Integrated Electrochemical Biosensors for Detection of Waterborne Pathogens in Low-Resource Settings
Abstract
1. Global Water Security
2. Conventional Techniques for Water Quality Monitoring
2.1. Mass Spectroscopy
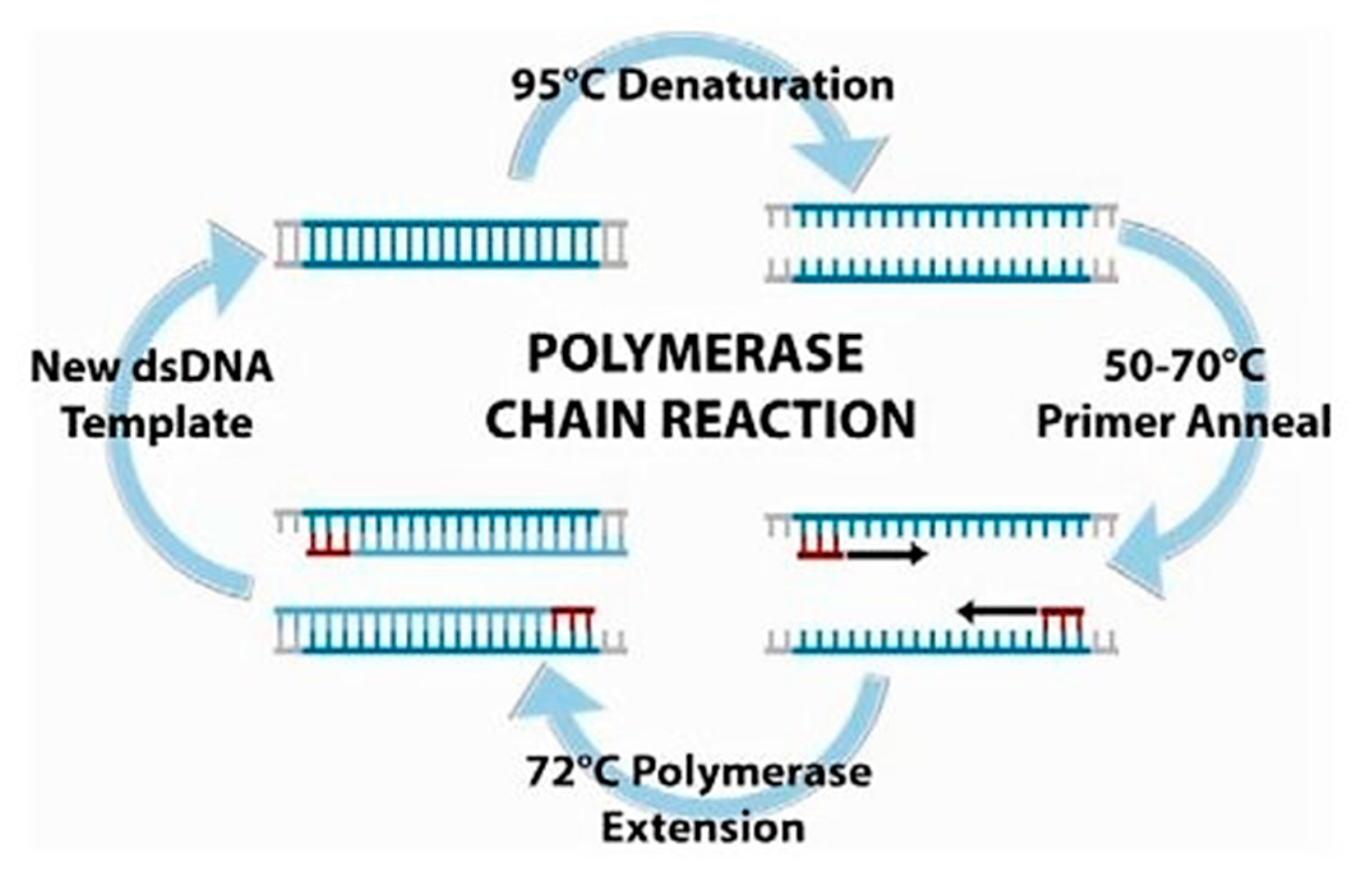
2.2. Polymerase Chain Reaction (PCR)
2.3. Microbiological Laboratory Methods
2.4. Microscopy Techniques
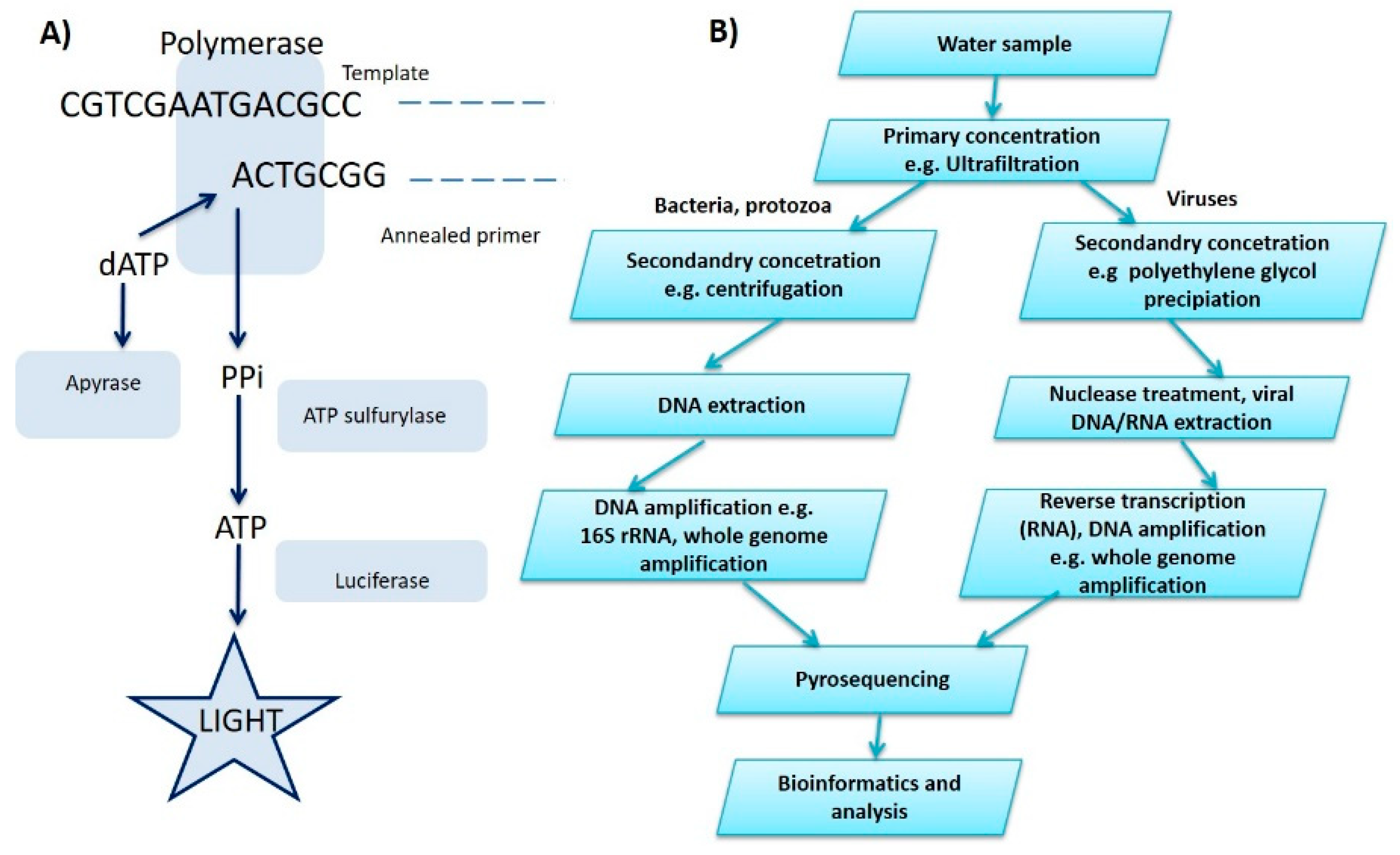
2.5. Pyrosequencing
2.6. DNA Microarrays
2.7. Fluorescence In Situ Hybridization (FISH)
2.8. Immunological-Based Methods
3. Challenges of Monitoring Fresh and Wastewater Quality in Low-Resource Settings
3.1. Ease of Use
3.2. Cost of Device Manufacture and Implementation
3.3. In Situ Measurement Capability
3.4. Real-Time Quantitative Data
4. On-Site Biosensors, Monitoring, and Implementation of Integrated Devices in LMICs
4.1. What is an Electrochemical Biosensor?
4.2. Advantages and Disadvantages of Electrochemical Biosensing
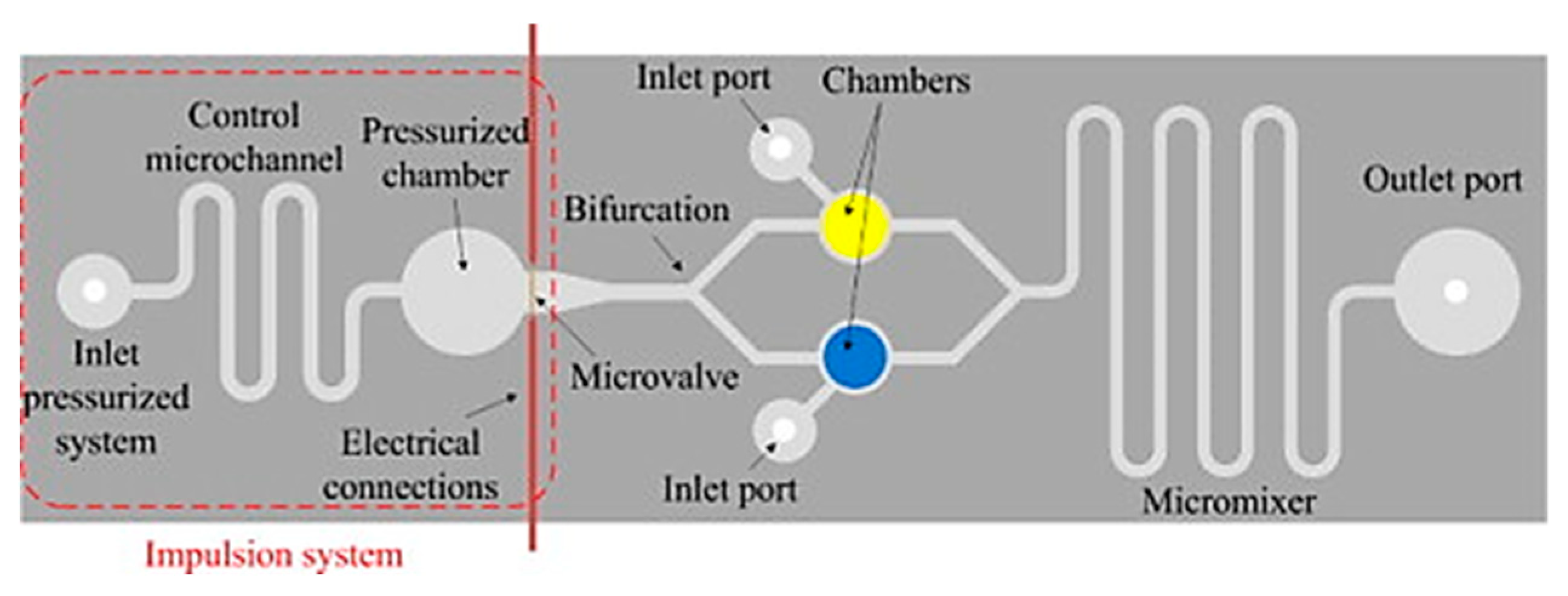
4.3. How to Detect On-Site
5. Conclusions and Future Perspectives
Funding
Conflicts of Interest
References
- How Much Water is there on Earth? Available online: https://www.usgs.gov/special-topic/water-science-school/science/how-much-water-there-earth?qt-science_center_objects=0#qt-science_center_objects (accessed on 28 October 2019).
- Where is All of the Earth’s Water? Available online: https://oceanservice.noaa.gov/facts/wherewater.html (accessed on 28 October 2019).
- Use of Freshwater Resources. Available online: https://www.eea.europa.eu/data-and-maps/indicators/use-of-freshwater-resources/use-of-freshwater-resources-assessment-2 (accessed on 28 October 2019).
- de Bell, S.; Graham, H.; Jarvis, S.; White, P. The importance of nature in mediating social and psychological benefits associated with visits to freshwater blue space. Landsc. Urban Plan. 2017, 167, 118–127. [Google Scholar] [CrossRef]
- The Top 10 Causes of Death. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 29 October 2019).
