Engineered Nanoparticle-Protein Interactions Influence Protein Structural Integrity and Biological Significance
Abstract
:1. Introduction
| Nanoparticle Synthesis Approach | Techniques | Description | Reference |
|---|---|---|---|
| Top-down approach | Synthesis is initiated by systematic leaching of bulk counterpart leading to the generation of nano-scaled particles. | ||
| Lithography | Categorized as masked and maskless lithography. In masked lithography transfer of nano-patterns over a large surface area is done using a specific mask e.g., photolithography and soft lithography. In maskless lithography, arbitrary nanopattern is written without using any mask e.g., electron beam lithography and focussed ion beam lithography. | [5] | |
| Mechanical milling | Formation of nano-scaled material by elastic, plastic and shear deformation followed by fracture, amorphization and chemical reactions. It helps in nanocomposite production. | [6] | |
| Electrospinning | Used for nanofibres production from various materials, typically polymers. | [7] | |
| Sputtering | Nanomaterials are produced by the bombardment of high-energy particles such as gas or plasma on the solid surface. Used for the production of thin films of nanomaterials. | [8] | |
| Arc discharge method | Used for the production of carbon-based materials such as fullerenes, carbon nanotubes etc. | [9] | |
| Laser ablation | Nanomaterials synthesized using a powerful beam of laser that hits the target. | [10] | |
| Bottom-up approach | Synthesis is by coalescence or assembling of atoms and molecules to produce various nanoparticles. | ||
| Sol-gel method | Metal oxide nanoparticles are synthesized by the transformation of liquid precursor to a sol followed by its conversion into a gel. | [11] | |
| Chemical vapour deposition | Nanomaterials are synthesized by thin film formation over the surface of the substrate due to the chemical reaction of vapor-phase precursors. | [12] | |
| Hydrothermal and solvothermal methods | Nanomaterials are synthesized in either aqueous medium (hydrothermal method) or non-aqueous medium (solvothermal method) by heterogeneous reaction under high pressure and temperature near the critical point in an enclosed vessel. | [13] | |
| Template methods | These are used to synthesize nanoporous materials either by using a soft template such as block polymers and surfactants or by using a hard template such as carbon nanotubes, carbon black, wood shells, silica and colloidal crystals. | [14,15] | |
| Reverse micelle methods | Nanoparticles are synthesized by the formation of reverse micelle which is created in the case of water-in-oil emulsion where hydrophilic heads point towards the core. This core act as a nanoreactor for nanoparticle synthesis. | [16] |
| ENPs Types | Size (nm) | Characteristics | Applications | References |
|---|---|---|---|---|
|
Organic ENPs | ||||
| Dendrimers | <10 nm | Radially symmetric molecules with highly branched structures made of one or more cores. These are homogeneous and monodispersed. | Controlled and targeted bioactive delivery to macrophages, liver targeting, transdermal drug delivery, gene delivery | [21,22,23] |
| Liposome | 50–100 nm | Vesicles of phospholipid with superior entrapment ability. These are biocompatible and versatile. | Passive and active gene delivery, can be used for peptides, proteins, and cell interactions studies, anti-cancer therapy | [24] |
|
Polymeric ENPs | 10–1000 nm | Biodegradable and biocompatible. | Controlled and sustained drug delivery carriers, protein carriers, intra-arterial localization of therapeutic agents | [25] |
| Micelles | 10–100 nm | Formed of amphiphilic molecules like polymers and lipids. | Targeted delivery of siRNA and anticancer drug, diagnosis | [26,27] |
|
Inorganic ENPs | ||||
| Metallic | <100 nm | Metal colloids with a high surface-to-volume ratio. These are stable and have better mechanical strength, optical and magnetic properties. | Delivery of genes and drugs, ultrasensitive diagnostic assays, radiotherapy, and thermal ablation | [28,29,30,31] |
| Metal oxide | <100 nm | Oxides of metals with antioxidant activities, chemical stability, catalytic and optical properties, and biocompatibility. | Medical implants, drug delivery, biological antioxidant, bioimaging, biosensors | [32,33,34] |
| Ceramic | <50 nm | Non-metallic solids of non-metallic and metallic compounds with the property of heat resistance. | Bone repair, drug delivery vehicles, photocatalysis, imaging, photodegradation of dyes | [35] |
|
Nanocrystal Quantum dots | 2–9.5 nm | Semiconductive material consists of a semiconductor core, a shell, and a cap. These have high photostability, broad UV excitation, narrow emission, bright fluorescence, and resistance to photobleaching. | Long-term multi-colour imaging of hepatocytes, DNA hybridization, immunoassays, receptor-mediated endocytosis, disease marker labeling | [36,37,38] |
| Fullerenes | 1–2 nm | High strength, electrical conductivity, electron affinity, and versatile structure. | Gene and drug delivery, antiviral activity | [39,40] |
|
Carbon nanotubes | 0.5–3 nm in diameter, 20–1000 nm in length | These are single or multi-walled nanotubes with unique strength and electrical properties. Found in crystalline form. | Can penetrate inside cell and nucleus, gene and peptide carrier, used in imaging, drug delivery, tissue engineering | [41] |
|
Hybrid ENPs | ||||
| Hydrogels | 0.1 to 100 μm | These are also known as polymeric nanogels and macromolecules micelles. These are polymeric networks having a three-dimensional structure with high water or biological fluid absorbing capacity owing to the hydrophilic groups present in the polymer chains. | These are used in the delivery of drugs of small molecular weight, peptides, proteins, nucleic acids, oligosaccharides and vaccines. | [42,43] |
