Development of a Gold Nanoparticle-Based Sensor for Authentication of Organic Milk Based on Differential Levels of miRNA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
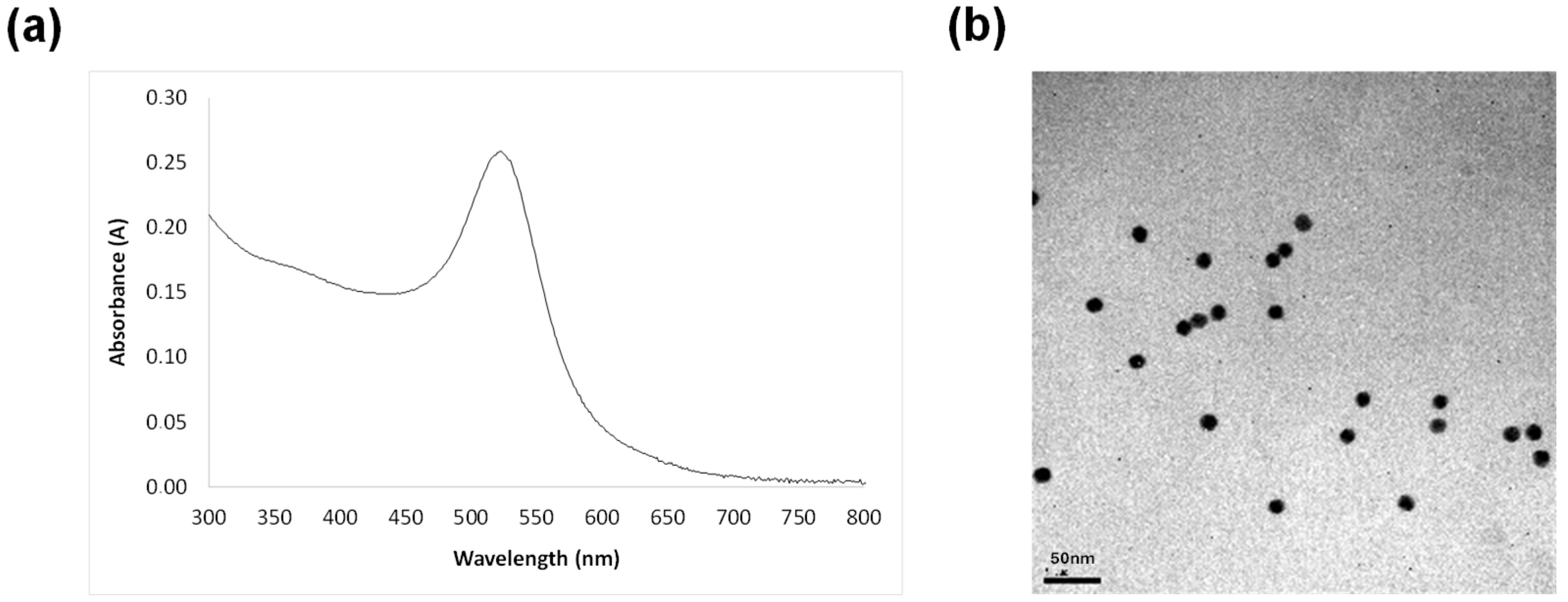
2.2. Synthesis of Gold Nanoparticles
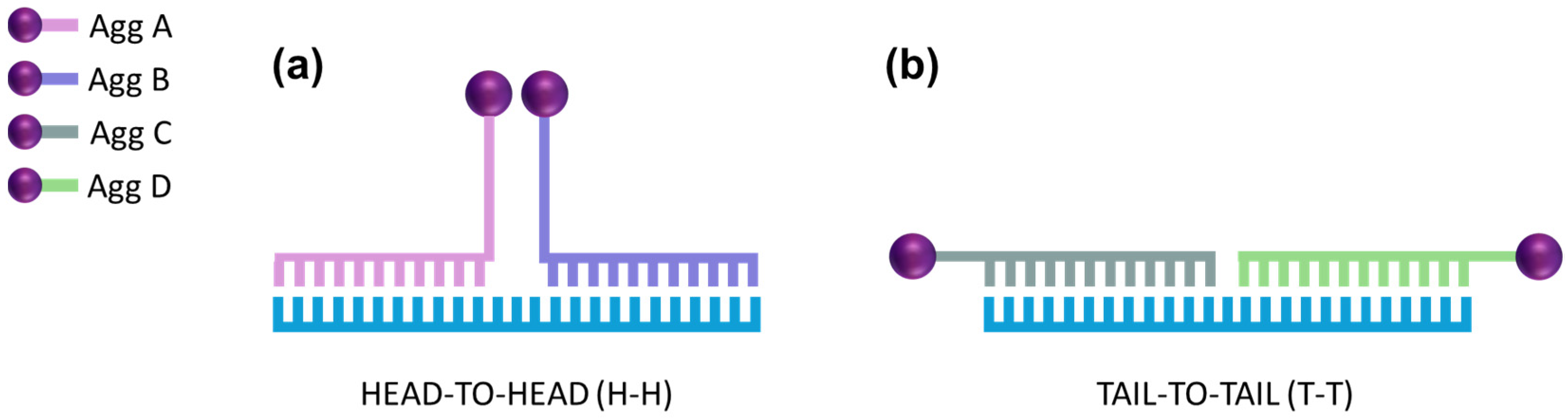
2.3. Functionalization of AuNPs
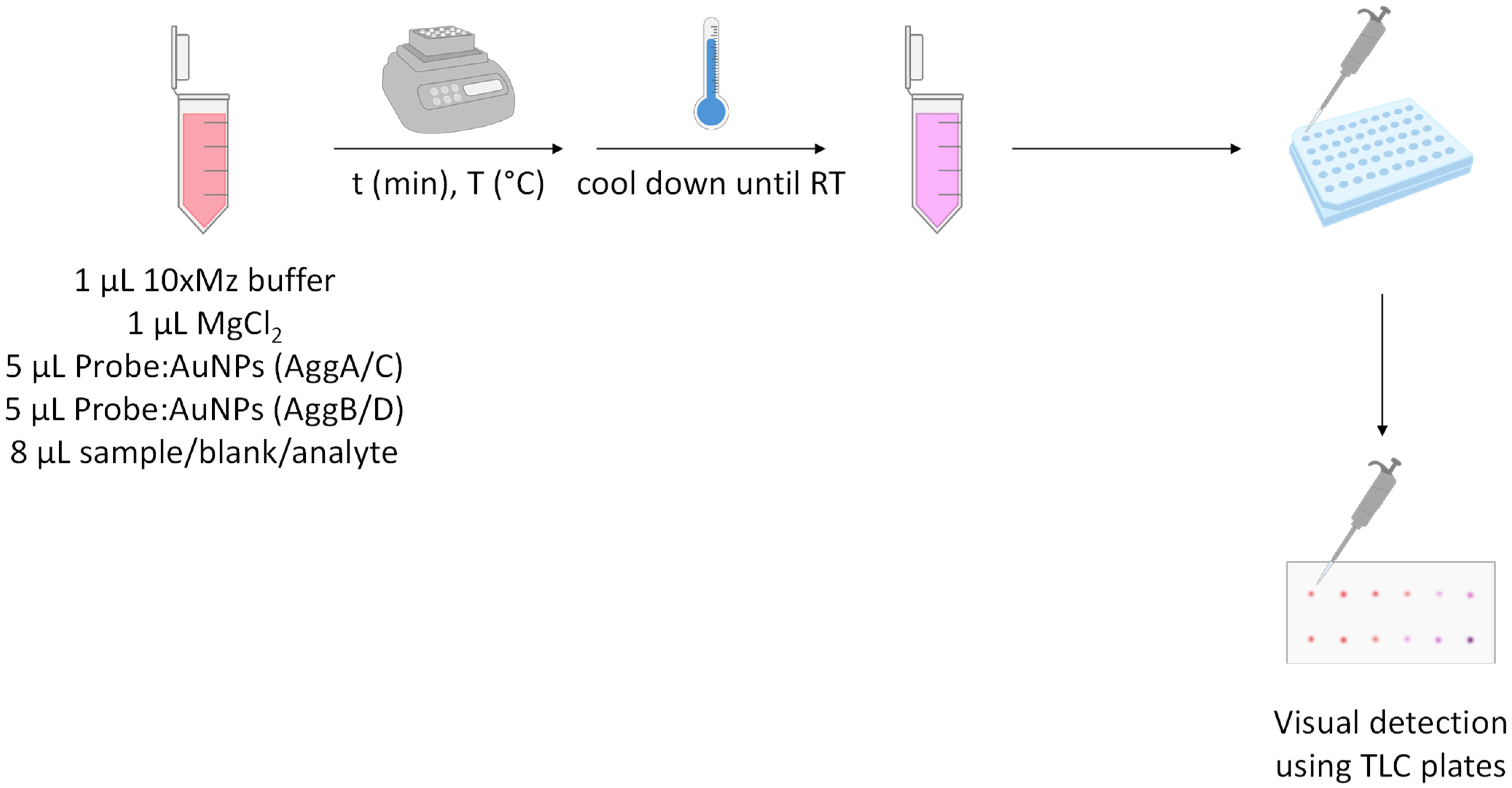
2.4. Method Development
2.5. Sample Pre-Treatment
3. Results and Discussion
3.1. Method Development
3.1.1. Assay Optimization
Agg Probe-AuNP Ratio Optimization
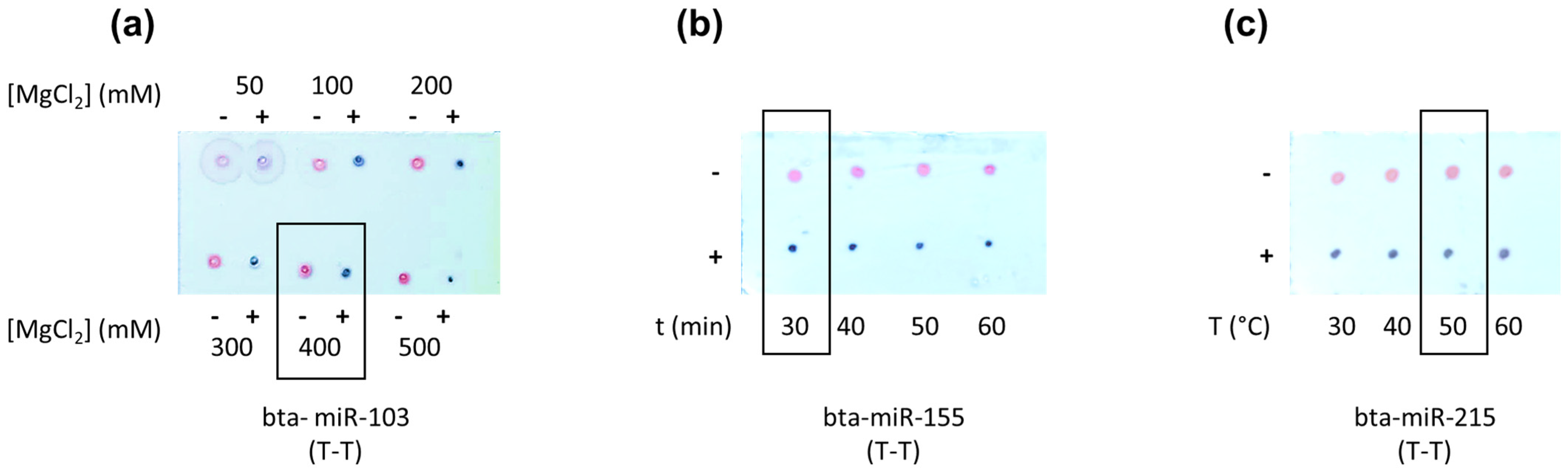
MgCl2 Concentration, Hybridization Temperature, and Time Optimization
3.1.2. Analytical Characterization of the Method
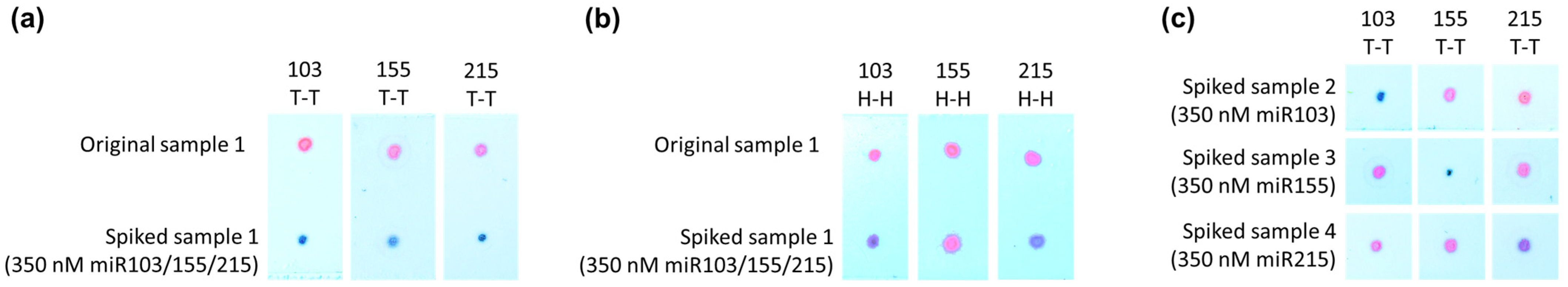
3.2. Application of the Developed Sensor
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Luykx, D.M.A.M.; van Ruth, S.M. An Overview of Analytical Methods for Determining the Geographical Origin of Food Products. Food Chem. 2008, 107, 897–911. [Google Scholar] [CrossRef]
- Tao, S.; Bubolz, J.W.; Do Amaral, B.C.; Thompson, I.M.; Hayen, M.J.; Johnson, S.E.; Dahl, G.E. Effect of heat stress during the dry period on mammary gland development. J. Dairy. Sci. 2011, 94, 5976–5986. [Google Scholar] [CrossRef]
- Ariza, A.B.; Juaristi, J.L. Sistemas y prácticas de manejo en rebaños de vacuno lechero en España. Tierras De Castilla Y León: Ganad. 2008, 151, 56–60. [Google Scholar]