- Childhood Diseases. Available online: https://www.unicef.org/health/index_51412.html (accessed on 29 October 2019).
- Ashipala, N.; Armitage, N.P. Impediments to the adoption of alternative sewerage in South African urban informal settlements. Water Sci. Technol. 2011, 64, 1781–1789. [Google Scholar] [CrossRef] [PubMed]
- Chirisa, I.; Bandauko, E.; Matamanda, A.; Mandisvika, G. Decentralized domestic wastewater systems in developing countries: The case study of Harare (Zimbabwe). Appl. Water Sci. 2017, 7, 1069–1078. [Google Scholar] [CrossRef]
- Palanca-Tan, R. Health and water quality benefits of alternative sewerage systems in Metro Manila, Philippines. Environ. Urban. 2017, 29, 567–580. [Google Scholar] [CrossRef]
- Karpouzoglou, T.; Zimmer, A. Ways of knowing the wastewaterscape: Urban political ecology and the politics of wastewater in Delhi, India. Habitat Int. 2016, 54, 150–160. [Google Scholar] [CrossRef]
- Raschid-Sally, L. The role and place of global surveys for assessing wastewater irrigation. Irrig. Drain. Syst. 2010, 24, 5–21. [Google Scholar] [CrossRef]
- World Health Organization. Excreta and greywater in agriculture. In Guidelines for the Safe Use of Wastewater, Excreta, Greywater; World Health Organization: Geneva, Switzerland, 2006; Volume IV, p. 204. [Google Scholar]
- Ejeian, F.; Etedali, P.; Mansouri-Tehrani, H.A.; Soozanipour, A.; Low, Z.X.; Asadnia, M.; Taheri-Kafrani, A.; Razmjou, A. Biosensors for wastewater monitoring: A review. Biosens. Bioelectron. 2018, 118, 66–79. [Google Scholar] [CrossRef]
- Water, Sanitation and Hygiene: Fact Sheet March 2016. Available online: http://www.wpro.who.int/entity/apac_rfhe/wash_factsheet_rfhe.pdf (accessed on 29 October 2019).
- World Health Organization; UNICEF. Progress on Drinking Water and Sanitation; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Osiemo, M.; Ogendi, G.; M’Erimba, C. Microbial quality of drinking water and prevalence of water-related diseases in Marigat Urban Centre, Kenya. Environ. Health Insights 2019, 13, 117863021983698. [Google Scholar] [CrossRef]
- Van Derslice, J. Drinking water infrastructure and environmental disparities: Evidence and methodological considerations. Am. J. Public Health 2011, 101, S109–S114. [Google Scholar] [CrossRef]
- Qin, H.; Su, Q.; Khu, S.; Tang, N. Water quality changes during rapid urbanization in the Shenzhen River catchment: An integrated view of socio-economic and infrastructure development. Sustainability 2014, 6, 7433–7451. [Google Scholar] [CrossRef]
- Cabral, J. Water microbiology. Bacterial pathogens and water. Int. J. Environ. Res. Public Health 2010, 7, 3657–3703. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Kass, P.; Soupir, M.; Biswas, S.; Singh, V. Contamination of water resources by pathogenic bacteria. AMB Express 2014, 4, 51. [Google Scholar] [CrossRef] [PubMed]
- Ellis, K.; Gowdy, C.; Jakomis, N.; Ryan, B.; Thom, C.; Biggs, C.; Speight, V. Understanding the costs of investigating coliform and E. Coli detections during routine drinking water quality monitoring. Urban Water J. 2017, 15, 101–108. [Google Scholar] [CrossRef]
- Drinking-Water. Available online: https://www.who.int/news-room/fact-sheets/detail/drinking-water (accessed on 4 January 2020).
- Bridle, H. Waterborne Pathogens: Detection Methods and Applications; Academic Press: London, UK, 2013. [Google Scholar]
- Maynard, C.; Berthiaume, F.; Lemarchand, K.; Harel, J.; Payment, P.; Bayardelle, P.; Masson, L.; Brousseau, R.J.A.E.M. Waterborne pathogen detection by use of oligonucleotide-based microarrays. Appl. Environ. Microbiol. 2005, 71, 8548–8557. [Google Scholar] [CrossRef]
- Hurst, G.B.; Doktycz, M.J.; Vass, A.A.; Buchanan, M.V. Detection of bacterial DNA polymerase chain reaction products by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 1996, 10, 377–382. [Google Scholar] [CrossRef]
- He, Y.; Chang, T.C.; Li, H.; Shi, G.; Tang, Y.-W. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry and database for identification of Legionella species. Can. J. Microbiol. 2011, 57, 533–538. [Google Scholar] [CrossRef]
- Donohue, M.J.; Smallwood, A.W.; Pfaller, S.; Rodgers, M.; Shoemaker, J.A. The development of a matrix-assisted laser desorption/ionization mass spectrometry-based method for the protein fingerprinting and identification of Aeromonas species using whole cells. J. Microbiol. Methods 2006, 65, 380–389. [Google Scholar] [CrossRef]
- Erler, R.; Wichels, A.; Heinemeyer, E.-A.; Hauk, G.; Hippelein, M.; Reyes, N.T.; Gerdts, G. VibrioBase: A MALDI-TOF MS database for fast identification of Vibrio spp. that are potentially pathogenic in humans. Syst. Appl. Microbiol. 2015, 38, 16–25. [Google Scholar] [CrossRef]
- Toze, S.J.W.R. PCR and the detection of microbial pathogens in water and wastewater. Water Res. 1999, 33, 3545–3556. [Google Scholar] [CrossRef]
- Kong, R.Y.C.; Lee, S.K.Y.; Law, T.W.F.; Law, S.H.W.; Wu, R.S.S. Rapid detection of six types of bacterial pathogens in marine waters by multiplex PCR. Water Res. 2002, 36, 2802–2812. [Google Scholar] [CrossRef]
- Marangi, M.; Giangaspero, A.; Lacasella, V.; Lonigro, A.; Gasser, R.B. Multiplex PCR for the detection and quantification of zoonotic taxa of Giardia, Cryptosporidium and Toxoplasma in wastewater and mussels. Mol. Cell. Probes 2015, 29, 122–125. [Google Scholar] [CrossRef] [PubMed]
- Parshionikar, S.U.; Cashdollar, J.; Fout, G.S. Development of homologous viral internal controls for use in RT-PCR assays of waterborne enteric viruses. J. Virol. Methods 2004, 121, 39–48. [Google Scholar] [CrossRef]
- Shannon, K.; Lee, D.-Y.; Trevors, J.; Beaudette, L.J. Application of real-time quantitative PCR for the detection of selected bacterial pathogens during municipal wastewater treatment. Sci. Total Environ. 2007, 382, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Bhagwat, A.A. Simultaneous detection of Escherichia coli O157:H7, Listeria monocytogenes and Salmonella strains by real-time PCR. Int. J. Food Microbiol. 2003, 84, 217–224. [Google Scholar] [CrossRef]
- Saxena, G.; Bharagava, R.N.; Kaithwas, G.; Raj, A. Microbial indicators, pathogens and methods for their monitoring in water environment. J. Water Health 2014, 13, 319–339. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.T.; Keevil, C.W. Study of microbial biofilms using light microscope techniques. Int. Biodeterior. Biodegrad. 1994, 34, 223–236. [Google Scholar] [CrossRef]
- Mazumder, S.; Sarkar, J.; Dey, R.; Mitra, M.K.; Mukherjee, S.; Das, G.C. Biofunctionalised quantum dots for sensing and identification of waterborne bacterial pathogens. J. Exp. Nanosci. 2010, 5, 438–446. [Google Scholar] [CrossRef]
- Aw, T.G.; Rose, J.B. Detection of pathogens in water: From phylochips to qPCR to pyrosequencing. Curr. Opin. Biotechnol. 2012, 23, 422–430. [Google Scholar] [CrossRef]
- Shen, S.; Qin, D. Pyrosequencing data analysis software: A useful tool for EGFR, KRAS, and BRAF mutation analysis. Diagn. Pathol. 2012, 7, 56. [Google Scholar] [CrossRef]
- Brinkman, N.; Francisco, R.; Nichols, T.; Robinson, D.; Schaefer, F., III; Schaudies, R.; Villegas, E.N. Detection of multiple waterborne pathogens using microsequencing arrays. J. Appl. Microbiol. 2013, 114, 564–573. [Google Scholar] [CrossRef]
- Lee, D.-Y.; Seto, P.; Korczak, R. DNA microarray-based detection and identification of waterborne protozoan pathogens. J. Microbiol. Methods 2010, 80, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Wen, S.; Liu, Y.; Li, R.; Zhong, X.; Feng, L.; Wang, L.; Cao, B. Development of a DNA microarray for detection and identification of Legionella pneumophila and ten other pathogens in drinking water. Int. J. Food Microbiol. 2011, 145, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Shah, J.; Weltman, H.; Narciso, P.; Murphy, C.; Poruri, A.; Baliga, S.; Sharon, L.; York, M.; Cunningham, G.; Miller, S.; et al. Dual color fluorescence in situ hybridization (FISH) assays for detecting Mycobacterium tuberculosis and Mycobacterium avium complexes and related pathogens in cultures. PLoS ONE 2017, 12, e0174989. [Google Scholar] [CrossRef] [PubMed]
- Almeida, C.; Azevedo, N.F.; Fernandes, R.; Keevil, C.W.; Vieira, M.J. Fluorescence in situ hybridization method using a peptide nucleic acid probe for identification of Salmonella spp. in a broad spectrum of samples. Appl. Environ. Microbiol. 2010, 76, 4476–4485. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Chattopadhyay, S.; Jackeray, R.; Abid, C.K.V.Z.; Kohli, G.S.; Singh, H. Highly sensitive detection of Salmonella typhi using surface aminated polycarbonate membrane enhanced-ELISA. Biosens. Bioelectron. 2012, 31, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Roser, M.; Ritchie, H.; Ortiz-Ospina, E. Internet. Available online: https://ourworldindata.org/internet#internet-access (accessed on 28 November 2019).