2. ENPs Interaction with Proteins
2.1. Protein Corona
2.2. Role of ENPs in Protein Folding Pathway
2.2.1. Role of ENPs in Protein Unfolding
2.2.2. Role of ENPs in Protein Folding
2.2.3. Role of ENPs in Protein Aggregation
2.2.4. Role of ENPs in Protein Fibrillation
3. Factors Responsible for ENP-Protein Interactions
3.1. Size and Radius of Curvature
3.2. Surface Charge
3.3. Shape
3.4. Affinity and Exposure Time
3.5. Thermodynamic Parameters
3.6. Biofluid
4. Chaperoning Functions of ENPs
4.1. ENPs Have Been Categorized as Chaperone-Mimicking ENPs and Chaperone-Aiding ENPs Based on Their Structure and Functions
4.1.1. Chaperone-Mimicking ENPs
4.1.2. Chaperone-Aiding ENPs
4.2. ENPs with Chaperone-like Activity
4.2.1. ENPs Assisted Refolding of Proteins
4.2.2. ENPs Assisted Modulation of Protein Misfolding
4.2.3. ENPs Assisted Clearance of Amyloid Proteins
4.2.4. ENPs Assisted Fibril Degradation
5. Therapeutics Based on ENPs-Protein Interactions
5.1. ENPs Controlled Protein-Ligand Binding Efficiency
5.2. Anti-Amyloidogenic Activity of ENPs
5.3. Nanozymes
5.4. ENPs for Drug Delivery
| ENPs | Protein/Model Used | Disease | Outcome | Reference |
|---|---|---|---|---|
| Zinc oxide nanoparticles (ZnO NPs), short ZnO nanorods (s-ZnO NRs), and long ZnO nanorods (l-ZnO NRs) | Human and zebrafish larvae neuroblastoma cells SH-SY5Y | Parkinson’s disease (PD) | PD like symptoms developed | [120] |
| CuO nanoparticles (CuONP), Fe2O3 nanoparticles (Fe2O3NP), ZnONP | Rat cell lines (PC12) and human SH-SY5Y and H4 cells | Alzheimer’s disease (AD) | Concentration-based neurotoxicity of CuONP but not Fe2O3NP and ZnONP. CuONP as an environmental risk factor for AD | [121] |
| Poly(trehalose) nanoparticles | HD150Q cells, HD transgenic mice [B6CBA-Tg (HDexon1) 62Gpb/3Jstrain] | Huntington’s disease (HD) | Inhibition of amyloid aggregation and prevention of polyglutamine aggregation. | [122] |
| Carbon nanoparticles (graphene and carbon nanotubes) | Mouse prion protein (moPrP117−231) | Prions disease | Carbon nanoparticles inhibited Prion fibrillation in In vitro studies SWCNT and graphene reduced interaction of peptide and caused the formation of β-structure | [123] |
| Nanoliposomes (NL) | Purified AL light chain proteins, ex-vivo human arteriole model, Human aortic artery endothelial cells (HAEC) | Light chain amyloidosis (AL) | Increased folded protein amount, reduced cell internalization. | [80] |
| Dendrimer–tesaglitazar | BV2 murine microglial cell line | AD and PD | Microglial phynotype shift, increased β-amyloid phagocytosis | [114] |
| Graphene QDs | 10 DIV mouse cortical neurons, C57BL/6 mice | PD | Inhibition of α-syn fibril formation, trigger fibril disaggregation, protects against dopaminergic neuron loss and Lewy body pathology | [124] |
| N-methyl D-aspartic acid functionalized gold nanoparticle (GNP-NMDA) | Low molecular weight (LMW) amyloid oligomers | AD | Inhibition of LMW tetramer of amyloid oligomer towards nontoxic aggregation path | [125] |
| Protein capped Fe3O4 (PC-Fe3O4) and PC-CdS | Tau protein | AD | Inhibition of Tau aggregation | [126] |
6. Techniques to Study ENP-Protein Interactions
| Techniques Applied | ENPs | Proteins | Parameters Analyzed | References |
|---|---|---|---|---|
| FTIR spectroscopy, UV–vis spectrophotometry, TEM, fluorescence and, CD spectroscopy | AuNPs | Bovine serum albumin (BSA) | Amount of α-helical structure Conformational change in proteins, secondary and tertiary structural alterations in proteins | [130,131] |
| DLS, TEM, Far-UV spectra, CD spectroscopy | Unmodified TiO2 | α-chymotrypsin, RNase A, and papain | Protein refolding | [132] |
| UV/vis spectrophotometry, Raman spectroscopy, electronic paramagnetic resonance (EPR) spectroscopy | Two amorphous pyrogenic silica ENPs | Bovine serum albumin (BSA), hen egg lysozyme (HEL), bovine pancreatic ribonuclease A, RNase and bovine lactoperoxidase (LPO) | Quantify adsorbed protein, surface-driven structural modification, protein orientation on the nanoparticle surface | [129] |
| SDS-PAGE, densitometry, AFM, analytical ultracentrifugation (AUC) | SiO2 and CeO2 nanoparticles | Serum proteins and BSA | Adsorption behaviour of proteins | [133] |
| Affinity chromatography, UV-visible spectroscopy, TEM, SANS, CD | Silica nanoparticles | Green fluorescent protein (GFP) | Relationship between unfolded proteins, silica nanoparticles and chaperonin | [134] |
| TEM, CD | Ultra-small AuNPs | Amyloid β | Inhibition of fibrillation process, disruption of peptide folding process | [135] |
| MD simulation | Monolayer-capped AuNPs | Amyloid β fibrils | Location and binding affinity of nanoparticles with proteins | [118] |
| ThT, Congo red assay, FTIR, CD, AFM | Silica nanoparticles (SiNPs) | Hen egg-white lysozyme (HEWL) | Aggregation behavior | [136] |
6.1. Spectroscopic Techniques
6.2. Microscopic Techniques
6.3. Thermodynamic Techniques
6.4. Separation Techniques
6.5. In Silico Techniques
7. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jeyaraj, M.; Gurunathan, S.; Qasim, M.; Kang, M.-H.; Kim, J.-H. A Comprehensive Review on the Synthesis, Characterization, and Biomedical Application of Platinum Nanoparticles. Nanomaterials 2019, 9, 1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auffan, M.; Rose, J.; Bottero, J.-Y.; Lowry, G.V.; Jolivet, J.-P.; Wiesner, M.R. Towards a definition of inorganic nanoparticles from an environmental, health and safety perspective. Nat. Nanotechnol. 2009, 4, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Keat, C.L.; Aziz, A.; Eid, A.M.; Elmarzugi, N.A. Biosynthesis of nanoparticles and silver nanoparticles. Bioresour. Bioprocess. 2015, 2, 47. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Xia, Y. Bottom-Up and Top-Down Approaches to the Synthesis of Monodispersed Spherical Colloids of Low Melting-Point Metals. Nano Lett. 2004, 4, 2047–2050. [Google Scholar] [CrossRef]