- Van Dijk, H.; Schukking, S.; Van der Berg, R. Fifty years of forage supply on dairy farms in the Netherlands. Grassl. Sci. 2015, 20, 12–20. [Google Scholar]
- Steinfeld, H.; Gerber, P.; Wassenaar, T.D.; Castel, V.; Rosales, M.; de Haan, C. Livestock’s long shadow: Environmental issues and options; Food and Agriculture Organization: Rome, Italy, 2006; Available online: https://www.fao.org/3/a0701e/a0701e00.htm (accessed on 30 June 2024).
- Santa, A.; Mierlita, D.; Dărăban, S.; Socol, C.T.; Vicas, S.I.; Șuteu, M.; Maerescu, C.M.; Stanciu, A.S.; Pop, I.M. The Effect of Sustainable Feeding Systems, Combining Total Mixed Rations and Pasture, on Milk Fatty Acid Composition and Antioxidant Capacity in Jersey Dairy Cows. Animals 2022, 12, 908. [Google Scholar] [CrossRef]
- De La Torre-Santos, S.; Royo, L.J.; Martínez-Fernández, A.; Menéndez-Miranda, M.; Rosa-García, R.; Vicente-Mainar, F. Influence of the Type of Silage in the Dairy Cow Ration, with or without Grazing, on the Fatty Acid and Antioxidant Profiles of Milk. Dairy 2021, 2, 716–728. [Google Scholar] [CrossRef]
- De La Torre-Santos, D.; Royo, L.J.; Martínez-Fernández, A.; Chocarro, C.; Vicente-Mainar, F. The mode of grass supply to dairy cows impacts on fatty acid and antioxidant profile of milk. Foods 2020, 9, 1256. [Google Scholar] [CrossRef] [PubMed]