- Yetisen, A.; Akram, M.; Lowe, C. Paper-based microfluidic point-of-care diagnostic devices. Lab Chip 2013, 13, 2210. [Google Scholar] [CrossRef]
- Jolly, P.; Rainbow, J.; Regoutz, A.; Estrela, P.; Moschou, D. A PNA-based Lab-on-PCB diagnostic platform for rapid and high sensitivity DNA quantification. Biosens. Bioelectron. 2019, 123, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Castillo, F.; Loera-Muro, A.; Jacques, M.; Garneau, P.; Avelar-González, F.; Harel, J.; Guerrero-Barrera, A. Waterborne pathogens: Detection methods and challenges. Pathogens 2015, 4, 307–334. [Google Scholar] [CrossRef]
- Alhamlan, F.; Al-Qahtani, A.; Al-Ahdal, M. Recommended advanced techniques for waterborne pathogen detection in developing countries. J. Infect. Dev. Ctries. 2015, 9, 128–135. [Google Scholar] [CrossRef]
- Turle, R. Expensive equipment—A guide to purchasing. Anal. Bioanal. Chem. 2008, 393, 13–14. [Google Scholar] [CrossRef][Green Version]
- Chouler, J.; Di Lorenzo, M. Water quality monitoring in developing countries; Can microbial fuel cells be the answer? Biosensors 2015, 5, 450–470. [Google Scholar] [CrossRef] [PubMed]
- Soleymani, L.; Li, F. Mechanistic challenges and advantages of biosensor miniaturization into the nanoscale. ACS Sens. 2017, 2, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Sinha, A.; Dhanjai; Mugo, S.; Zhao, H.; Chen, J.; Jain, R. Electrochemical Immunosensors for Rapid Detection of Breast Cancer Biomarkers. Adv. Biosens. Health Care Appl. 2019, 147–169. [Google Scholar]
- Hammond, J.; Gross, A.; Giroud, F.; Travelet, C.; Borsali, R.; Cosnier, S. Solubilized Enzymatic Fuel Cell (SEFC) for quasi-continuous operation exploiting carbohydrate block copolymer glyconanoparticle mediators. Acs Energy Lett. 2018, 4, 142–148. [Google Scholar] [CrossRef]
- Martinez, S.; Di Lorenzo, M. Electricity generation from untreated fresh digestate with a cost-effective array of floating microbial fuel cells. Chem. Eng. Sci. 2019, 198, 108–116. [Google Scholar] [CrossRef]
- Moschou, D.; Tserepi, A. The lab-on-PCB approach: Tackling the μTAS commercial upscaling bottleneck. Lab Chip 2017, 17, 1388–1405. [Google Scholar] [CrossRef] [PubMed]
- Jang, A.; Zou, Z.; Lee, K.; Ahn, C.; Bishop, P. State-of-the-art lab chip sensors for environmental water monitoring. Meas. Sci. Technol. 2011, 22, 032001. [Google Scholar] [CrossRef]
- St John, A.; Price, C. Existing and emerging technologies for point-of-care testing. Clin. Biochem. Rev. 2014, 35, 155–167. [Google Scholar]
- Sher, M.; Zhuang, R.; Demirci, U.; Asghar, W. Paper-based analytical devices for clinical diagnosis: Recent advances in the fabrication techniques and sensing mechanisms. Expert Rev. Mol. Diagn. 2017, 17, 351–366. [Google Scholar] [CrossRef]
- Aracil, C.; Perdigones, F.; Moreno, J.; Luque, A.; Quero, J. Portable Lab-on-PCB platform for autonomous micromixing. Microelectron. Eng. 2015, 131, 13–18. [Google Scholar] [CrossRef]
- Aniyikaiye, T.; Oluseyi, T.; Odiyo, J.; Edokpayi, J. Physico-chemical analysis of wastewater discharge from selected paint industries in Lagos, Nigeria. Int. J. Environ. Res. Public Health 2019, 16, 1235. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.; Reinhard, M.; Khan, E.; Chen, H.; Nguyen, V.; Li, Y.; Goh, S.; Nguyen, Q.; Saeidi, N.; Gin, K. Emerging contaminants in wastewater, stormwater runoff, and surface water: Application as chemical markers for diffuse sources. Sci. Total Environ. 2019, 676, 252–267. [Google Scholar] [CrossRef] [PubMed]
- Lew, S.; Lew, M.; Biedunkiewicz, A.; Szarek, J. Impact of pesticide contamination on aquatic microorganism populations in the littoral zone. Arch. Environ. Contam. Toxicol. 2012, 64, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Blettler, M.; Ulla, M.; Rabuffetti, A.; Garello, N. Plastic pollution in freshwater ecosystems: Macro-, meso-, and microplastic debris in a floodplain lake. Environ. Monit. Assess. 2017, 189, 581. [Google Scholar] [CrossRef]
- Crocker, J.; Bartram, J. Comparison and cost analysis of drinking water quality monitoring requirements versus practice in seven developing countries. Int. J. Environ. Res. Public Health 2014, 11, 7333–7346. [Google Scholar] [CrossRef]
- Farkas, K.; McDonald, J.; Malham, S.; Jones, D. Two-step concentration of complex water samples for the detection of viruses. Methods Protoc. 2018, 1, 35. [Google Scholar] [CrossRef]
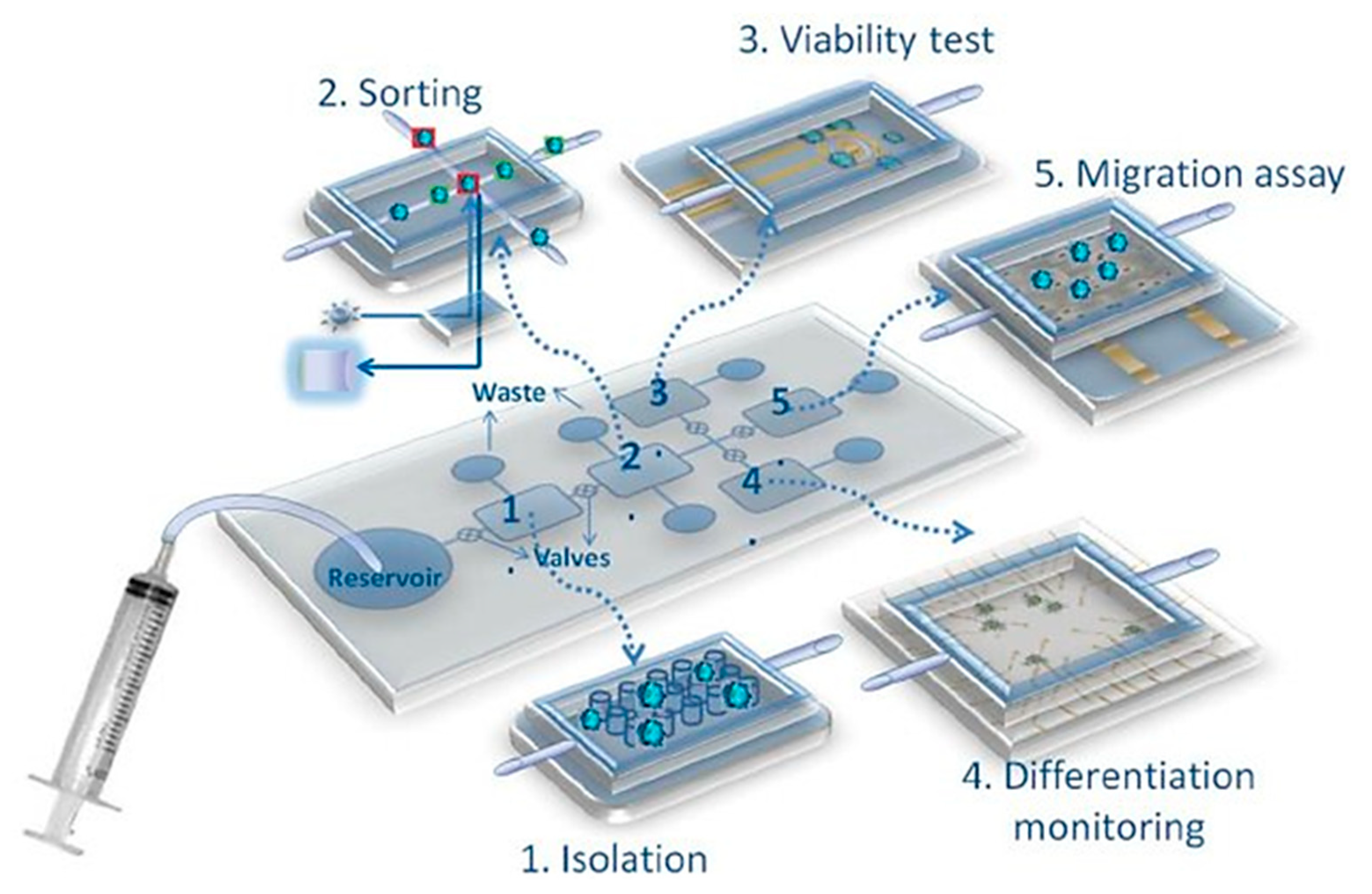
- Dhar, B.; Lee, N. Lab-on-a-Chip technology for environmental monitoring of microorganisms. BioChip J. 2018, 12, 173–183. [Google Scholar] [CrossRef]
- Worako, A. Physicochemical and biological water quality assessment of lake hawassa for multiple designated water uses. J. Urban Environ. Eng. 2015, 9, 146–157. [Google Scholar] [CrossRef]