- Pimpin, A.; Srituravanich, W. Review on Micro- and Nanolithography Techniques and their Applications. Eng. J. 2012, 16, 37–56. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, J.; Bhattacharyya, S. Application of graphene and graphene-based materials in clean energy-related devices Minghui. Arch. Thermodyn. 2012, 33, 23–40. [Google Scholar] [CrossRef]
- Ostermann, R.; Cravillon, J.; Weidmann, C.; Wiebcke, M.; Smarsly, B.M. Metal–organic framework nanofibers viaelectrospinning. Chem. Commun. 2010, 47, 442–444. [Google Scholar] [CrossRef] [Green Version]
- Ayyub, P.; Chandra, R.; Taneja, P.; Sharma, A.; Pinto, R. Synthesis of nanocrystalline material by sputtering and laser ablation at low temperatures. Appl. Phys. A 2001, 73, 67–73. [Google Scholar] [CrossRef]
- Zhang, D.; Ye, K.; Yao, Y.; Liang, F.; Qu, T.; Ma, W.; Yang, B.; Dai, Y.; Watanabe, T. Controllable synthesis of carbon nanomaterials by direct current arc discharge from the inner wall of the chamber. Carbon 2018, 142, 278–284. [Google Scholar] [CrossRef]
- Amendola, V.; Meneghetti, M. Laser ablation synthesis in solution and size manipulation of noble metal nanoparticles. Phys. Chem. Chem. Phys. 2009, 11, 3805–3821. [Google Scholar] [CrossRef]
- Danks, A.E.; Hall, S.R.; Schnepp, Z. The evolution of ‘sol–gel’ chemistry as a technique for materials synthesis. Mater. Horiz. 2015, 3, 91–112. [Google Scholar] [CrossRef] [Green Version]
- Jones, A.C.; Aspinall, H.C.; Chalker, P.R. Chapter 8. Chemical Vapour Deposition of Metal Oxides for Microelectronics Applications; RSC Publishing: London, UK, 2008; ISBN 9781847558794. [Google Scholar] [CrossRef]
- Chen, A.; Holt-hindle, P. Platinum-Based Nanostructured Materials: Synthesis, Properties, and Applications. Chem. Rev. 2010, 110, 3767–3804. [Google Scholar] [CrossRef]
- Li, W.; Zhao, D. An overview of the synthesis of ordered mesoporous materials. Chem. Commun. 2012, 49, 943–946. [Google Scholar] [CrossRef]
- Szczęśniak, B.; Choma, J.; Jaroniec, M. Major advances in the development of ordered mesoporous materials. Chem. Commun. 2020, 56, 7836–7848. [Google Scholar] [CrossRef]
- Malik, M.A.; Wani, M.Y.; Hashim, M.A. Microemulsion method: A novel route to synthesize organic and inorganic nanomaterials. Arab. J. Chem. 2010, 5, 397–417. [Google Scholar] [CrossRef] [Green Version]
- Inshakova, E.; Inshakov, O. World market for nanomaterials: Structure and trends. MATEC Web Conf. 2017, 129, 02013. [Google Scholar] [CrossRef]
- Hawthorne, J.; Roche, R.D.L.T.; Xing, B.; Newman, L.A.; Ma, X.; Majumdar, S.; Gardea-Torresdey, J.; White, J.C. Particle-Size Dependent Accumulation and Trophic Transfer of Cerium Oxide through a Terrestrial Food Chain. Environ. Sci. Technol. 2014, 48, 13102–13109. [Google Scholar] [CrossRef] [PubMed]
- De la Torre Roche, R.; Servin, A.; Hawthorne, J.; Xing, B.; Newman, L.A.; Ma, X.; Chen, G.; White, J.C. Terrestrial Trophic Transfer of Bulk and Nanoparticle La2O3 Does Not Depend on Particle Size. Environ. Sci. Technol. 2015, 49, 11866–11874. [Google Scholar] [CrossRef]
- Tangaa, S.R.; Selck, H.; Winther-Nielsen, M.; Khan, F.R. Trophic transfer of metal-based nanoparticles in aquatic environments: A review and recommendations for future research focus. Environ. Sci. Nano 2016, 3, 966–981. [Google Scholar] [CrossRef] [Green Version]
- Tomalia, D.A.; Frechet, J.M.J. Discovery of dendrimers and dendritic polymers: A brief historical perspective. J. Polym. Sci. Part A Polym. Chem. 2002, 40, 2719–2728. [Google Scholar] [CrossRef]
- Master, A.M.; Rodriguez, M.E.; Kenney, M.E.; Oleinick, N.L.; Sen Gupta, A. Delivery of the photosensitizer Pc 4 in PEG–PCL micelles for in vitro PDT studies. J. Pharm. Sci. 2010, 99, 2386–2398. [Google Scholar] [CrossRef]
- Fu, H.-L.; Cheng, S.-X.; Zhang, X.-Z.; Zhuo, R.-X. Dendrimer/DNA complexes encapsulated functional biodegradable polymer for substrate-mediated gene delivery. J. Gene Med. 2008, 10, 1334–1342. [Google Scholar] [CrossRef]
- Banerjee, R.; Tyagi, P.; Li, S.; Huang, L. Anisamide-targeted stealth liposomes: A potent carrier for targeting doxorubicin to human prostate cancer cells. Int. J. Cancer 2004, 112, 693–700. [Google Scholar] [CrossRef]
- Des Rieux, A.; Fievez, V.; Garinot, M.; Schneider, Y.-J.; Préat, V. Nanoparticles as potential oral delivery systems of proteins and vaccines: A mechanistic approach. J. Control. Release 2006, 116, 1–27. [Google Scholar] [CrossRef]
- Duong, H.H.P.; Yung, L.-Y.L. Synergistic co-delivery of doxorubicin and paclitaxel using multi-functional micelles for cancer treatment. Int. J. Pharm. 2013, 454, 486–495. [Google Scholar] [CrossRef]
- Xiong, X.-B.; Lavasanifar, A. Traceable Multifunctional Micellar Nanocarriers for Cancer-Targeted Co-delivery of MDR-1 siRNA and Doxorubicin. ACS Nano 2011, 5, 5202–5213. [Google Scholar] [CrossRef] [PubMed]
- Dobson, J.M. Gene therapy progress and prospects: Magnetic nanoparticle-based gene delivery. Gene Ther. 2006, 13, 283–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudge, S.; Peterson, C.; Vessely, C.; Koda, J.; Stevens, S.; Catterall, L. Adsorption and desorption of chemotherapeutic drugs from a magnetically targeted carrier (MTC). J. Control. Release 2001, 74, 335–340. [Google Scholar] [CrossRef]
- Chen, H.; Shao, L.; Ming, T.; Sun, Z.; Zhao, C.; Yang, B.; Wang, J. Understanding the Photothermal Conversion Efficiency of Gold Nanocrystals. Small 2010, 6, 2272–2280. [Google Scholar] [CrossRef]