- Pacífico, C.; Ricci, S.; Sajovitz, F.; Castillo-Lopez, E.; Rivera-Chacon, R.; Petri, R.M.; Zebeli, Q.; Reisinger, N.; Kreuzer-Redmer, S. Bovine rumen epithelial miRNA-mRNA dynamics reveals post-transcriptional regulation of gene expression upon transition to high-grain feeding and phytogenic supplementation. Genomics 2022, 114, 110333. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Sanjurjo, M.; Úbeda, N.; Fernández-García, B.; Del Valle, M.; Ramírez de Molina, A.; Crespo, M.C.; Martín-Hernández, R.; Casas-Agustench, P.; Martínez-Camblor, P.; de Gonzalo-Calvo, D.; et al. Exercise dose affects the circulating microRNA profile in response to acute endurance exercise in male amateur runners. Scand. J. Med. Sci. Sports 2020, 30, 1896–1907. [Google Scholar] [CrossRef]
- Mobuchon, L.; Le Guillou, S.; Marthey, S.; Laubier, J.; Laloe, D.; Bes, S.; Le Provost, F.; Leroux, C. Sunflower oil supplementation affects the expression of miR-20a-5p and miR-142-5p in the lactating bovine mammary gland. PLoS ONE 2017, 12, e0185511. [Google Scholar] [CrossRef]
- Muroya, S.; Shibata, M.; Hayashi, M.; Oe, M.; Ojima, K. Differences in circulating microRNAs between Grazing and Grain-Fed Wagyu Cattle Are Associated with Altered Expression of Intramuscular microRNA, the Potential Target PTEN, and Lipogenic Genes. PLoS ONE 2016, 11, e0162496. [Google Scholar] [CrossRef]