- Rahmanian, N.; Ali, S.; Homayoonfard, M.; Ali, N.; Rehan, M.; Sadef, Y.; Nizami, A. Analysis of physiochemical parameters to evaluate the drinking water quality in the state of Perak, Malaysia. J. Chem. 2015, 2015, 1–10. [Google Scholar] [CrossRef]
- Hamaidi-Chergui, F.; Brahim Errahmani, M. Water quality and physicochemical parameters of outgoing waters in a pharmaceutical plant. Appl. Water Sci. 2019, 9, 165. [Google Scholar] [CrossRef]
- Domínguez, I.; Oviedo-Ocaña, E.; Hurtado, K.; Barón, A.; Hall, R. Assessing sustainability in rural water supply systems in developing countries using a novel tool based on multi-criteria analysis. Sustainability 2019, 11, 5363. [Google Scholar] [CrossRef]
- Mabey, D.; Peeling, R.; Ustianowski, A.; Perkins, M. Diagnostics for the developing world. Nat. Rev. Microbiol. 2004, 2, 231–240. [Google Scholar] [CrossRef]
- Land, K.; Boeras, D.; Chen, X.; Ramsay, A.; Peeling, R. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 2018, 4, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Kelly, V.; Codispoti, L.; Suttles, S. Autonomous Device with Biofouling Control and Method for Monitoring Aquatic Environment; United. States Patent Application 20090041621, 2014. [Google Scholar]
- Sabaté del Río, J.; Henry, O.; Jolly, P.; Ingber, D. An antifouling coating that enables affinity-based electrochemical biosensing in complex biological fluids. Nat. Nanotechnol. 2019, 14, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Leng, C.; Hung, H.; Sun, S.; Wang, D.; Li, Y.; Jiang, S.; Chen, Z. Probing the surface hydration of nonfouling Zwitterionic and PEG materials in contact with proteins. ACS Appl. Mater. Interfaces 2015, 7, 16881–16888. [Google Scholar] [CrossRef] [PubMed]
- White, A.; Jiang, S. Local and bulk hydration of Zwitterionic glycine and its analogues through molecular simulations. J. Phys. Chem. B 2011, 115, 660–667. [Google Scholar] [CrossRef] [PubMed]
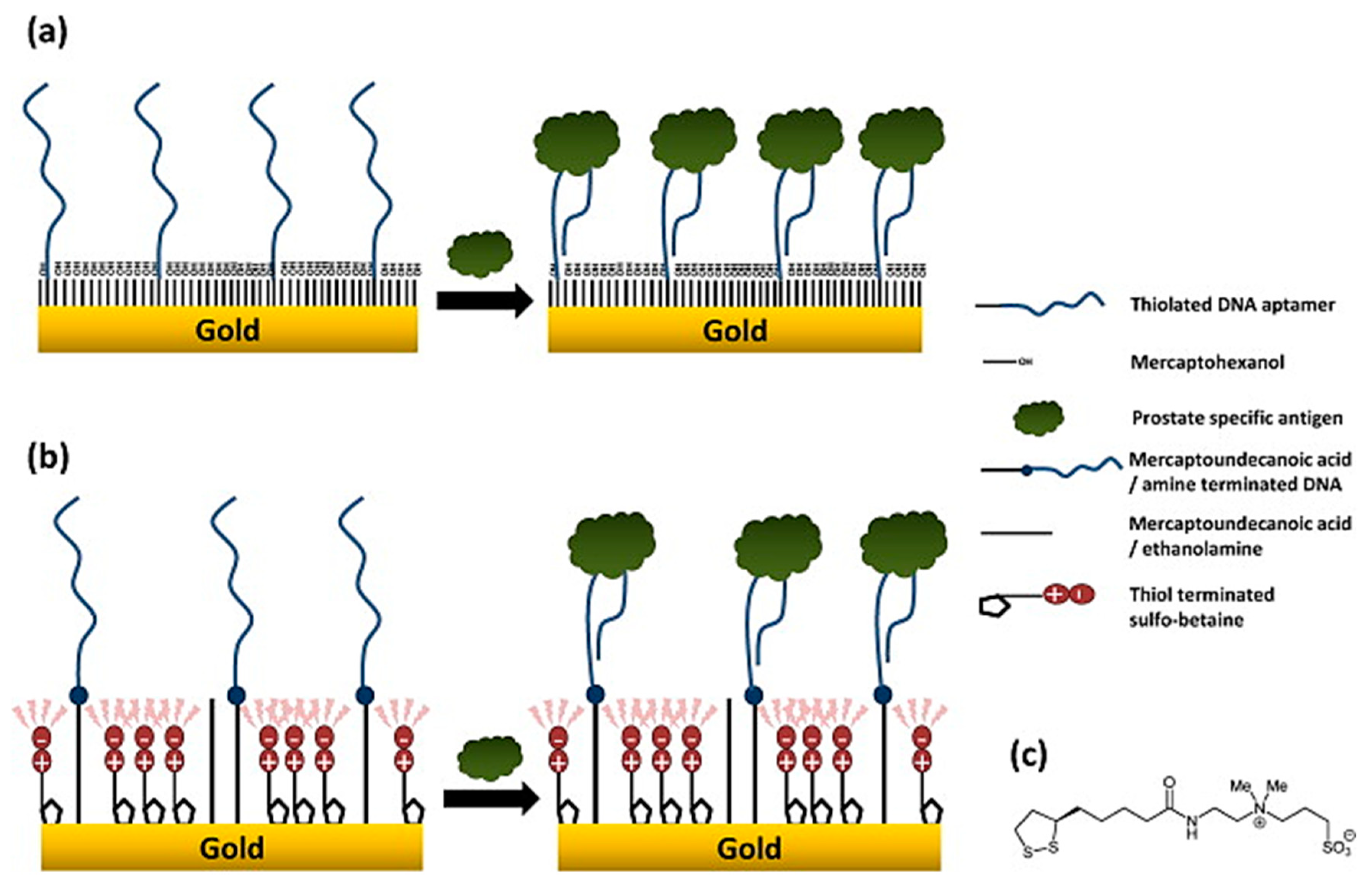
- Jolly, P.; Formisano, N.; Tkáč, J.; Kasák, P.; Frost, C.; Estrela, P. Label-free impedimetric aptasensor with antifouling surface chemistry: A prostate specific antigen case study. Sens. Actuators B Chem. 2015, 209, 306–312. [Google Scholar] [CrossRef]
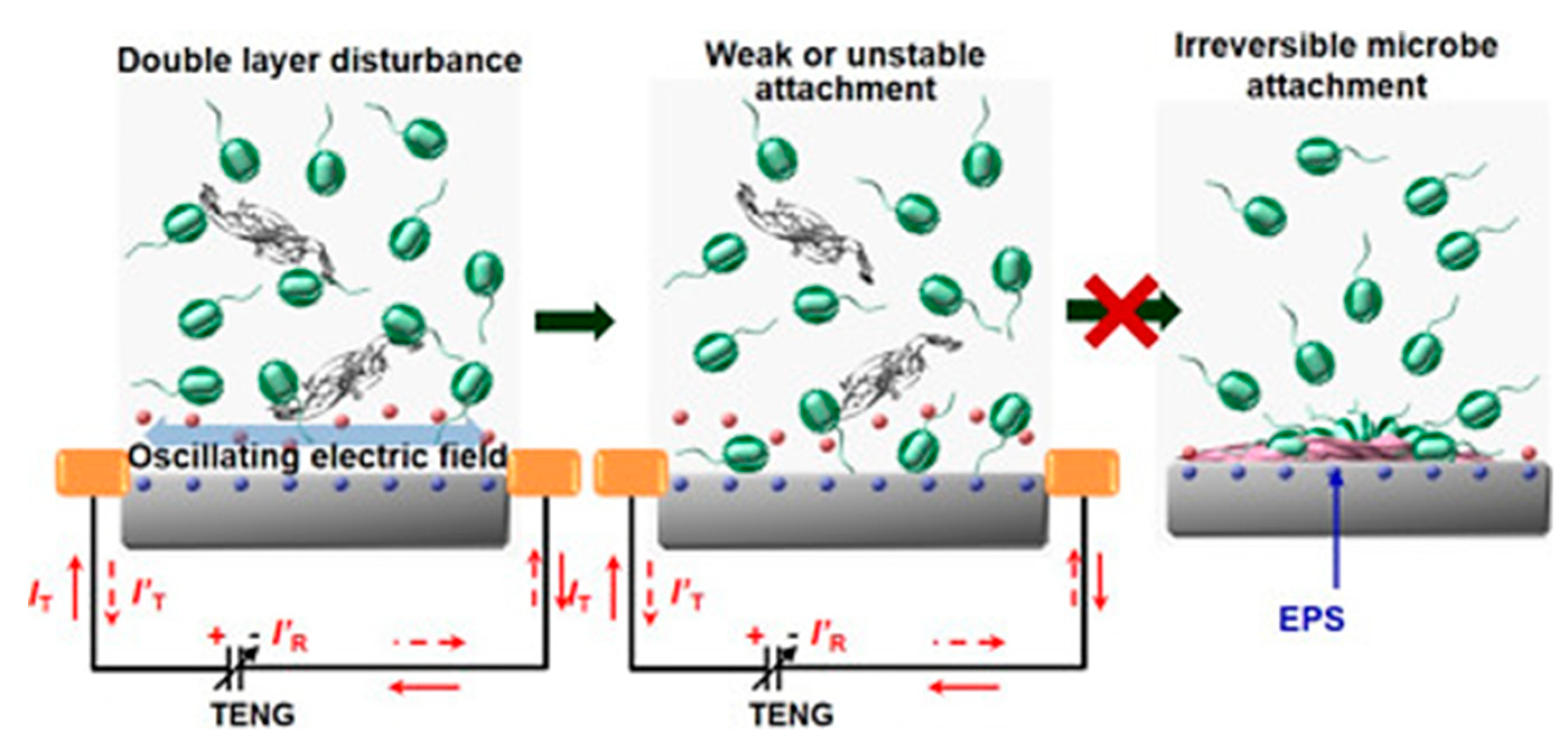
- Long, Y.; Yu, Y.; Yin, X.; Li, J.; Carlos, C.; Du, X.; Jiang, Y.; Wang, X. Effective anti-biofouling enabled by surface electric disturbance from water wave-driven nanogenerator. Nano Energy 2019, 57, 558–565. [Google Scholar] [CrossRef]
- Hafer, M. Quantity and electricity consumption of plug load equipment on a university campus. Energy Effic. 2016, 10, 1013–1039. [Google Scholar] [CrossRef]
- Peixoto, L.; Min, B.; Martins, G.; Brito, A.; Kroff, P.; Parpot, P.; Angelidaki, I.; Nogueira, R. In situ microbial fuel cell-based biosensor for organic carbon. Bioelectrochemistry 2011, 81, 99–103. [Google Scholar] [CrossRef]
- Jiang, Y.; Liang, P.; Liu, P.; Wang, D.; Miao, B.; Huang, X. A novel microbial fuel cell sensor with biocathode sensing element. Biosens. Bioelectron. 2017, 94, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Chouler, J.; Cruz-Izquierdo, Á.; Rengaraj, S.; Scott, J.; Di Lorenzo, M. A screen-printed paper microbial fuel cell biosensor for detection of toxic compounds in water. Biosens. Bioelectron. 2018, 102, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Cunningham, B.; Boeras, D.; Mafaune, P.; Simbi, R.; Peeling, R. Data connectivity: A critical tool for external quality assessment. Afr. J. Lab. Med. 2016, 5, 535. [Google Scholar] [CrossRef] [PubMed]
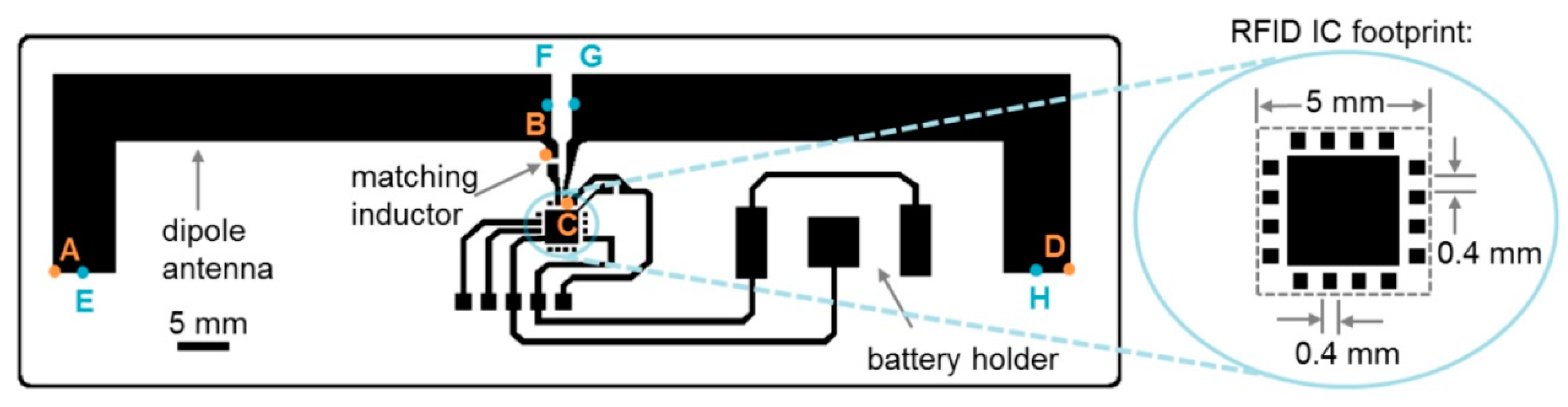
- Smith, S.; Oberholzer, A.; Land, K.; Korvink, J.; Mager, D. Functional screen-printed radio frequency identification tags on flexible substrates, facilitating low-cost and integrated point-of-care diagnostics. Flex. Print. Electron. 2018, 3, 025002. [Google Scholar] [CrossRef]
- Aboul-Enein, Y.; Zuhur, S. Islamic Rulings on Warfare; U.S. Army War College: Carlisle Barracks, PA, USA, 2004. [Google Scholar]
- Vallero, D.; Letcher, T. Regulation of wastes. In Waste: A Handbook for Management; Academic Press: London, UK, 2011; pp. 23–59. [Google Scholar]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar]
- Faridbod, F.; Gupta, V.; Zamani, H. Electrochemical sensors and biosensors. Int. J. Electrochem. 2011, 2011, 1–2. [Google Scholar] [CrossRef]
- Abdalla, N.S.; Amr, A.E.G.E.; El-Tantawy, A.S.M.; Al-Omar, M.A.; Kamel, A.H.; Khalifa, N.M. Tailor-made specific recognition of cyromazine pesticide integrated in a potentiometric strip cell for environmental and food analysis. Polym. (Basel) 2019, 11, 1526. [Google Scholar] [CrossRef]
- Fiel, W.A.; de Freitas Borges, P.A.; Lins, F.C.; de Faria, R.A.D. Recent advances on the electrochemical transduction techniques for the biosensing of pharmaceuticals in aquatic environments. Int. J. Biosens. Bioelectron. 2019, 5, 119–123. [Google Scholar] [CrossRef]