- Day, E.S.; Morton, J.G.; West, J.L. Nanoparticles for Thermal Cancer Therapy. J. Biomech. Eng. 2009, 131, 074001. [Google Scholar] [CrossRef]
- Celardo, I.; Pedersen, J.Z.; Traversa, E.; Ghibelli, L. Pharmacological potential of cerium oxide nanoparticles. Nanoscale 2011, 3, 1411–1420. [Google Scholar] [CrossRef] [PubMed]
- Gold, K.; Slay, B.; Knackstedt, M.; Gaharwar, A.K. Antimicrobial Activity of Metal and Metal-Oxide Based Nanoparticles. Adv. Ther. 2018, 1, 1700033. [Google Scholar] [CrossRef]
- Jeong, H.; Yoo, J.; Park, S.; Lu, J.; Park, S.; Lee, J. Non-Enzymatic Glucose Biosensor Based on Highly Pure TiO2 Nanoparticles. Biosensors 2021, 11, 149. [Google Scholar] [CrossRef] [PubMed]
- Mauricio, M.D.; Guerra-Ojeda, S.; Marchio, P.; Valles, S.L.; Aldasoro, M.; Escribano-Lopez, I.; Herance, J.R.; Rocha, M.; Vila, J.M.; Victor, V.M. Nanoparticles in Medicine: A Focus on Vascular Oxidative Stress. Oxidative Med. Cell. Longev. 2018, 2018, 6231482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voura, E.B.; Jaiswal, J.K.; Mattoussi, H.; Simon, S.M. Tracking metastatic tumor cell extravasation with quantum dot nanocrystals and fluorescence emission-scanning microscopy. Nat. Med. 2004, 10, 993–998. [Google Scholar] [CrossRef]
- Yu, W.W.; Qu, L.; Guo, W.; Peng, X. Experimental Determination of the Extinction Coefficient of CdTe, CdSe, and CdS Nanocrystals. Chem. Mater. 2003, 15, 2854–2860. [Google Scholar] [CrossRef]
- Derfus, A.M.; Chan, W.C.W.; Bhatia, S.N. Probing the Cytotoxicity of Semiconductor Quantum Dots. Nano Lett. 2003, 4, 11–18. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, A.P.; Rohiwal, S.S. Synthesis and Bioconjugation of Hybrid Nanostructures for Biomedical Applications; Elsevier: Amsterdam, The Netherlands, 2018; ISBN 9780128139073. [Google Scholar]
- Bakry, R.; Vallant, R.M.; Najam-Ul-Haq, M.; Rainer, M.; Szabo, Z.; Huck, C.W.; Bonn, G.K. Medicinal applications of fullerenes. Int. J. Nanomed. 2007, 2, 639–649. [Google Scholar]
- Edwards, S.L.; Church, J.S.; Werkmeister, J.A.; Ramshaw, J.A. Tubular micro-scale multiwalled carbon nanotube-based scaffolds for tissue engineering. Biomaterials 2009, 30, 1725–1731. [Google Scholar] [CrossRef]
- Gonçalves, C.; Pereira, P.; Gama, M. Self-Assembled Hydrogel Nanoparticles for Drug Delivery Applications. Materials 2010, 3, 1420–1460. [Google Scholar] [CrossRef] [Green Version]
- Hamidi, M.; Azadi, A.; Rafiei, P. Hydrogel nanoparticles in drug delivery. Adv. Drug Deliv. Rev. 2008, 60, 1638–1649. [Google Scholar] [CrossRef] [PubMed]
- Adamcik, J.; Mezzenga, R. Amyloid Polymorphism in the Protein Folding and Aggregation Energy Landscape. Angew. Chem. Int. Ed. 2018, 57, 8370–8382. [Google Scholar] [CrossRef] [PubMed]
- Javed, I.; Peng, G.; Xing, Y.; Yu, T.; Zhao, M.; Kakinen, A.; Faridi, A.; Parish, C.L.; Ding, F.; Davis, T.P.; et al. Inhibition of amyloid beta toxicity in zebrafish with a chaperone-gold nanoparticle dual strategy. Nat. Commun. 2019, 10, 3780. [Google Scholar] [CrossRef] [PubMed]
- Sylvester, A.; Sivaraman, B.; Deb, P.; Ramamurthi, A. Nanoparticles for localized delivery of hyaluronan oligomers towards regenerative repair of elastic matrix. Acta Biomater. 2013, 9, 9292–9302. [Google Scholar] [CrossRef] [Green Version]
- Fedeli, C.; Segat, D.; Tavano, R.; Bubacco, L.; De Franceschi, G.; de Laureto, P.P.; Lubian, E.; Selvestrel, F.; Mancin, F.; Papini, E. The functional dissection of the plasma corona of SiO2-NPs spots histidine rich glycoprotein as a major player able to hamper nanoparticle capture by macrophages. Nanoscale 2015, 7, 17710–17728. [Google Scholar] [CrossRef]
- Engin, A.B.; Nikitovic, D.; Neagu, M.; Henrich-Noack, P.; Docea, A.O.; Shtilman, M.I.; Golokhvast, K.; Tsatsakis, A.M. Mechanistic understanding of nanoparticles’ interactions with extracellular matrix: The cell and immune system. Part. Fibre Toxicol. 2017, 14, 22. [Google Scholar] [CrossRef]
- Brown, D.M.; Hutchison, L.; Donaldson, K.; Stone, V. The effects of PM10 particles and oxidative stress on macrophages and lung epithelial cells: Modulating effects of calcium-signaling antagonists. Am. J. Physiol. Cell. Mol. Physiol. 2007, 292, L1444–L1451. [Google Scholar] [CrossRef] [Green Version]
- Park, S.J. Protein–Nanoparticle Interaction: Corona Formation and Conformational Changes in Proteins on Nanoparticles. Int. J. Nanomed. 2020, 15, 5783–5802. [Google Scholar] [CrossRef]
- Chen, M.; Von Mikecz, A. Formation of nucleoplasmic protein aggregates impairs nuclear function in response to SiO2 nanoparticles. Exp. Cell Res. 2005, 305, 51–62. [Google Scholar] [CrossRef]
- Cedervall, T.; Lynch, I.; Foy, M.; Berggård, T.; Donnelly, S.C.; Cagney, G.; Linse, S.; Dawson, K.A. Detailed Identification of Plasma Proteins Adsorbed on Copolymer Nanoparticles. Angew. Chem. 2007, 119, 5856–5858. [Google Scholar] [CrossRef]
- Landsiedel, R.; Ma-Hock, L.; Kroll, A.; Hahn, D.; Schnekenburger, J.; Wiench, K.; Wohlleben, W. Testing Metal-Oxide Nanomaterials for Human Safety. Adv. Mater. 2010, 22, 2601–2627. [Google Scholar] [CrossRef] [PubMed]
- Lundqvist, M.; Sethson, I.; Jonsson, B.-H. Protein Adsorption onto Silica Nanoparticles: Conformational Changes Depend on the Particles’ Curvature and the Protein Stability. Langmuir 2004, 20, 10639–10647. [Google Scholar] [CrossRef] [PubMed]
- Lundqvist, M.; Sethson, I.; Jonsson, B.-H. Transient Interaction with Nanoparticles “Freezes” a Protein in an Ensemble of Metastable Near-Native Conformations.Biochemistry (Mosc). In Computational Nanotoxicology: Challenges and Perspectives; Springer: Berlin/Heidelberg, Germany, 2005; pp. 10093–10099. [Google Scholar]