- Huang, J.; Wang, F.; Argyris, E.; Chen, K.; Liang, Z.; Tian, H.; Huang, W.; Squires, K.; Verlinghieri, G.; Zhang, H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 2007, 13, 1241–1247. [Google Scholar] [CrossRef]
- Izumi, H.; Kosaka, N.; Shimizu, T.; Sekine, K.; Ochiya, T.; Takase, M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J. Dairy. Sci. 2012, 95, 4831–4841. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gao, C.; Li, H.; Huang, L.; Sun, Q.; Dong, Y.; Tian, C.; Gao, S.; Dong, H.; Guan, D.; et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010, 20, 1128–1137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Lozano, E.; Sebastián-Valles, F.; Knott-Torcal, C. Circulating microRNAs in Breast Milk and Their Potential Impact on the Infant. Nutrients 2020, 12, 3066. [Google Scholar] [CrossRef] [PubMed]
- Cammaerts, S.; Strazisar, M.; De Rijk, P.; Del Favero, J. Genetic variants in microRNA genes: Impact on microRNA expression, function, and disease. Front. Genet. 2015, 6, 186. [Google Scholar] [CrossRef]
- Colitti, M.; Sgorlon, S.; Licastro, D.; Stefanon, B. Differential expression of miRNAs in milk exosomes of cows subjected to group relocation. Res. Vet. Sci. 2019, 122, 148–155. [Google Scholar] [CrossRef]
- Lai, Y.C.; Fujikawa, T.; Maemura, T.; Ando, T.; Kitahara, G.; Endo, Y.; Yamato, O.; Koiwa, M.; Kubota, C.; Miura, N. Inflammation-related microRNA expression level in the bovine milk is affected by mastitis. PLoS ONE 2017, 12, e0177182. [Google Scholar] [CrossRef] [PubMed]