- Chinnappan, R.; Eissa, S.; Alotaibi, A.; Siddiqua, A.; Alsager, O.A.; Zourob, M. In vitro selection of DNA aptamers and their integration in a competitive voltammetric biosensor for azlocillin determination in waste water. Anal. Chim. Acta 2020, 1101, 149–156. [Google Scholar] [CrossRef]
- Fekry, A.M.; Shehata, M.; Azab, S.M.; Walcarius, A. Voltammetric detection of caffeine in pharmacological and beverages samples based on simple nano- Co (II, III) oxide modified carbon paste electrode in aqueous and micellar media. Sens. Actuators B Chem. 2020, 302, 127172. [Google Scholar] [CrossRef]
- Yang, Z.; Castrignanò, E.; Estrela, P.; Frost, C.G.; Kasprzyk-Hordern, B. Community sewage sensors towards evaluation of drug use trends: Detection of cocaine in wastewater with DNA-directed immobilization aptamer sensors. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mayedwa, N.; Ajayi, R.F.; Mongwaketsi, N.; Matinise, N.; Mulaudzi-Masuku, T.; Hendricks, K.; Maaza, M. Development of a novel tyrosinase amperometric biosensor based on tin nanoparticles for the detection of bisphenol A (4.4-isopropylidenediphenol) in water. J. Phys. Conf. Ser. 2019, 1310, 12005. [Google Scholar] [CrossRef]
- Ding, R.; Lisak, G. Sponge-based microfluidic sampling for potentiometric ion sensing. Anal. Chim. Acta 2019, 1091, 103–111. [Google Scholar] [CrossRef] [PubMed]
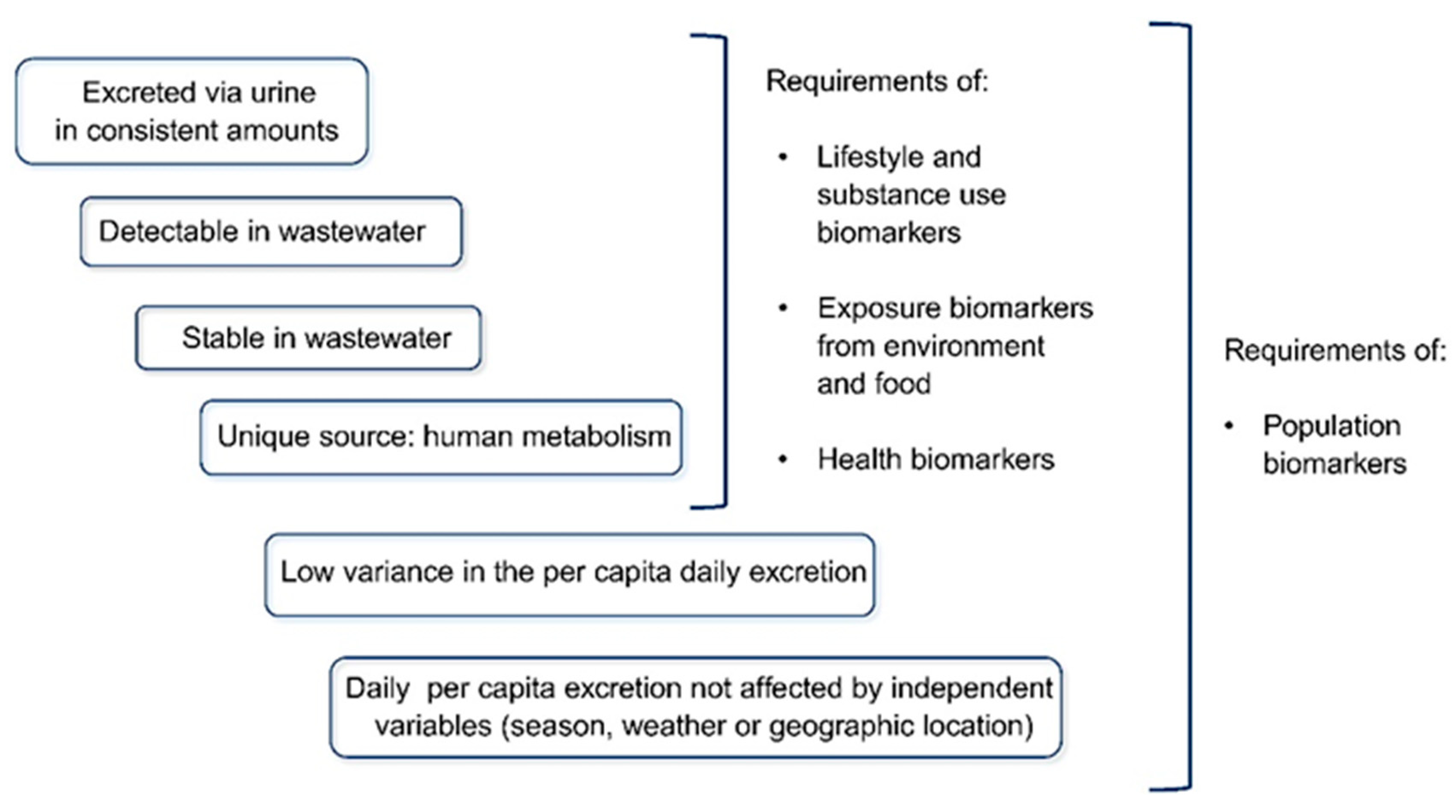
- Senta, I.; Gracia-Lor, E.; Borsotti, A.; Zuccato, E.; Castiglioni, S. Wastewater analysis to monitor use of caffeine and nicotine and evaluation of their metabolites as biomarkers for population size assessment. Water Res. 2015, 74, 23–33. [Google Scholar] [CrossRef]
- Gracia-Lor, E.; Castiglioni, S.; Bade, R.; Been, F.; Castrignanò, E.; Covaci, A.; González-Mariño, I.; Hapeshi, E.; Kasprzyk-Hordern, B.; Kinyua, J.; et al. Measuring biomarkers in wastewater as a new source of epidemiological information: Current state and future perspectives. Environ. Int. 2017, 99, 131–150. [Google Scholar] [CrossRef]
- Khalil, M.M.; Moaty, S.A.A.; Korany, M.A. Carbon nanotubes based potentiometric sensor for determination of bambuterol hydrochloride: Electrochemical and morphology study. Sens. Actuators B Chem. 2018, 273, 429–438. [Google Scholar] [CrossRef]
- Tran, T.B.; Son, S.J.; Min, J. Nanomaterials in label-free impedimetric biosensor: Current process and future perspectives. Biochip J. 2016, 10, 318–330. [Google Scholar] [CrossRef]
- Li, F.; Yu, Z.; Han, X.; Lai, R.Y. Electrochemical aptamer-based sensors for food and water analysis: A review. Anal. Chim. Acta 2019, 1051, 1–23. [Google Scholar] [CrossRef]
- Evans, R.J. Encyclopedia of Biophysics; Springer: Berlin, Germany, 2013. [Google Scholar]
- Gaiji, H.; Jolly, P.; Ustuner, S.; Goggins, S.; Abderrabba, M.; Frost, C.G.; Estrela, P. A Peptide Nucleic Acid (PNA)-DNA ferrocenyl intercalator for electrochemical sensing. Electroanalysis 2017, 29, 917–922. [Google Scholar] [CrossRef]
- Singh, A.; Sinsinbar, G.; Choudhary, M.; Kumar, V.; Pasricha, R.; Verma, H.; Singh, S.; Arora, K. Graphene oxide-chitosan nanocomposite based electrochemical DNA biosensor for detection of typhoid. Sens. Actuators B Chem. 2013, 185, 675–684. [Google Scholar] [CrossRef]
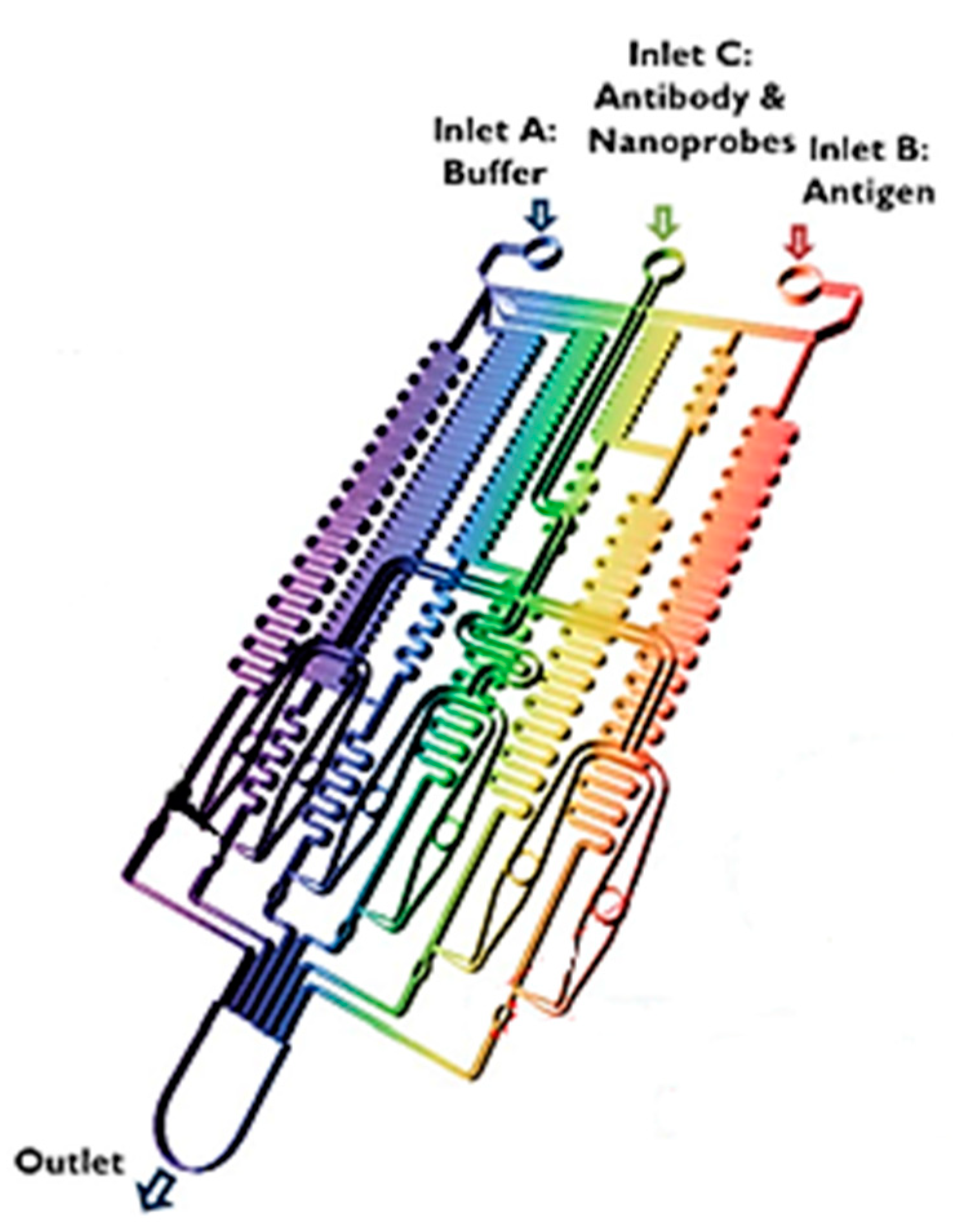
- Altintas, Z.; Akgun, M.; Kokturk, G.; Uludag, Y. A fully automated microfluidic-based electrochemical sensor for real-time bacteria detection. Biosens. Bioelectron. 2018, 100, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, J.; Chen, P.; Zhou, X.; Yang, Y.; Wu, S.; Niu, L.; Han, Y.; Wang, L.; Chen, P.; et al. Label-free, electrochemical detection of methicillin-resistant staphylococcus aureus DNA with reduced graphene oxide-modified electrodes. Biosens. Bioelectron. 2011, 26, 3881–3886. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, N.; Pandey, C.; Soni, A.; Tiwari, I.; Rosillo-Lopez, M.; Salzmann, C.; Malhotra, B.; Sumana, G. Electrochemical genosensor based on carboxylated graphene for detection of water-borne pathogen. Sens. Actuators B Chem. 2018, 275, 312–321. [Google Scholar] [CrossRef]
- Liang, R.; Ding, J.; Gao, S.; Qin, W. Mussel-inspired surface-imprinted sensors for potentiometric label-free detection of biological species. Angew. Chem. Int. Ed. Engl. 2017, 56, 6833–6837. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Seong, T.; Jeun, M.; Lee, K. Field-effect biosensors for on-site detection: Recent advances and promising targets. Adv. Healthc. Mater. 2017, 6, 1700796. [Google Scholar] [CrossRef] [PubMed]
- Primiceri, E.; Chiriacò, M.; Rinaldi, R.; Maruccio, G. Cell chips as new tools for cell biology—Results, perspectives and opportunities. Lab Chip 2013, 13, 3789. [Google Scholar] [CrossRef]
- Chiriacò, M.; Parlangeli, I.; Sirsi, F.; Poltronieri, P.; Primiceri, E. Impedance sensing platform for detection of the food pathogen listeria monocytogenes. Electronics 2018, 7, 347. [Google Scholar] [CrossRef]