- Saptarshi, S.R.; Duschl, A.; Lopata, A.L. Interaction of nanoparticles with proteins: Relation to bio-reactivity of the nanoparticle. J. Nanobiotechnology 2013, 11, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doorley, G.W.; Payne, C.K. Cellular binding of nanoparticles in the presence of serum proteins. Chem. Commun. 2011, 47, 466–468. [Google Scholar] [CrossRef]
- Lesniak, A.; Campbell, A.; Monopoli, M.P.; Lynch, I.; Salvati, A.; Dawson, K.A. Serum heat inactivation affects protein corona composition and nanoparticle uptake. Biomaterials 2010, 31, 9511–9518. [Google Scholar] [CrossRef]
- Fleischer, C.C.; Kumar, U.; Payne, C.K. Cellular binding of anionic nanoparticles is inhibited by serum proteins independent of nanoparticle composition. Biomater. Sci. 2013, 1, 975–982. [Google Scholar] [CrossRef] [Green Version]
- Jedlovszky-Hajdú, A.; Bombelli, F.B.; Monopoli, M.P.; Tombácz, E.; Dawson, K.A. Surface Coatings Shape the Protein Corona of SPIONs with Relevance to Their Application In Vivo. Langmuir 2012, 28, 14983–14991. [Google Scholar] [CrossRef]
- Hirsch, V.; Kinnear, C.; Moniatte, M.; Rothen-Rutishauser, B.; Clift, M.J.D.; Fink, A. Surface charge of polymer coated SPIONs influences the serum protein adsorption, colloidal stability and subsequent cell interaction in vitro. Nanoscale 2012, 5, 3723–3732. [Google Scholar] [CrossRef] [Green Version]
- Podila, R.; Chen, R.; Ke, P.C.; Brown, J.M.; Rao, A.M. Effects of surface functional groups on the formation of nanoparticle-protein corona. Appl. Phys. Lett. 2012, 101, 263701. [Google Scholar] [CrossRef] [Green Version]
- Lacerda, S.H.D.P.; Park, J.J.; Meuse, C.; Pristinski, D.; Becker, M.L.; Karim, A.; Douglas, J.F. Interaction of Gold Nanoparticles with Common Human Blood Proteins. ACS Nano 2010, 4, 365–379. [Google Scholar] [CrossRef]
- Mohammad-Beigi, H.; Hayashi, Y.; Zeuthen, C.M.; Eskandari, H.; Scavenius, C.; Juul-Madsen, K.; Vorup-Jensen, T.; Enghild, J.J.; Sutherland, D.S. Mapping and identification of soft corona proteins at nanoparticles and their impact on cellular association. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Fleischer, C.C.; Payne, C.K. Nanoparticle–Cell Interactions: Molecular Structure of the Protein Corona and Cellular Outcomes. Accounts Chem. Res. 2014, 47, 2651–2659. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.R.; Andrieux, K.; Gil, S.; Taverna, M.; Chacun, H.; Desmaële, D.; Taran, F.; Georgin, D.; Couvreur, P. Translocation of Poly(ethylene glycol-co-hexadecyl)cyanoacrylate Nanoparticles into Rat Brain Endothelial Cells: Role of Apolipoproteins in Receptor-Mediated Endocytosis. Biomacromolecules 2007, 8, 793–799. [Google Scholar] [CrossRef] [PubMed]
- Michaelis, K.; Hoffmann, M.M.; Dreis, S.; Herbert, E.; Alyautdin, R.N.; Michaelis, M.; Kreuter, J.; Langer, K. Covalent linkage of apolipoprotein e to albumin nanoparticles strongly enhances drug transport into the brain. J. Pharmacol. Exp. Ther. 2006, 317, 1246–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, I.; Dawson, K.A. Protein-Nanoparticle Interactions; Springer: Berlin/Heidelberg, Germany, 2008; Volume 3. [Google Scholar] [CrossRef]
- Calzolai, L.; Franchini, F.; Gilliland, D.; Rossi, F. Protein-nanoparticle interaction: Identification of the ubiquitin-gold nanoparticle interaction site. Nano Lett. 2010, 10, 3101–3105. [Google Scholar] [CrossRef]
- Sukhanova, A.; Poly, S.; Bozrova, S.; Lambert, É.; Ewald, M.; Karaulov, A.; Molinari, M.; Nabiev, I. Nanoparticles With a Specific Size and Surface Charge Promote Disruption of the Secondary Structure and Amyloid-Like Fibrillation of Human Insulin Under Physiological Conditions. Front. Chem. 2019, 7, 480. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.; Ahmad, R.; Khare, S. Refolding of thermally denatured cholesterol oxidases by magnetic nanoparticles. Int. J. Biol. Macromol. 2019, 138, 958–965. [Google Scholar] [CrossRef]
- Onoda, A.; Kawasaki, T.; Tsukiyama, K.; Takeda, K.; Umezawa, M. Carbon nanoparticles induce endoplasmic reticulum stress around blood vessels with accumulation of misfolded proteins in the developing brain of offspring. Sci. Rep. 2020, 10, 10028. [Google Scholar] [CrossRef]
- Khan, R.; Ahmad, E.; Zaman, M.; Qadeer, A.; Rabbani, G. Nanoparticles in relation to peptide and protein aggregation. Int. J. Nanomed. 2014, 9, 899–912. [Google Scholar] [CrossRef] [Green Version]
- Yarjanli, Z.; Ghaedi, K.; Esmaeili, A.; Rahgozar, S.; Zarrabi, A. Iron oxide nanoparticles may damage to the neural tissue through iron accumulation, oxidative stress, and protein aggregation. BMC Neurosci. 2017, 18, 51. [Google Scholar] [CrossRef] [Green Version]
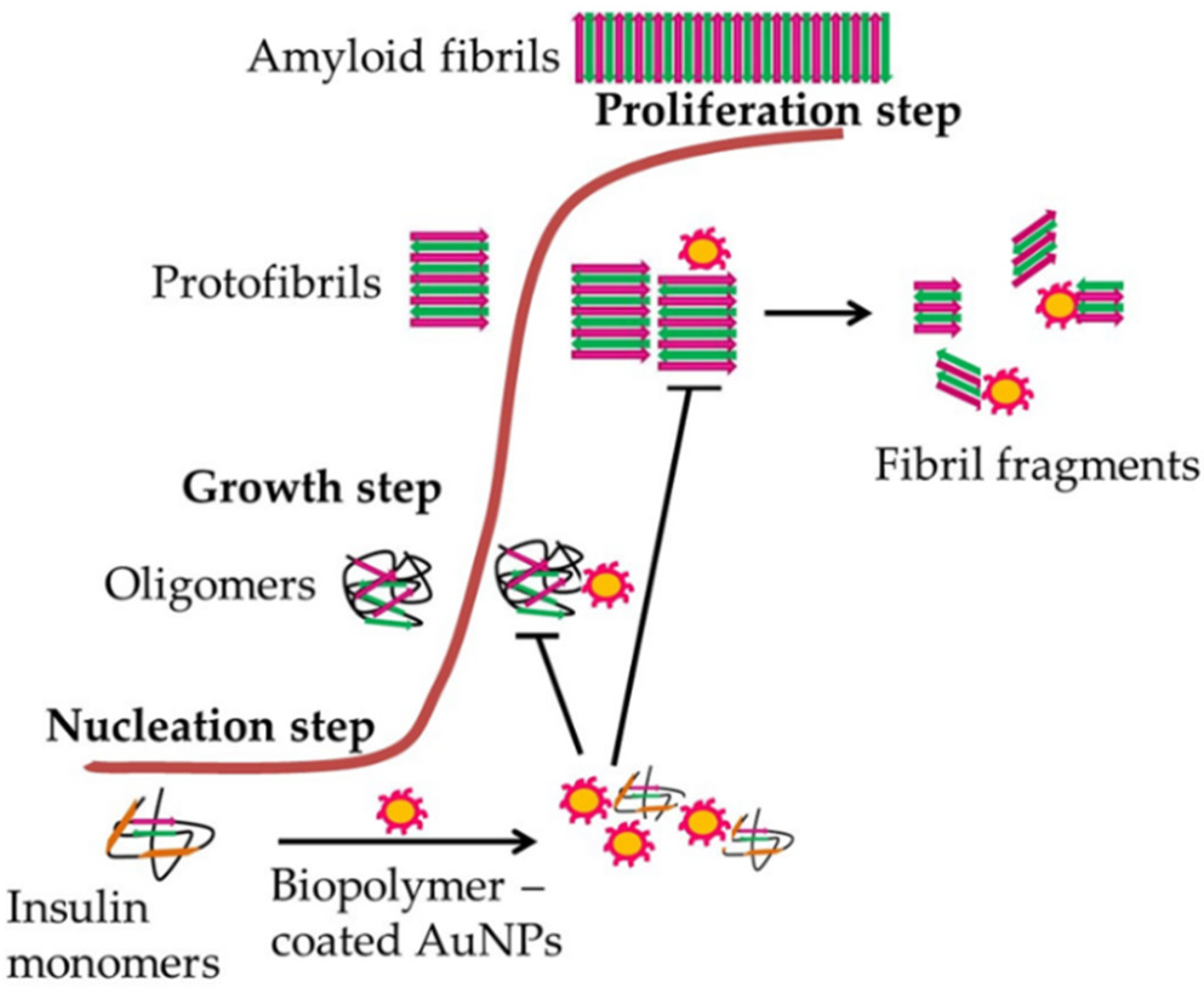
- Meesaragandla, B.; Karanth, S.; Janke, U.; Delcea, M. Biopolymer-coated gold nanoparticles inhibit human insulin amyloid fibrillation. Sci. Rep. 2020, 10, 7862. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, M.; Kalhor, H.R.; Laurent, S.; Lynch, I. Protein fibrillation and nanoparticle interactions: Opportunities and challenges. Nanoscale 2013, 5, 2570–2588. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Qin, M.; Meng, W.; Cao, Y.; Wang, W. How Do Proteins Unfold upon Adsorption on Nanoparticle Surfaces? Langmuir 2012, 28, 12779–12787. [Google Scholar] [CrossRef]
- Pandurangan, M.; Zamany, A.J.; Kim, D.H. ZnO nanoparticles assist the refolding of denatured green fluorescent protein. J. Mol. Recognit. 2015, 29, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Volodina, K.V.; Avnir, D.; Vinogradov, V.V. Alumina nanoparticle-assisted enzyme refolding: A versatile methodology for proteins renaturation. Sci. Rep. 2017, 7, 1458. [Google Scholar] [CrossRef] [Green Version]
