approximate calculation of the potential profile in a graphene-based device. IET Circuits Devices Syst. 2015, 9, 30–38. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.Y.; Pham, T.D.; Zand, K.; Li, J.; Burke, P.J. Charging the Quantum Capacitance of Graphene with a Single Biological Ion Channel. ACS Nano 2014, 8, 4228–4238. [Google Scholar] [CrossRef]
- Massobrio, P.; Massobrio, G.; Martinoia, S. Interfacing cultured neurons to microtransducers arrays: A review of the neuro-electronic junction models. Front. Neurosci. 2016, 10, 282. [Google Scholar] [CrossRef]
- Chen, C.-W.; Yip, B.-S.; Pan, F.-M.; Sheu, J.-T. Optimization of Nanobelt Field Effect Transistor with a Capacitive Extended Gate for Use as a Biosensor. ECS J. Solid State Sci. Technol. 2018, 7, Q3172–Q3179. [Google Scholar] [CrossRef]
- Park, Y.; Kim, S.H.; Matalon, S.; Wang, N.-H.L.; Franses, E.I. Effect of phosphate salts concentrations, supporting electrolytes, and calcium phosphate salt precipitation on the pH of phosphate buffer solutions. Fluid Phase Equilibria 2009, 278, 76–84. [Google Scholar] [CrossRef]
- Walker, D.A.; Kowalczyk, B.; De La Cruz, M.O.; Grzybowski, B.A. Electrostatics at the nanoscale. Nanoscale 2011, 3, 1316–1344. [Google Scholar] [CrossRef]
- Aliakbarinodehi, N.; Jolly, P.; Bhalla, N.; Miodek, A.; De Micheli, G.; Estrela, P.; Carrara, S. Aptamer-based field-effect biosensor for tenofovir detection. Sci. Rep. 2017, 7, 44409. [Google Scholar] [CrossRef] [PubMed]
- Jantz, D.; Berg, J. Probing the DNA-binding affinity and specificity of designed zinc finger proteins. Biophys. J. 2010, 98, 852–860. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kretsinger, R. Calcium-Modulated Proteins (EF-Hand). Ref. Modul. Biomed. Sci. Encycl. Biol. Chem. 2013, 12, 316–321. [Google Scholar]
- Swinney, D.C. Molecular Mechanism of Action (MMoA) in Drug Discovery. In Annual Reports in Medicinal Chemistry; Elsevier: Amsterdam, The Netherlands, 2011; Volume 46, pp. 301–317. [Google Scholar]
- Nguyen, T.H.; Elimelech, M. Adsorption of plasmid DNA to a natural organic matter-coated silica surface: Kinetics, conformation, and reversibility. Langmuir 2007, 23, 3273–3279. [Google Scholar] [CrossRef]
- Wu, J.; Wang, H.; Zhu, A.; Long, F. Adsorption Kinetics of Single-Stranded DNA on Functional Silica Surfaces and Its Influence Factors: An Evanescent-Wave Biosensor Study. ACS Omega 2018, 3, 5605–5614. [Google Scholar] [CrossRef]
| Buffer. | µ (mM) | λD (nm) | Hill’s Function | Linear Area Fitting | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R2 | SD Blank | LoB | SD Lowest Conc. | LoD ΔV | LoD (aM) | R2 | Linear Region (M) | Sensitivity (mV/dec.) | |||
| 0.1 mM PB | 0.1 | 21.00 | 0.98 | 0.0054 | 0.0088 | 0.0042 | 0.015 | 4.17 | 0.96 | 10−18–10−15 | 16.47 |
| 1 mM PB | 1 | 6.65 | 0.96 | 0.0052 | 0.0090 | 0.0029 | 0.013 | 3.44 | 0.95 | 10−18−10−15 | 11.30 |
| 10 mM PB | 10 | 2.11 | 0.89 | 0.0022 | 0.0037 | 0.0044 | 0.011 | 10.15 | 0.98 | 10−18–10−14 | 9.22 |
| PBS 0.01× | 1.627 | 7.61 | 0.99 | 0.0010 | 0.0016 | 0.0027 | 0.006 | 5.01 | 0.97 | 10−18–10−15 | 26.69 |
| PBS 0.1× | 16.27 | 2.41 | 0.97 | 0.0019 | 0.0031 | 0.0034 | 0.008 | 7.58 | 0.93 | 10−18–10−14 | 15.95 |
| PBS 1× | 162.7 | 0.76 | 0.95 | 0.0026 | 0.0042 | 0.0037 | 0.010 | 22.60 | 0.99 | 10−18–10−14 | 12.67 |
| Background Solution | Variables | Regression Coefficient | ||
|---|---|---|---|---|
| a | b | c | ||
| PB | 0.468 | 0.049 | 0.044 | 0.972 |
| PBS | 0.372 | 4.124 × 10−4 | −0.916 | 0.881 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Purwidyantri, A.; Domingues, T.; Borme, J.; Guerreiro, J.R.; Ipatov, A.; Abreu, C.M.; Martins, M.; Alpuim, P.; Prado, M. Influence of the Electrolyte Salt Concentration on DNA Detection with Graphene Transistors. Biosensors 2021, 11, 24. https://doi.org/10.3390/bios11010024
Purwidyantri A, Domingues T, Borme J, Guerreiro JR, Ipatov A, Abreu CM, Martins M, Alpuim P, Prado M. Influence of the Electrolyte Salt Concentration on DNA Detection with Graphene Transistors. Biosensors. 2021; 11(1):24. https://doi.org/10.3390/bios11010024
Chicago/Turabian StylePurwidyantri, Agnes, Telma Domingues, Jérôme Borme, Joana Rafaela Guerreiro, Andrey Ipatov, Catarina M. Abreu, Marco Martins, Pedro Alpuim, and Marta Prado. 2021. "Influence of the Electrolyte Salt Concentration on DNA Detection with Graphene Transistors" Biosensors 11, no. 1: 24. https://doi.org/10.3390/bios11010024
APA StylePurwidyantri, A., Domingues, T., Borme, J., Guerreiro, J. R., Ipatov, A., Abreu, C. M., Martins, M., Alpuim, P., & Prado, M. (2021). Influence of the Electrolyte Salt Concentration on DNA Detection with Graphene Transistors. Biosensors, 11(1), 24. https://doi.org/10.3390/bios11010024