- Shoute, L.; Anwar, A.; MacKay, S.; Abdelrasoul, G.; Lin, D.; Yan, Z.; Nguyen, A.; McDermott, M.; Shah, M.; Yang, J.; et al. Immuno-impedimetric biosensor for onsite monitoring of ascospores and forecasting of sclerotinia stem rot of canola. Sci. Rep. 2018, 8, 12396. [Google Scholar] [CrossRef]
- Smith, J.P.; Foster, C.W.; Metters, J.P.; Sutcliffe, O.B.; Banks, C.E. Metallic impurities in graphene screen-printed electrodes can influence their electrochemical properties. Electroanalysis 2014, 26, 2429–2433. [Google Scholar] [CrossRef]
- Grant, N. Automated Sample Preparation Using Adaptive Digital Microfluidics for Lab-on-Chip Devices. Master’s Thesis, Colorado State University, Fort Collins, CO, USA, 2018. [Google Scholar]
- Lee, H.; Xu, L.; Koh, D.; Nyayapathi, N.; Oh, K. Various on-chip sensors with microfluidics for biological applications. Sensors 2014, 14, 17008–17036. [Google Scholar] [CrossRef]
- Lee, M.; Lee, K.; Kim, K.; Oh, K.; Choo, J. SERS-based immunoassay using a gold array-embedded gradient microfluidic chip. Lab Chip 2012, 12, 3720. [Google Scholar] [CrossRef] [PubMed]
- Shanko, E.; van de Burgt, Y.; Anderson, P.; den Toonder, J. Microfluidic magnetic mixing at low reynolds numbers and in stagnant fluids. Micromachines 2019, 10, 731. [Google Scholar] [CrossRef] [PubMed]
- Owen, D.; Mao, W.; Alexeev, A.; Cannon, J.; Hesketh, P. Microbeads for sampling and mixing in a complex sample. Micromachines 2013, 4, 103–115. [Google Scholar] [CrossRef]
- Salafi, T.; Zeming, K.; Zhang, Y. Advancements in microfluidics for nanoparticle separation. Lab Chip 2017, 17, 11–33. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Lombardo, J.; Westerhof, T.; Pennell, M.; Ng, A.; Alshetaiwi, H.; Luna, B.; Nelson, E.; Kessenbrock, K.; Hui, E.; et al. Microfluidic filter device with nylon mesh membranes efficiently dissociates cell aggregates and digested tissue into single cells. Lab Chip 2018, 18, 2776–2786. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, Y.; Lin, Q. A microfluidic device for continuous-flow magnetically controlled capture and isolation of microparticles. J. Microelectromechanical Syst. 2010, 19, 743–751. [Google Scholar] [CrossRef]
- Ou, X.; Chen, P.; Huang, X.; Li, S.; Liu, B. Microfluidic chip electrophoresis for biochemical analysis. J. Sep. Sci. 2019, 43, 258–270. [Google Scholar] [CrossRef]
- Yu, S.; Xu, J.; Huang, K.; Chen, J.; Duan, J.; Xu, Y.; Qing, H.; Geng, L.; Deng, Y. A novel method to predict protein aggregations using two-dimensional native protein microfluidic chip electrophoresis. Anal. Methods 2016, 8, 8306–8313. [Google Scholar] [CrossRef]
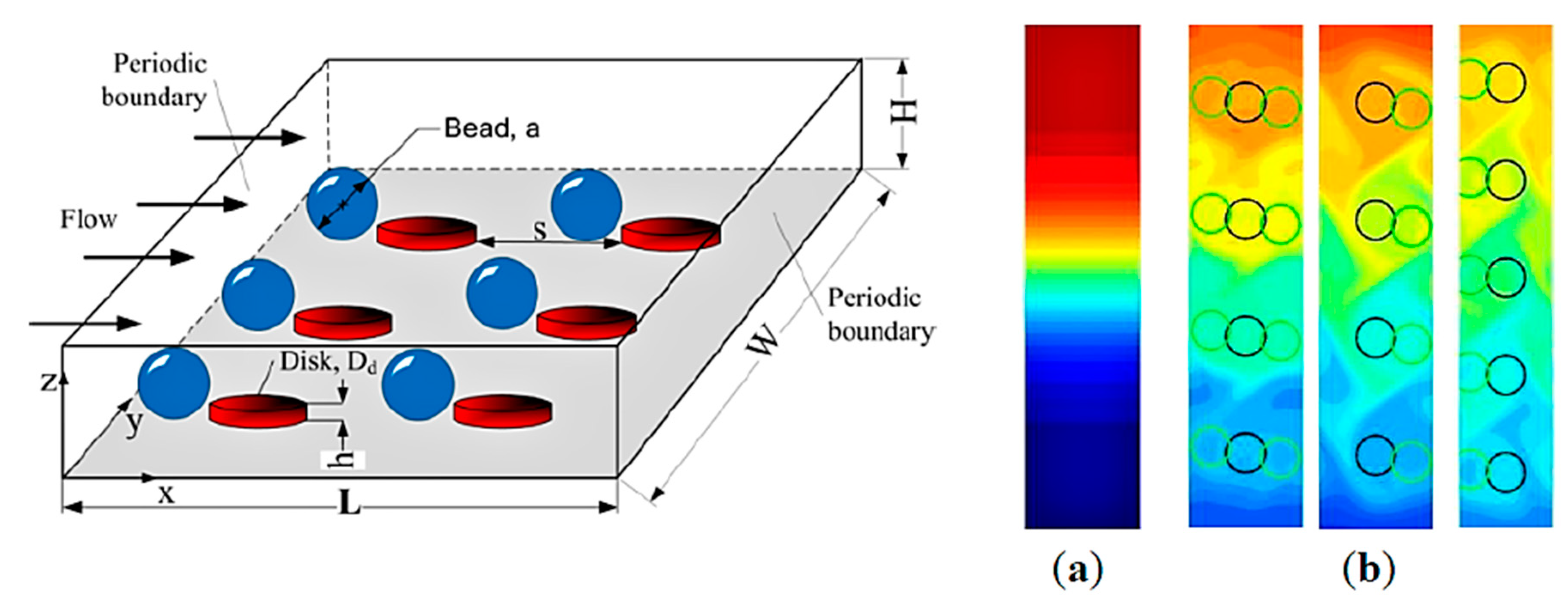
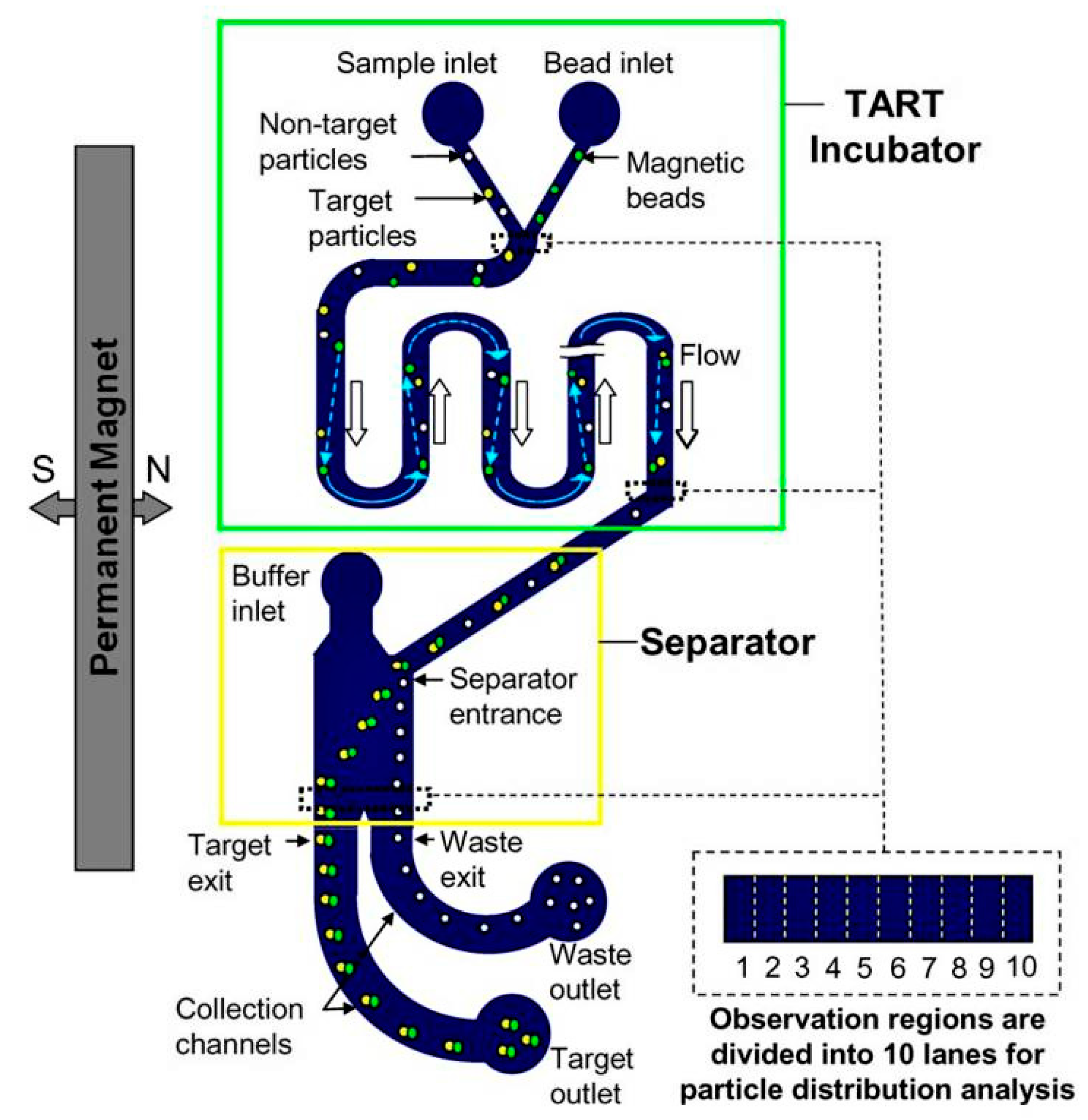
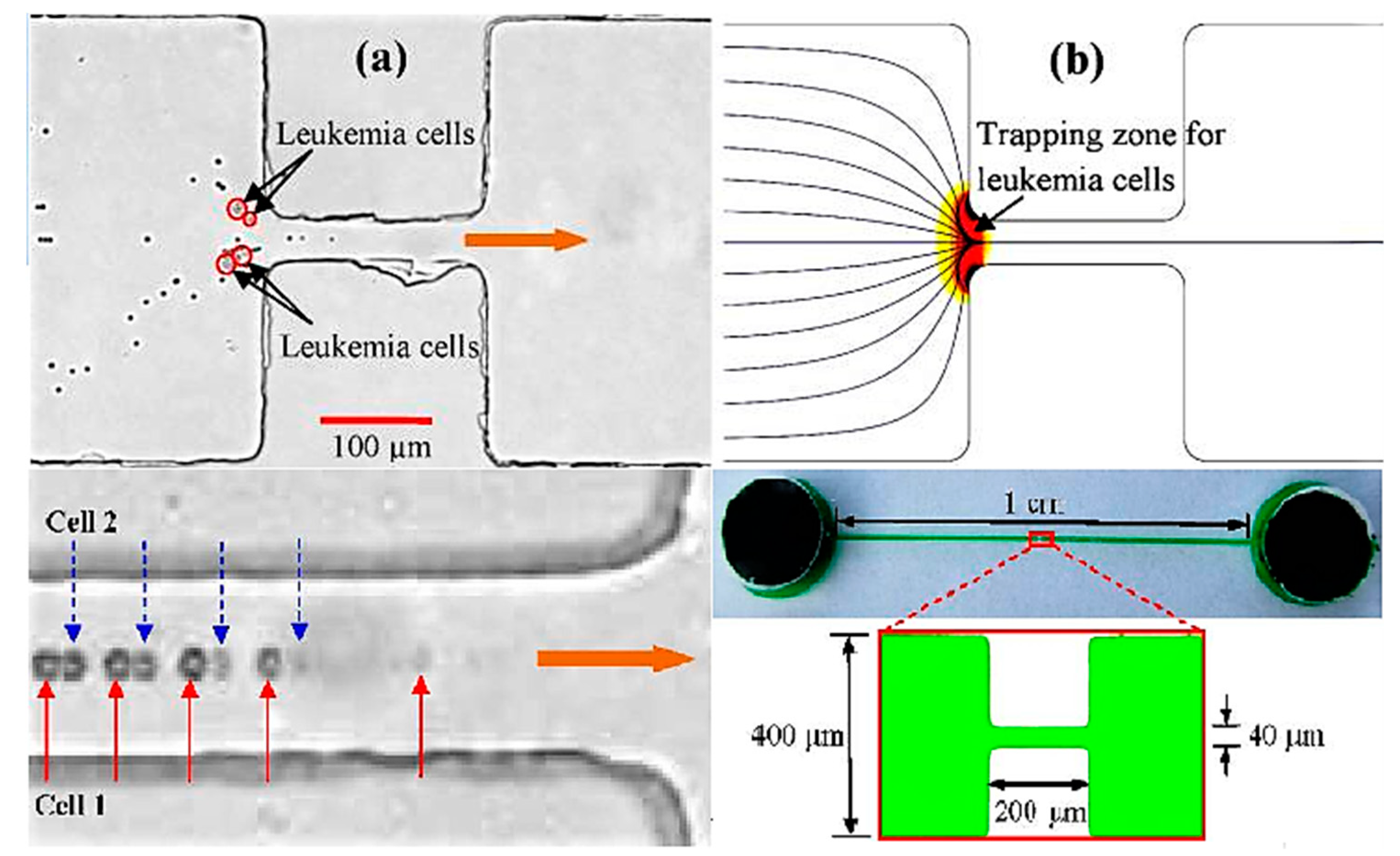
- Zhang, H.; Chang, H.; Neuzil, P. DEP-on-a-Chip: Dielectrophoresis applied to microfluidic platforms. Micromachines 2019, 10, 423. [Google Scholar] [CrossRef]
- Ali, N.; Rampazzo, R.; Costa, A.; Krieger, M. Current nucleic acid extraction methods and their implications to point-of-care diagnostics. BioMed Res. Int. 2017, 2017, 9306564. [Google Scholar] [CrossRef]
- Dutta, G.; Rainbow, J.; Zupancic, U.; Papamatthaiou, S.; Estrela, P.; Moschou, D. Microfluidic devices for label-free dna detection. Chemosensors 2018, 6, 43. [Google Scholar] [CrossRef]
- Bhattacharyya, A.; Klapperich, C. On-chip cell lysis. In Encyclopedia of Microfluidics and Nanofluidics; Springer: Berlin, Germany, 2008; pp. 1513–1515. [Google Scholar]
- Nan, L.; Jiang, Z.; Wei, X. Emerging microfluidic devices for cell lysis: A review. Lab Chip 2014, 14, 1060. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Yoo, J. Lab-on-a-disc platform for automated chemical cell lysis. Sensors 2018, 18, 687. [Google Scholar]
- Church, C.; Zhu, J.; Huang, G.; Tzeng, T.; Xuan, X. Integrated electrical concentration and lysis of cells in a microfluidic chip. Biomicrofluidics 2010, 4, 044101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wu, X.; Peng, R.; Li, D. Electromagnetically controlled microfluidic chip for DNA extraction. Measurement 2015, 75, 23–28. [Google Scholar] [CrossRef]
- Hook, S.; Gallagher, E.; Batley, G. The role of biomarkers in the assessment of aquatic ecosystem health. Integr. Environ. Assess. Manag. 2014, 10, 327–341. [Google Scholar] [CrossRef]
- Contreras-Naranjo, J.; Aguilar, O. Suppressing non-specific binding of proteins onto electrode surfaces in the development of electrochemical immunosensors. Biosensors 2019, 9, 15. [Google Scholar] [CrossRef]
- Lichtenberg, J.; Ling, Y.; Kim, S. Non-specific adsorption reduction methods in biosensing. Sensors 2019, 19, 2488. [Google Scholar] [CrossRef]
- Whiteaker, J.R.; Zhao, L.; Zhang, H.Y.; Feng, L.C.; Piening, B.D.; Anderson, L.; Paulovich, A.G. Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers. Anal. Biochem. 2007, 362, 44–54. [Google Scholar] [CrossRef]