- Truran, S.; Weissig, V.; Ramirez-Alvarado, M.; Franco, D.A.; Burciu, C.; Georges, J.; Murarka, S.; Okoth, W.A.; Schwab, S.; Hari, P.; et al. Nanoliposomes protect against AL amyloid light chain protein-induced endothelial injury. J. Liposome Res. 2013, 24, 69–73. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Han, Y.; Fan, Y.; Wang, Y. Effects of Gold Nanospheres and Nanocubes on Amyloid-β Peptide Fibrillation. Langmuir 2019, 35, 2334–2342. [Google Scholar] [CrossRef]
- Barkhade, T.; Mahapatra, S.K.; Banerjee, I. A Protein and Membrane Integrity Study of TiO2 Nanoparticles-Induced Mitochondrial Dysfunction and Prevention by Iron Incorporation. J. Membr. Biol. 2021, 254, 217–237. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.; Ghormade, V.; Kolge, H.; Paknikar, K.M. Dual effect of chitosan-based nanoparticles on the inhibition of β-amyloid peptide aggregation and disintegration of the preformed fibrils. J. Mater. Chem. B 2019, 7, 3362–3373. [Google Scholar] [CrossRef]
- Álvarez, Y.D.; Fauerbach, J.A.; Pellegrotti, J.V.; Jovin, T.M.; Jares-Erijman, E.A.; Stefani, F.D. Influence of Gold Nanoparticles on the Kinetics of α-Synuclein Aggregation. Nano Lett. 2013, 13, 6156–6163. [Google Scholar] [CrossRef] [Green Version]
- Kopp, M.; Kollenda, S.; Epple, M. Nanoparticle–Protein Interactions: Therapeutic Approaches and Supramolecular Chemistry. Accounts Chem. Res. 2017, 50, 1383–1390. [Google Scholar] [CrossRef]
- De, M.; You, C.-C.; Srivastava, S.; Rotello, V.M. Biomimetic Interactions of Proteins with Functionalized Nanoparticles: A Thermodynamic Study. J. Am. Chem. Soc. 2007, 129, 10747–10753. [Google Scholar] [CrossRef] [PubMed]
- Roach, P.; Farrar, D.; Perry, C.C. Interpretation of Protein Adsorption: Surface-Induced Conformational Changes. J. Am. Chem. Soc. 2005, 127, 8168–8173. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Laurent, S.; Tawil, N.; Yahia, L.H.; Mahmoudi, M. Nanoparticle and protein corona. In Protein-Nanoparticle Interactions; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Maiorano, G.; Sabella, S.; Sorce, B.; Brunetti, V.; Malvindi, M.A.; Cingolani, R.; Pompa, P.P. Effects of Cell Culture Media on the Dynamic Formation of Protein−Nanoparticle Complexes and Influence on the Cellular Response. ACS Nano 2010, 4, 7481–7491. [Google Scholar] [CrossRef] [PubMed]
- Sabuncu, A.C.; Grubbs, J.; Qian, S.; Abdel-Fattah, T.M.; Stacey, M.W.; Beskok, A. Probing nanoparticle interactions in cell culture media. Colloids Surf. B Biointerfaces 2012, 95, 96–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudhakar, S.; Kalipillai, P.; Santhosh, P.B.; Mani, E. Role of Surface Charge of Inhibitors on Amyloid Beta Fibrillation. J. Phys. Chem. C 2017, 121, 6339–6348. [Google Scholar] [CrossRef]
- Moustaoui, H.; Saber, J.; Djeddi, I.; Liu, Q.; Movia, D.; Prina-Mello, A.; Spadavecchia, J.; De La Chapelle, M.L.; Djaker, N. A protein corona study by scattering correlation spectroscopy: A comparative study between spherical and urchin-shaped gold nanoparticles. Nanoscale 2019, 11, 3665–3673. [Google Scholar] [CrossRef]
- Stradner, A.; Sedgwick, H.; Cardinaux, F.; Poon, W.C.K.; Egelhaaf, S.U.; Schurtenberger, P. Equilibrium cluster formation in concentrated protein solutions and colloids. Nature 2004, 432, 492–495. [Google Scholar] [CrossRef] [Green Version]
- Baimanov, D.; Cai, R.; Chen, C. Understanding the Chemical Nature of Nanoparticle–Protein Interactions. Bioconjug. Chem. 2019, 30, 1923–1937. [Google Scholar] [CrossRef]
- Precupas, A.; Gheorghe, D.; Botea-Petcu, A.; Leonties, A.R.; Sandu, R.; Popa, V.T.; Mariussen, E.; Naouale, E.Y.; Rundén-Pran, E.; Dumit, V.; et al. Thermodynamic Parameters at Bio–Nano Interface and Nanomaterial Toxicity: A Case Study on BSA Interaction with ZnO, SiO2, and TiO2. Chem. Res. Toxicol. 2020, 33, 2054–2071. [Google Scholar] [CrossRef]
- Monopoli, M.P.; Walczyk, D.; Campbell, A.; Elia, G.; Lynch, I.; Baldelli Bombelli, F.; Dawson, K.A. Physical−Chemical Aspects of Protein Corona: Relevance to in Vitro and in Vivo Biological Impacts of Nanoparticles. J. Am. Chem. Soc. 2011, 133, 2525–2534. [Google Scholar] [CrossRef] [PubMed]
- Tenzer, S.; Docter, D.; Kuharev, J.; Musyanovych, A.; Fetz, V.; Hecht, R.; Schlenk, F.; Fischer, D.; Kiouptsi, K.; Reinhardt, C.; et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 2013, 8, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Moyano, D.F.; Liu, Y.; Peer, D.; Rotello, V.M. Modulation of Immune Response Using Engineered Nanoparticle Surfaces. Small 2015, 12, 76–82. [Google Scholar] [CrossRef] [Green Version]
- Caballero, A.B.; Gamez, P. Nanochaperone-Based Strategies to Control Protein Aggregation Linked to Conformational Diseases. Angew. Chem. Int. Ed. 2020, 60, 41–52. [Google Scholar] [CrossRef] [PubMed]
- De, M.; Rotello, V.M. Synthetic “chaperones”: Nanoparticle-mediated refolding of thermally denatured proteins. Chem. Commun. 2008, 3504–3506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, F.H.; Li, C.; Liu, Y.; Shi, L. Mimicking Molecular Chaperones to Regulate Protein Folding. Adv. Mater. 2020, 32, e1805945. [Google Scholar] [CrossRef]
- Nakamoto, M.; Nonaka, T.; Shea, K.J.; Miura, Y.; Hoshino, Y. Design of Synthetic Polymer Nanoparticles That Facilitate Resolubilization and Refolding of Aggregated Positively Charged Lysozyme. J. Am. Chem. Soc. 2016, 138, 4282–4285. [Google Scholar] [CrossRef]
- Rozema, D.; Gellman, S.H. Artificial Chaperone-Assisted Refolding of Denatured-Reduced Lysozyme: Modulation of the Competition between Renaturation and Aggregation. Biochemistry 1996, 35, 15760–15771. [Google Scholar] [CrossRef]
- Boridy, S.; Maysinger, D. Nano-Chaperones: Nanoparticles Acting as Artificial Chaperones. Sel. Top. Nanomed. 2013, 3, 449. [Google Scholar] [CrossRef]
- Yang, H.; Li, X.; Zhu, L.; Wu, X.; Zhang, S.; Huang, F.; Feng, X.; Shi, L. Heat Shock Protein Inspired Nanochaperones Restore Amyloid-β Homeostasis for Preventative Therapy of Alzheimer‘s Disease. Adv. Sci. 2019, 6, 1901844. [Google Scholar] [CrossRef] [Green Version]