- Muroya, S.; Ogasawara, H.; Hojito, M. Grazing affects exosomal circulating microRNAs in cattle. PLoS ONE 2015, 10, e0136475. [Google Scholar] [CrossRef]
- Bauman, D.E.; Harvatine, K.J.; Lock, A.L. Nutrigenomics, rumen-derived bioactive fatty acids, and the regulation of milk fat synthesis. Annu. Rev. Nutr. 2011, 31, 299–319. [Google Scholar] [CrossRef]
- Padovani, M.; Lavigne, J.A.; Chandramouli, G.V.; Perkins, S.N.; Barrett, J.C.; Hursting, S.D.; Bennett, L.M.; Berrigan, D. Distinct effects of calorie restriction and exercise on mammary gland gene expression in C57BL/6 mice. Cancer Prev. Res. 2009, 2, 1076–1087. [Google Scholar] [CrossRef]
- Abou El Qassim, L.; Alonso, J.; Zhao, K.; Le Guillou, S.; Diez, J.; Vicente, F.; Fernández-Sanjurjo, M.; Iglesias-Gutiérrez, E.; Guan, L.; Royo, L.J. Differences in the microRNAs Levels of Raw Milk from Dairy Cattle Raised under Extensive or Intensive Production Systems. Vet. Sci. 2022, 9, 661. [Google Scholar] [CrossRef] [PubMed]
- Abou El Qassim, L.; Vicente, F.; de la Torre, S.; Jiménez-Calderón, J.D.; Baizán, S.; Soldado, A.; Martínez-Fernández, A.; Royo, L.J. ¿Es posible certificar la leche producida en ecológico utilizando microARN? 2018. XIX Reunión Nacional de Mejora Genética Animal. Available online: https://www.acteon.webs.upv.es/CONGRESOS/Z-XIX_Reunion_MG_LEON_2018/053_AbouelQassim.pdf (accessed on 30 June 2024).