- Katoba, J.; Kuupiel, D.; Mashamba-Thompson, T.P. Toward improving accessibility of point-of-care diagnostic services for maternal and child health in low-and middle-income countries. Point Care 2019, 18, 17–25. [Google Scholar] [CrossRef]
- Sommer, G.J.; Hatch, A.V. IEF in microfluidic devices. Electrophoresis 2009, 30, 742–757. [Google Scholar] [CrossRef] [PubMed]
- Fakruddin, M.; Mannan, K.; Hossain, M.; Islam, S.; Mazumdar, R.; Chowdhury, A.; Chowdhury, M. Nucleic acid amplification: Alternative methods of polymerase chain reaction. J. Pharm. Bioallied Sci. 2013, 5, 245. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Jin, H.; Xia, D.; Shao, H.; Peng, K.; Liu, X.; Huang, H.; Zhang, Q.; Guo, J.; Wang, Y.; et al. Biomimetic polymer-based method for selective capture of C-reactive protein in biological fluids. ACS Appl. Mater. Interfaces 2018, 10, 41999–42008. [Google Scholar] [CrossRef] [PubMed]
- Płotka-Wasylka, J.; Szczepańska, N.; de la Guardia, M.; Namieśnik, J. Modern trends in solid phase extraction: New sorbent media. TrAC Trends Anal. Chem. 2016, 77, 23–43. [Google Scholar] [CrossRef]
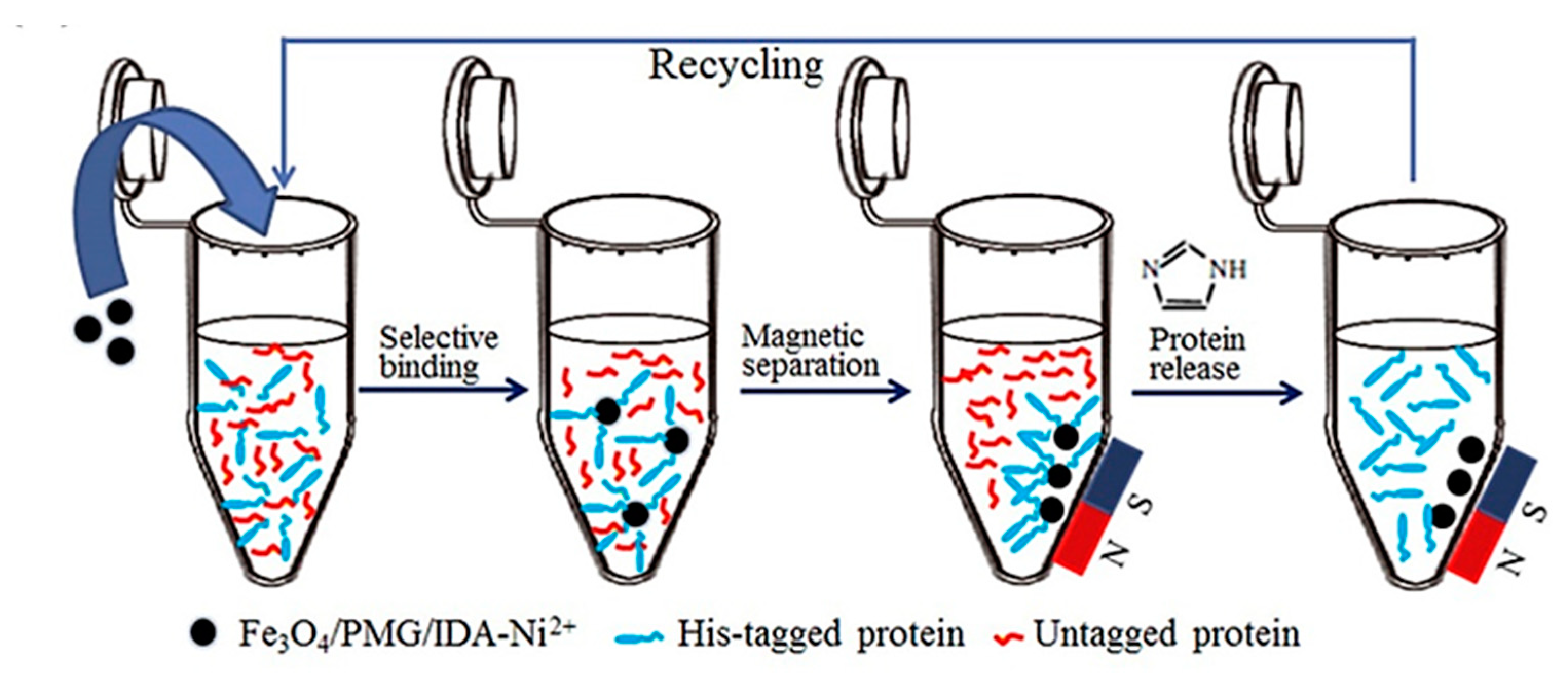
- Zhou, Y.; Yan, D.; Yuan, S.; Chen, Y.; Fletcher, E.E.; Shi, H.; Han, B. Selective binding, magnetic separation and purification of histidine-tagged protein using biopolymer magnetic core-shell nanoparticles. Protein Expr. Purif. 2018, 144, 5–11. [Google Scholar] [CrossRef]
- Safarik, I.; Safarikova, M. Magnetic techniques for the isolation and purification of proteins and peptides. Biomagn. Res. Technol. 2004, 2, 1–17. [Google Scholar] [CrossRef]
- Lin, C.-C.; Hsu, J.-L.; Lee, G.-B. Sample preconcentration in microfluidic devices. Microfluid. Nanofluidics 2011, 10, 481–511. [Google Scholar] [CrossRef]
- Wang, Z.; Ivory, C.; Minerick, A.R. Surface isoelectric focusing (sIEF) with carrier ampholyte pH gradient. Electrophoresis 2017, 38, 2565–2575. [Google Scholar] [CrossRef]
- Taylor, J.; Triggle, D. Comprehensive Medicinal Chemistry 2, 5th ed.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 357–397. [Google Scholar]
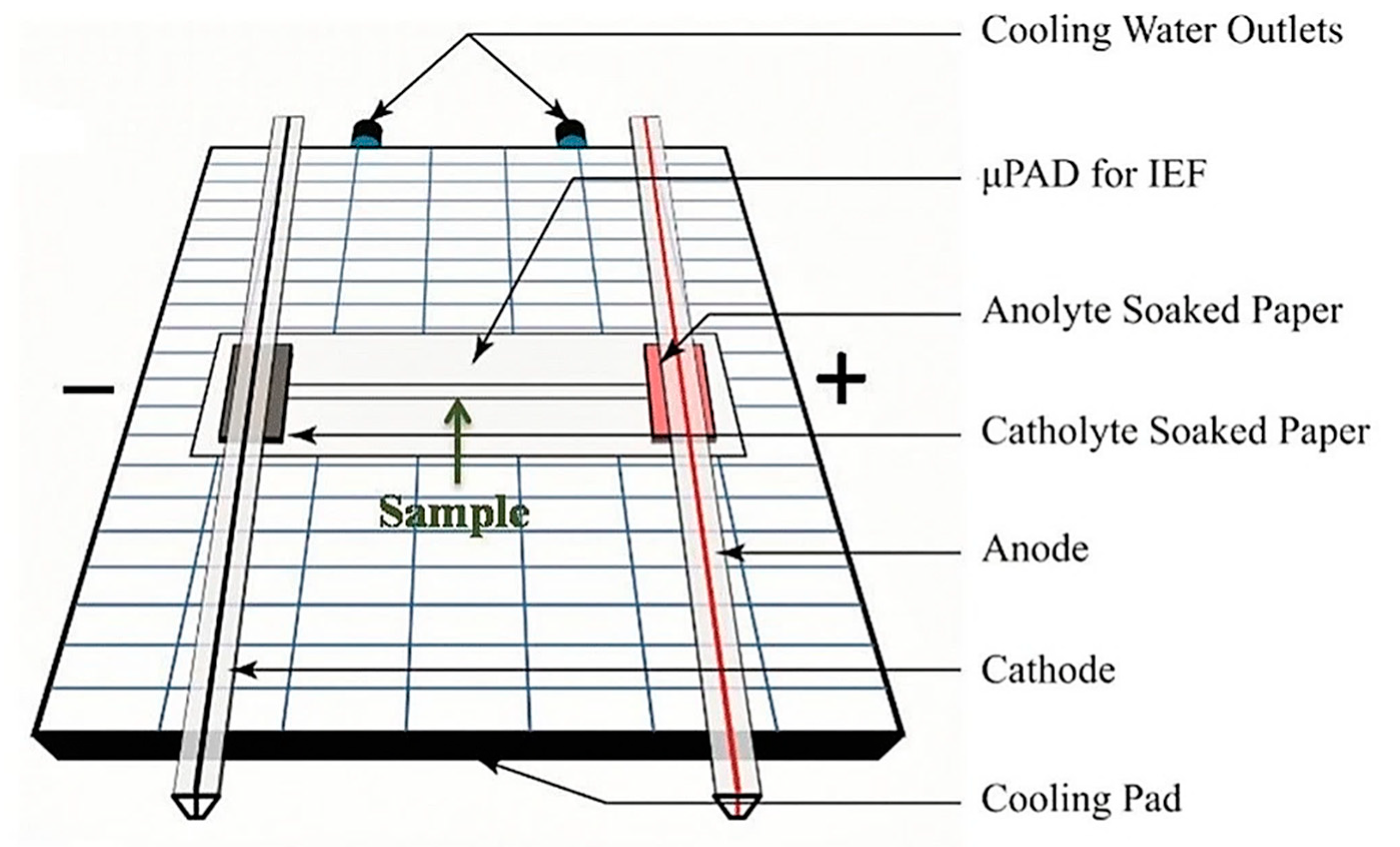
- Yu, S.; Yan, C.; Hu, X.; He, B.; Jiang, Y.; He, Q. Isoelectric focusing on microfluidic paper-based chips. Anal. Bioanal. Chem. 2019, 411, 5415–5422. [Google Scholar] [CrossRef]
- Zhang, Y.; Ozdemir, P. Microfluidic DNA amplification—A review. Anal. Chim. Acta 2009, 638, 115–125. [Google Scholar] [CrossRef]
- Mullis, K.; Faloona, F.; Scharf, S.; Saiki, R.; Horn, G.; Erlich, H. Specific enzymatic amplification of DNA in vitro: The polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 1986, 51, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Lui, C.; Cady, N.; Batt, C. Nucleic acid-based detection of bacterial pathogens using integrated microfluidic platform systems. Sensors 2009, 9, 3713–3744. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Kodzius, R.; Cao, W.; Wen, W. Extraction, amplification and detection of DNA in microfluidic chip-based assays. Microchim. Acta 2013, 181, 1611–1631. [Google Scholar] [CrossRef]
- Ahrberg, C.; Manz, A.; Chung, B. Polymerase chain reaction in microfluidic devices. Lab Chip 2016, 16, 3866–3884. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Kim, J.; Ku, J.; Kim, Y.; Song, H.; Kim, J. Printed circuit board-based polymerase chain reaction chip. Sens. Lett. 2012, 10, 1197–1202. [Google Scholar] [CrossRef]
- Moschou, D.; Vourdas, N.; Filippidou, M.; Tsouti, V.; Kokkoris, G.; Tsekenis, G.; Zergioti, I.; Chatzandroulis, S.; Tserepi, A. Integrated biochip for PCR-based DNA amplification and detection on capacitive biosensors. SPIE Proc. 2013, 8765. [Google Scholar]
- Hwang, J.; Kim, S.; Kim, Y.; Song, H.; Park, C.; Kim, J. Implementation of PCB-based PCR chip using double-sided tape. Int. J. Control Autom. 2015, 8, 117–124. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Itou, T.; Markotter, W.; Nel, L. Reverse transcription-loop-mediated isothermal amplification system for the detection of rabies virus. In Current Laboratory Techniques in Rabies Diagnosis, Research and Prevention; Academic Press: London, UK, 2014; pp. 85–95. [Google Scholar]
- Zhang, H.; Xu, Y.; Fohlerova, Z.; Chang, H.; Iliescu, C.; Neuzil, P. LAMP-on-a-chip: Revising microfluidic platforms for loop-mediated DNA amplification. TrAC Trends Anal. Chem. 2019, 113, 44–53. [Google Scholar] [CrossRef]
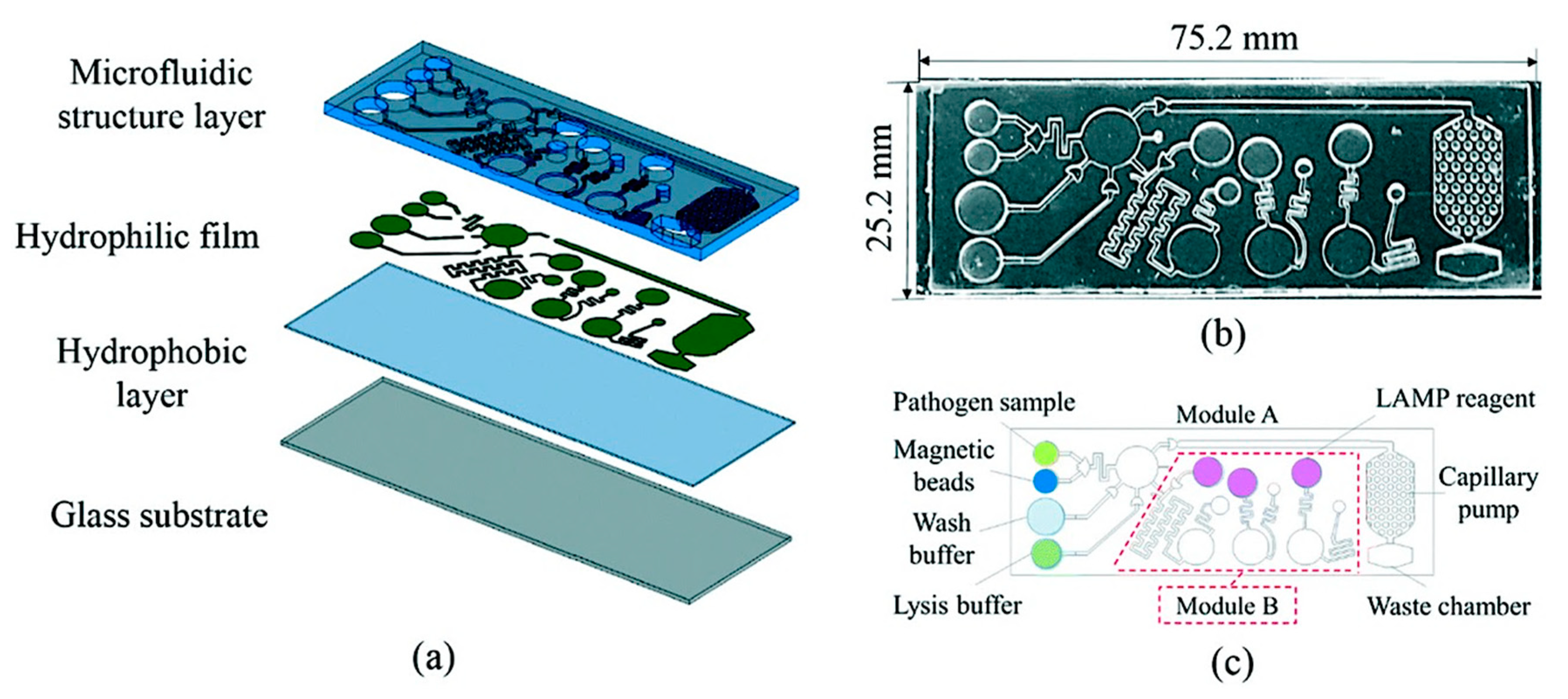
- Ma, Y.; Li, K.; Chen, Y.; Lee, Y.; Chou, S.; Lai, Y.; Huang, P.; Ma, H.; Lee, G. A sample-to-answer, portable platform for rapid detection of pathogens with a smartphone interface. Lab Chip 2019, 19, 3804–3814. [Google Scholar] [CrossRef]


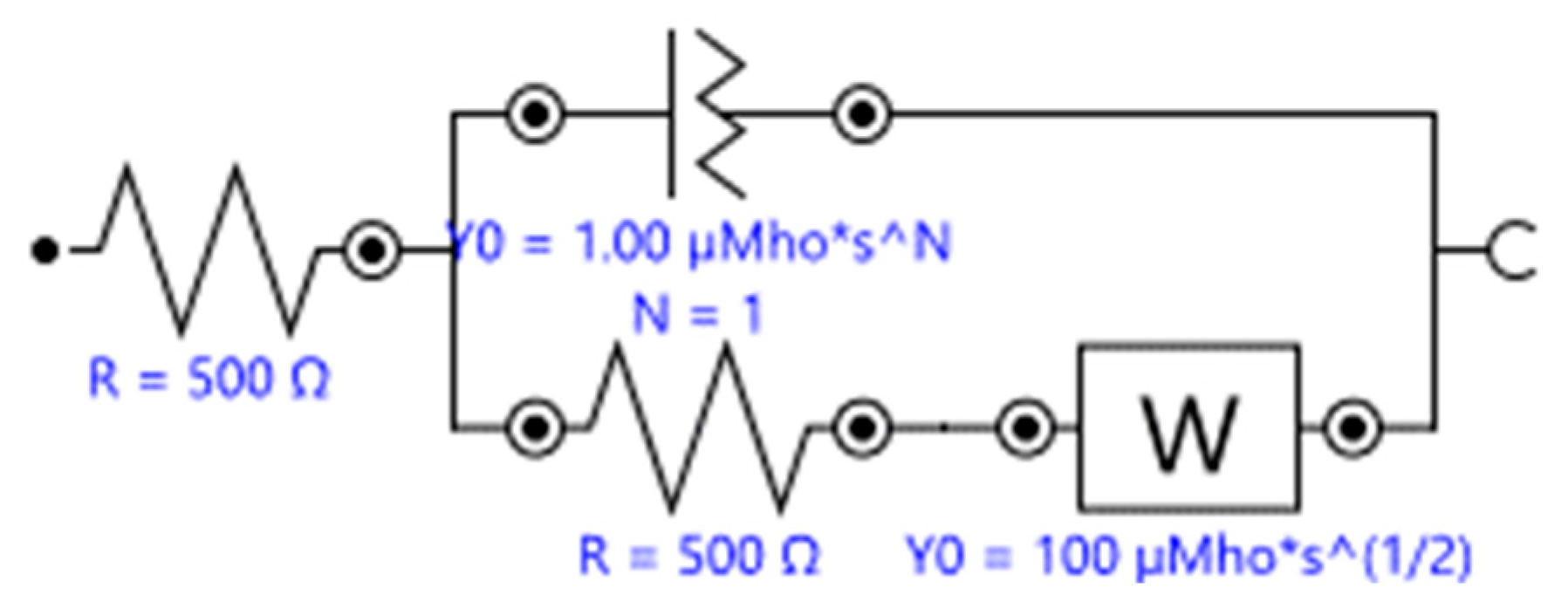



| Amperometric (Voltammetric) | Potentiometric | Impedimetric | |
|---|---|---|---|