- Fang, X.; Yang, T.; Wang, L.; Yu, J.; Wei, X.; Zhou, Y.; Wang, C.; Liang, W. Nano-cage-mediated refolding of insulin by PEG-PE micelle. Biomaterials 2016, 77, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Cabaleiro-Lago, C.; Lynch, I.; Dawson, K.A.; Linse, S. Inhibition of IAPP and IAPP(20−29) Fibrillation by Polymeric Nanoparticles. Langmuir 2010, 26, 3453–3461. [Google Scholar] [CrossRef]
- Boland, B.; Yu, W.H.; Corti, O.; Mollereau, B.; Henriques, A.; Bezard, E.; Pastores, G.M.; Rubinsztein, D.C.; Nixon, R.A.; Duchen, M.; et al. Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 2018, 17, 660–688. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Lin, Y.-X.; Yang, P.-P.; Wang, Y.; Qi, G.-B.; Qiao, Z.-Y.; Li, B.-N.; Zhang, K.; Zhang, J.; Wang, L.; et al. A self-destructive nanosweeper that captures and clears amyloid β-peptides. Nat. Commun. 2018, 9, 1802. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Z.-Y.; Lin, Y.-X.; Lai, W.-J.; Hou, C.-Y.; Wang, Y.; Qiao, S.-L.; Zhang, D.; Fang, Q.-J.; Wang, H. A General Strategy for Facile Synthesis and In Situ Screening of Self-Assembled Polymer-Peptide Nanomaterials. Adv. Mater. 2015, 28, 1859–1867. [Google Scholar] [CrossRef]
- Barbalinardo, M.; Antosova, A.; Gambucci, M.; Bednarikova, Z.; Albonetti, C.; Valle, F.; Sassi, P.; Latterini, L.; Gazova, Z.; Bystrenova, E. Effect of metallic nanoparticles on amyloid fibrils and their influence to neural cell toxicity. Nano Res. 2020, 13, 1081–1089. [Google Scholar] [CrossRef]
- Sudhakar, S.; Mani, E. Rapid Dissolution of Amyloid β Fibrils by Silver Nanoplates. Langmuir 2019, 35, 6962–6970. [Google Scholar] [CrossRef]
- Kamshad, M.; Talab, M.J.; Beigoli, S.; Rad, A.S.; Chamani, J. Use of spectroscopic and zeta potential techniques to study the interaction between lysozyme and curcumin in the presence of silver nanoparticles at different sizes. J. Biomol. Struct. Dyn. 2018, 37, 2030–2040. [Google Scholar] [CrossRef]
- DeRidder, L.; Sharma, A.; Liaw, K.; Sharma, R.; John, J.; Kannan, S.; Kannan, R.M. Dendrimer–tesaglitazar conjugate induces a phenotype shift of microglia and enhances β-amyloid phagocytosis. Nanoscale 2021, 13, 939–952. [Google Scholar] [CrossRef]
- Asthana, S.; Bhattacharyya, D.; Kumari, S.; Nayak, P.S.; Saleem, M.; Bhunia, A.; Jha, S. Interaction with zinc oxide nanoparticle kinetically traps α-synuclein fibrillation into off-pathway non-toxic intermediates. Int. J. Biol. Macromol. 2020, 150, 68–79. [Google Scholar] [CrossRef]
- Fei, L.; Perrett, S. Effect of nanoparticles on protein folding and fibrillogenesis. Int. J. Mol. Sci. 2009, 10, 646–655. [Google Scholar] [CrossRef] [Green Version]
- Kang, T.; Kim, Y.G.; Kim, D.; Hyeon, T. Inorganic nanoparticles with enzyme-mimetic activities for biomedical applications. Coord. Chem. Rev. 2020, 403, 213092. [Google Scholar] [CrossRef]
- Tavanti, F.; Pedone, A.; Menziani, M.C.; Alexander-Katz, A. Computational Insights into the Binding of Monolayer-Capped Gold Nanoparticles onto Amyloid-β Fibrils. ACS Chem. Neurosci. 2020, 11, 3153–3160. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, X.; Zhang, Y.; Lv, J.; Huang, F.; Wu, G.; Liu, Y.; Ma, R.; An, Y.; Shi, L. Nanochaperones Mediated Delivery of Insulin. Nano Lett. 2020, 20, 1755–1765. [Google Scholar] [CrossRef]
- Jin, M.; Li, N.; Sheng, W.; Ji, X.; Liang, X.; Kong, B.; Yin, P.; Li, Y.; Zhang, X.; Liu, K. Toxicity of different zinc oxide nanomaterials and dose-dependent onset and development of Parkinson’s disease-like symptoms induced by zinc oxide nanorods. Environ. Int. 2020, 146, 106179. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Pilozzi, A.R.; Huang, X. Exposure of CuO Nanoparticles Contributes to Cellular Apoptosis, Redox Stress, and Alzheimer’s Aβ Amyloidosis. Int. J. Environ. Res. Public Health 2020, 17, 1005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debnath, K.; Pradhan, N.; Singh, B.K.; Jana, N.R.; Jana, N.R. Poly(trehalose) Nanoparticles Prevent Amyloid Aggregation and Suppress Polyglutamine Aggregation in a Huntington’s Disease Model Mouse. ACS Appl. Mater. Interfaces 2017, 9, 24126–24139. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Zhu, Y.; Yao, X.; Liu, H. Carbon Nanoparticles Inhibit the Aggregation of Prion Protein as Revealed by Experiments and Atomistic Simulations. J. Chem. Inf. Model. 2018, 59, 1909–1918. [Google Scholar] [CrossRef]
- Kim, D.; Yoo, J.M.; Hwang, H.; Lee, J.; Lee, S.H.; Yun, S.P.; Park, M.J.; Lee, M.J.; Choi, S.; Kwon, S.H.; et al. Graphene quantum dots prevent α-synucleinopathy in Parkinson’s disease. Nat. Nanotechnol. 2018, 13, 812–818. [Google Scholar] [CrossRef]
- Mondal, S.; Chowdhury, S.R.; Shah, M.; Kumar, V.; Kumar, S.; Iyer, P.K. Nanoparticle Assisted Regulation of Nucleation Pathway of Amyloid Tetramer and Inhibition of Their Fibrillation Kinetics. ACS Appl. Bio. Mater. 2019, 2, 2137–2142. [Google Scholar] [CrossRef]
- Sonawane, S.K.; Ahmad, A.; Chinnathambi, S. Protein-Capped Metal Nanoparticles Inhibit Tau Aggregation in Alzheimer’s Disease. ACS Omega 2019, 4, 12833–12840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahmoudi, M.; Lynch, I.; Ejtehadi, M.R.; Monopoli, M.P.; Bombelli, F.B.; Laurent, S. Protein−Nanoparticle Interactions: Opportunities and Challenges. Chem. Rev. 2011, 111, 5610–5637. [Google Scholar] [CrossRef] [PubMed]
- Sapsford, K.E.; Tyner, K.M.; Dair, B.J.; Deschamps, J.R.; Medintz, I.L. Analyzing Nanomaterial Bioconjugates: A Review of Current and Emerging Purification and Characterization Techniques. Anal. Chem. 2011, 83, 4453–4488. [Google Scholar] [CrossRef] [PubMed]