- Abou El Qassim, L.; Le Guillou, S.; Royo, L.J. Variation of miRNA Content in Cow Raw Milk Depending on the Dairy Production System. Int. J. Mol. Sci. 2022, 23, 11681. [Google Scholar] [CrossRef]
- De La Torre-Santos, S. Identificación de Biomarcadores Específicos Para Autentificar El Origen y El Sistema de Alimentación Del Vacuno Lechero. Ph.D. Thesis, Universidad de Zaragoza, Zaragoza, Spain, 2021. [Google Scholar]
- Várallyay, É.; Burgyán, J.; Havelda, Z. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat. Protoc. 2008, 3, 190–196. [Google Scholar] [CrossRef]
- Mocellin, S.; Rossi, C.R.; Pilati, P.; Nitti, D.; Marincola, F.M. Quantitative real-time PCR: A powerful ally in cancer research. Trends Mol. Med. 2003, 9, 189–195. [Google Scholar] [CrossRef]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef]
- Mattie, M.D.; Benz, C.C.; Bowers, J.; Sensinger, K.; Wong, L.; Scott, G.K.; Fedele, V.; Ginzinger, D.; Getts, R.; Haqq, C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol. Cancer 2006, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.T.; Wilfred, B.R. In situ hybridization is a necessary experimental complement to microRNA (miRNA) expression profiling in the human brain. Neurosci. Lett. 2009, 466, 69–72. [Google Scholar] [CrossRef]
- De Planell-Saguer, M.; Rodicio, M.C. Analytical aspects of microRNA in diagnostics: A review. Anal. Chim. Acta 2011, 699, 134–152. [Google Scholar] [CrossRef]
- Hunt, E.A.; Goulding, A.M.; Deo, S.K. Direct detection and quantification of microRNAs. Anal. Biochem. 2009, 387, 1–12. [Google Scholar] [CrossRef]
- Pall, G.S.; Codony-Servat, C.; Byrne, J.; Ritchie, L.; Hamilton, A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007, 35, e60. [Google Scholar] [CrossRef] [PubMed]
- Espy, M.J.; Uhl, J.R.; Sloan, L.M.; Buckwalter, S.P.; Jones, M.F.; Vetter, E.A.; Yao, J.D.C.; Wengenack, N.L.; Rosenblatt, J.E.; Cockerill, F.R., III; et al. Real-time PCR in clinical microbiology: Applications for routine laboratory testing. Clin. Microbiol. Rev. 2006, 19, 165–256. [Google Scholar] [CrossRef]
- Wegman, D.W.; Krylov, S.N. Direct miRNA-hybridization assays and their potential in diagnostics. Trends Anal. Chem. 2013, 44, 121–130. [Google Scholar] [CrossRef]
- Russell, C.; Welch, K.; Jarvius, J.; Cai, Y.; Brucas, R.; Nikolajeff, F.; Svedlindh, P.; Nilsson, M. Gold nanowire based electrical DNA detection using rolling circle amplification. ACS Nano 2014, 8, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Saha, K.; Agasti, S.S.; Kim, C.; Li, X.; Rotello, V.M. Gold Nanoparticles in Chemical and Biological Sensing. Chem. Rev. 2012, 112, 2739–2779. [Google Scholar] [CrossRef]
- Woong Lee, J.; Choi, S.R.; Hyuk Heo, J. Simultaneous Stabilization and Functionalization of Gold Nanoparticles via Biomolecule Conjugation: Progress and Perspectives. ACS Appl. Mater. Interfaces 2021, 13, 42311–42328. [Google Scholar] [CrossRef]
- González-Morales, S.; López-Portugués, C.; Fernández-Sanjurjo, M.; Iglesias-Gutiérrez, E.; Montes-Bayón, M.; Corte-Rodríguez, M. Amplification-Free Strategy for miRNA Quantification in Human Serum Using Single Particle ICP–MS and Gold Nanoparticles as Labels. Anal. Chem. 2024, 96, 12414. [Google Scholar] [CrossRef] [PubMed]