| Input | Controlled constant voltage or a controlled series of voltages (e.g., linear potential sweep, cyclic voltammetry, differential pulse voltammetry, etc.). | A defined voltage at the reference electrode. Little to no current presence. | A DC bias potential and a wide range AC frequency scans (Faradaic impedance) or wide range AC frequency scan (non-Faradaic capacitance). |
| Output | Change in current against time or change in current against the applied voltages. | The potential difference between the WE and the RE (a constant potential). | Complex impedance or complex capacitance. |
| How to Improve | Utilize electron mediator or conductive nano-architectures (e.g., nanotubes) or employ interdigitated electrodes to amplify the signal and to improve the signal-to-noise ratio. | Incorporates CNT into the sensor to improve heterogeneous charge transfer and increase surface area [100]. | Immobilize nanoparticles as a secondary architecture to increase the surface area and improve charge transfer [101]. For capacitive sensors, implement single frequency monitoring to realize a faster and simpler system. |
| Principle of Detection | Measured Faradaic current is proportional to the concentration of the analyte. | The potential difference between the WE and RE is proportionally present in the ion activity in the sample. Widely applied with ion-selective electrodes (ISE). | Measured complex impedance contains the information of charge transfer resistance (if redox couple is present) and the capacitive property of the electrochemical double layer in the vicinity of the WE. |
| Advantages | System simplicity (easy to integrate and potentially low cost); fast detection [102]; Low detection limit and high sensitivity [103]; potential in intercalator based DNA detection using PNA probes [104]. | Suitable for mass-production. | Potentially label-free and easy to prepare the electrode. |
| Limitations | pH changes upon the introduction of a biological specimen may impede the enzyme activity. | High requirement to the reference electrode (stable and accurate) [102]; same drawbacks of using enzyme as a label or a BRE; inherently susceptible to the environment’s pH value during the measurements as the detecting method relies on the ion activity; limited applications due to difficulties in being employed in affinity-based detections. | Taking relatively more time to conduct one measurement due to the time-consuming frequency scan process; difficulties in miniaturization due to the more complicated input and output control; for its label-free nature, an adequate antifouling system and certain signal amplification modifications may be required. |
| Analyte | Analyte Type | Biosensing Method | Biorecognition Element | Labelling | Detection Limit (sample matrix) | Ref. |
|---|---|---|---|---|---|---|
| Salmonella typhi | DNA sequence | Voltammetry (differential pulse voltammetry) | Complementary DNA Probe | Methylene blue (MB) redox intercalator | 10 fM within 60 s hybridisation time, 100 fM in serum samples | [105] |
| Escherichia coli | Whole cell sandwich assay | Cyclic voltammetry | E. Coli surface antibody | Horseradish peroxidase antibody (electrochemical sandwich ELISA) | 50 cfu mL−1 in water samples | [106] |
| methicillin-resistant staphylococcus aureus (MRSA) | DNA sequence | Electrochemical impedance spectroscopy | DNA sequence | Label-free (in presence of ferrocene redox mediator) | 100 fM in buffer | [107] |
| Escherichia coli(O157:H7 serotype) | DNA sequence | Electrochemical impedance spectroscopy | DNA sequence | Label-free | 1 fM in buffer | [108] |
| Yeast cells | Whole cell microbe | Chronopotentiometry | Molecularly imprinted polymer (MIP)layer | Label-free | Cultured yeast cells: 50 CFU/ml (diluted in buffer) | [109] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rainbow, J.; Sedlackova, E.; Jiang, S.; Maxted, G.; Moschou, D.; Richtera, L.; Estrela, P. Integrated Electrochemical Biosensors for Detection of Waterborne Pathogens in Low-Resource Settings. Biosensors 2020, 10, 36. https://doi.org/10.3390/bios10040036
Rainbow J, Sedlackova E, Jiang S, Maxted G, Moschou D, Richtera L, Estrela P. Integrated Electrochemical Biosensors for Detection of Waterborne Pathogens in Low-Resource Settings. Biosensors. 2020; 10(4):36. https://doi.org/10.3390/bios10040036
Chicago/Turabian StyleRainbow, Joshua, Eliska Sedlackova, Shu Jiang, Grace Maxted, Despina Moschou, Lukas Richtera, and Pedro Estrela. 2020. "Integrated Electrochemical Biosensors for Detection of Waterborne Pathogens in Low-Resource Settings" Biosensors 10, no. 4: 36. https://doi.org/10.3390/bios10040036
APA StyleRainbow, J., Sedlackova, E., Jiang, S., Maxted, G., Moschou, D., Richtera, L., & Estrela, P. (2020). Integrated Electrochemical Biosensors for Detection of Waterborne Pathogens in Low-Resource Settings. Biosensors, 10(4), 36. https://doi.org/10.3390/bios10040036