- Lance Kelly, K.; Coronado, E.; Lin Zhao, L.; Schatz, G.C. The Optical Properties of Metal Nanoparticles: The Influence of Size, Shape, and Dielectric Environment. J. Phys. Chem. B 2003, 107, 668–677. [Google Scholar] [CrossRef]
- Li, R.D.; Yin, B.C.; Ye, B.C. Ultrasensitive, colorimetric detection of microRNAs based on isothermal exponential amplification reaction-assisted gold nanoparticle amplification. Biosens. Bioelectron. 2016, 86, 1011–1016. [Google Scholar] [CrossRef]
- Shi, H.Y.; Yang, L.; Zhou, X.Y.; Bai, J.; Gao, J.; Jia, H.X.; Li, Q.G. A gold nanoparticle-based colorimetric strategy coupled to duplex-specific nuclease signal amplification for the determination of microRNA. Microchim. Acta 2017, 184, 525–531. [Google Scholar] [CrossRef]
- Ren, M.; Wang, S.; Cai, C.; Chen, C.; Chen, X. A simple and sensitive resonance light scattering method based on aggregation of gold nanoparticles for selective detection of microRNA-21. RSC Adv. 2016, 6, 83078–83083. [Google Scholar] [CrossRef]
- Elghanian, R.; Storhoff, J.J.; Mucic, R.C.; Letsinger, R.L.; Mirkin, C.A. Selective colorimetric detection of polynucleotides based on the distancedependent optical properties of gold nanoparticles. Science 1997, 277, 1078–1081. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Visedo, A.; Gallego, B.; Royo, L.J.; Soldado, A.; Valledor, M.; Ferrero, F.J.; Campo, J.C.; Costa-Fernández, J.M.; Fernández-Argüelles, M.T. Visual detection of microRNA146a by using RNA-functionalized gold nanoparticles. Microchim. Acta 2020, 187, 192. [Google Scholar] [CrossRef]
- Sánchez-Visedo, A.; Gallego-Martínez, B.; Royo, L.J.; Soldado, A.; Valledor, M.; Campo, J.C.; Ferrero, F.J.; Costa-Fernández, J.M.; Fernández-Argüelles, M.T. MNAzymes and gold nanoparticles as isothermal signal amplification strategy for visual detection of miRNA. Microchim. Acta 2023, 190, 292. [Google Scholar] [CrossRef]
- Jin, R.; Wu, G.; Li, Z.; Mirkin, C.A.; Schatz, G.C. What Controls the Melting Properties of DNA-Linked Gold Nanoparticle Assemblies? have been used as probes in a variety of DNA detection methods. J. Am. Chem. Soc. 2003, 125, 1643–1654. [Google Scholar] [CrossRef]
- Shin, J.; Zhang, X.; Liu, J. DNA-Functionalized Gold Nanoparticles in Macromolecularly Crowded Polymer Solutions. J. Phys. Chem. B 2012, 116, 13396–13402. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Kudlay, A.; Schatz, G.C. Molecular Dynamics Studies of Ion Distributions for DNA Duplexes and DNA Clusters: Salt Effects and Connection to DNA Melting. J. Phys. Chem. B 2006, 110, 2918–2926. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.F.; Record, M.T. Polyelectrolyte Theories and their Applications to DNA. Annu. Rev. Phys. Chem. 1982, 33, 191–222. [Google Scholar] [CrossRef]
- Cai, J.; Ding, L.; Gong, P.; Huang, J. A colorimetric detection of microRNA-148a in gastric cancer by gold nanoparticle-RNA conjugates. Nanotechnology 2020, 31, 095501. [Google Scholar] [CrossRef]
- Persano, S.; Guevara, M.L.; Wolfram, J.; Blanco, E.; Shen, H.; Ferrari, M.; Pompa, P.P. Label-free isothermal amplification assay for specific and highly sensitive colorimetric miRNA detection. ACS Omega 2016, 1, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Zhen, G.X. Sensitive colorimetric detection of microrna based on target catalyzed double-arm hairpin DNA assembling. Anal. Sci. 2016, 32, 751–756. [Google Scholar] [CrossRef] [PubMed]


| Conformation | Oligo Name | Sequence (5′→3′) |
|---|---|---|
| - | bta-miR-103 | AGC AGC AUU GUA CAG GGG UAU GA |
| H-H | Agg/A m103 | Thiol-C6-AAA AAA AAA AAA CAA TGC TGC T |
| Agg/B m103 | TCA TAG CCC TGT AAA AAA AAA-C3-Thiol | |
| T-T | Agg/C m103 | Thiol-C6-AAA AAA AAA ATC ATA GCC CTG |
| Agg/D m103 | TAC AAT GCT GCT AAA AAA AAA AA-C3-Thiol | |
| - | bta-miR-155 | UUA AUG CUA AUC GUG AUA GGG GU |
| H-H | Agg/A m155 | Thiol-C6-AAA AAA AAA AGA TTA GCA TTA A |
| Agg/B m155 | ACC CCT ATC ACA AAA AAA AAA-C3-Thiol | |
| T-T | Agg/C m155 | Thiol-C6-AAA AAA AAA ACC CCT ATC ACG |
| Agg/D m155 | ATT AGC ATT AAA AAA AAA AAA-C3-Thiol | |
| - | bta-miR-215 | AUG ACC UAU GAA UUG ACA GAC A |
| H-H | Agg/A m215 | Thiol-C6-AAA AAA AAA ATC ATA GGT CAT |
| Agg/B m215 | TGT CTG TCA ATA AAA AAA AAA-C3-Thiol | |
| T-T | Agg/C m215 | Thiol-C6-AAA AAA AAA ATG TCT GTC AAT |
| Agg/D m215 | TCA TAG GTC ATA AAA AAA AAA A-C3-Thiol |
| Oligo Name | (Probe:AuNPs) Ratio |
|---|---|
| Agg/A m103 | 250:1 |
| Agg/B m103 | 350:1 |
| Agg/C m103 | 250:1 |
| Agg/D m103 | 500:1 |
| Agg/A m155 | 300:1 |
| Agg/B m155 | 350:1 |
| Agg/C m155 | 350:1 |
| Agg/D m155 | 400:1 |
| Agg/A m215 | 400:1 |
| Agg/B m215 | 350:1 |
| Agg/C m215 | 400:1 |
| Agg/D m215 | 400:1 |
| miRNA | Sample 1 (Ct) | Sample 2 (Ct) | Sample 3 (Ct) | Sample 4 (Ct) | ||||
|---|---|---|---|---|---|---|---|---|
| Original | Spiked | Original | Spiked | Original | Spiked | Original | Spiked | |
| 103 | 33.7 | 22 | 28.7 | 27.5 | 24.5 | 16 | 27.3 | 27.5 |
| 155 | 29.1 | 22 | 20.1 | 21.2 | 27.3 | 28.5 | 25.7 | 18.5 |
| 215 | 32.7 | 15.8 | 28.5 | 13.8 | 27.6 | 27.9 | 28.6 | 27.6 |
| 30a-5p | 30.9 | 27.4 | 23.7 | 22.9 | 22.7 | 25.5 | 23.5 | 22.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopez-Benitez, K.; Alcazar-Gonzalez, P.; el qassim, L.A.; Fernandez-Argüelles, M.T.; Vicente, F.; Royo, L.J.; Menendez-Miranda, M. Development of a Gold Nanoparticle-Based Sensor for Authentication of Organic Milk Based on Differential Levels of miRNA. Nanomaterials 2024, 14, 1364. https://doi.org/10.3390/nano14161364
Lopez-Benitez K, Alcazar-Gonzalez P, el qassim LA, Fernandez-Argüelles MT, Vicente F, Royo LJ, Menendez-Miranda M. Development of a Gold Nanoparticle-Based Sensor for Authentication of Organic Milk Based on Differential Levels of miRNA. Nanomaterials. 2024; 14(16):1364. https://doi.org/10.3390/nano14161364
Chicago/Turabian StyleLopez-Benitez, Karelmar, Patricia Alcazar-Gonzalez, Loubna Abou el qassim, Mª Teresa Fernandez-Argüelles, Fernando Vicente, Luis J. Royo, and Mario Menendez-Miranda. 2024. "Development of a Gold Nanoparticle-Based Sensor for Authentication of Organic Milk Based on Differential Levels of miRNA" Nanomaterials 14, no. 16: 1364. https://doi.org/10.3390/nano14161364
APA StyleLopez-Benitez, K., Alcazar-Gonzalez, P., el qassim, L. A., Fernandez-Argüelles, M. T., Vicente, F., Royo, L. J., & Menendez-Miranda, M. (2024). Development of a Gold Nanoparticle-Based Sensor for Authentication of Organic Milk Based on Differential Levels of miRNA. Nanomaterials, 14(16), 1364. https://doi.org/10.3390/nano14161364