- Turci, F.; Ghibaudi, E.; Colonna, M.; Boscolo, B.; Fenoglio, I.; Fubini, B. An Integrated Approach to the Study of the Interaction between Proteins and Nanoparticles. Langmuir 2010, 26, 8336–8346. [Google Scholar] [CrossRef] [PubMed]
- Shang, L.; Wang, Y.; Jiang, A.J.; Dong, S. pH-Dependent Protein Conformational Changes in Albumin: Gold Nanoparticle Bioconjugates: A Spectroscopic Study. Langmuir 2007, 23, 2714–2721. [Google Scholar] [CrossRef] [PubMed]
- Wangoo, N.; Suri, C.R.; Shekhawat, G. Interaction of gold nanoparticles with protein: A spectroscopic study to monitor protein conformational changes. Appl. Phys. Lett. 2008, 92, 133104. [Google Scholar] [CrossRef]
- Raghava, S.; Singh, P.K.; Rao, A.R.; Dutta, V.; Gupta, M.N. Nanoparticles of unmodified titanium dioxide facilitate protein refolding. J. Mater. Chem. 2009, 19, 2830–2834. [Google Scholar] [CrossRef]
- Schaefer, J.; Schulze, C.; Marxer, E.E.J.; Schaefer, U.F.; Wohlleben, W.; Bakowsky, U.; Lehr, C.-M. Atomic Force Microscopy and Analytical Ultracentrifugation for Probing Nanomaterial Protein Interactions. ACS Nano 2012, 6, 4603–4614. [Google Scholar] [CrossRef]
- Klein, G.; Devineau, S.; Aude, J.C.; Boulard, Y.; Pasquier, H.; Labarre, J.; Pin, S.; Renault, J.-P. Interferences of Silica Nanoparticles in Green Fluorescent Protein Folding Processes. Langmuir 2015, 32, 195–202. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Li, Y.; Wei, L.; Liu, H.; Qiu, J.; Xiao, L. Attenuation of the Aggregation and Neurotoxicity of Amyloid Peptides with Neurotransmitter-Functionalized Ultra-Small-Sized Gold Nanoparticles. Eng. Sci. 2019, 6, 53–63. [Google Scholar] [CrossRef]
- Konar, M.; Mathew, A.; Dasgupta, S. Effect of Silica Nanoparticles on the Amyloid Fibrillation of Lysozyme. ACS Omega 2019, 4, 1015–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aubin-Tam, M.-E.; Hamad-Schifferli, K. Structure and function of nanoparticle–protein conjugates. Biomed. Mater. 2008, 3, 034001. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Haris, P.I.; Severcan, F. FTIR spectroscopic characterization of protein structure in aqueous and non-aqueous media. J. Mol. Catal. B Enzym. 1999, 7, 207–221. [Google Scholar] [CrossRef]
- Wojnarowska-Nowak, R.; Polit, J.; Sheregii, E.M. Interaction of gold nanoparticles with cholesterol oxidase enzyme in bionanocomplex—determination of the protein structure by Fourier transform infrared spectroscopy. J. Nanoparticle Res. 2020, 22, 1–14. [Google Scholar] [CrossRef]
- Spinozzi, F.; Ceccone, G.; Moretti, P.; Campanella, G.; Ferrero, C.; Combet, S.; Ojea-Jimenez, I.; Ghigna, P. Structural and Thermodynamic Properties of Nanoparticle-Protein Complexes: A Combined SAXS and SANS Study. Langmuir 2017, 33, 2248–2256. [Google Scholar] [CrossRef]
- Käkinen, A.; Ding, F.; Chen, P.; Mortimer, M.; Kahru, A.; Ke, P.C. Interaction of firefly luciferase and silver nanoparticles and its impact on enzyme activity. Nanotechnology 2013, 24, 345101. [Google Scholar] [CrossRef]
- Tonigold, M.; Simon, J.; Estupiñán, D.; Kokkinopoulou, M.; Reinholz, J.; Kintzel, U.; Kaltbeitzel, A.; Renz, P.; Domogalla, M.P.; Steinbrink, K.; et al. Pre-adsorption of antibodies enables targeting of nanocarriers despite a biomolecular corona. Nat. Nanotechnol. 2018, 13, 862–869. [Google Scholar] [CrossRef]
- Kokkinopoulou, M.; Simon, J.; Landfester, K.; Mailänder, V.; Lieberwirth, I. Visualization of the protein corona: Towards a biomolecular understanding of nanoparticle-cell-interactions. Nanoscale 2017, 9, 8858–8870. [Google Scholar] [CrossRef] [Green Version]
- Serpooshan, V.; Mahmoudi, M.; Zhao, M.; Wei, K.; Sivanesan, S.; Motamedchaboki, K.; Malkovskiy, A.V.; Goldstone, A.B.; Cohen, J.E.; Yang, P.C.; et al. Protein Corona Influences Cell-Biomaterial Interactions in Nanostructured Tissue Engineering Scaffolds. Adv. Funct. Mater. 2015, 25, 4379–4389. [Google Scholar] [CrossRef]
- García-Campaña, A.M.; Lara, F.J.; Gámiz-Gracia, L.; Huertas-Pérez, J.F. Chemiluminescence detection coupled to capillary electrophoresis. TrAC Trends Anal. Chem. 2009, 28, 973–986. [Google Scholar] [CrossRef]
- Cedervall, T.; Lynch, I.; Lindman, S.; Berggard, T.; Thulin, E.; Nilsson, H.; Dawson, K.A.; Linse, S. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. USA 2007, 104, 2050–2055. [Google Scholar] [CrossRef] [Green Version]
- Stalmach, A.; Albalat, A.; Mullen, W.; Mischak, H. Recent advances in capillary electrophoresis coupled to mass spectrometry for clinical proteomic applications. Electrophoresis 2013, 34, 1452–1464. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, S.; Dasgupta, N.; Chinnappan, S.; Ramalingam, C.; Kumar, A. Titanium dioxide nanoparticle–protein interaction explained by docking approach. Int. J. Nanomed. 2018, 13, 47–50. [Google Scholar] [CrossRef] [Green Version]
- Chibber, S.; Ahmed, I. Molecular docking, a tool to determine interaction of CuO and TiO2 nanoparticles with human serum albumin. Biochem. Biophys. Rep. 2016, 6, 63–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebrahim-Habibi, M.B.; Ghobeh, M.; Aghakhani Mahyari, F.; Rafii-Tabar, H.; Sasanpour, P. Protein G selects two binding sites for carbon nanotube with dissimilar behavior; a molecular dynamics study. J. Mol. Graph. Model. 2019, 87, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Fardanesh, A.; Zibaie, S.; Shariati, B.; Attar, F.; Rouhollah, F.; Akhtari, K.; Shahpasand, K.; Saboury, A.A.; Falahati, M. Amorphous aggregation of tau in the presence of titanium dioxide nanoparticles: Biophysical, computational, and cellular studies. Int. J. Nanomed. 2019, 14, 901–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tavanti, F.; Pedone, A.; Menziani, M.C. Disclosing the Interaction of Gold Nanoparticles with Aβ(1–40) Monomers through Replica Exchange Molecular Dynamics Simulations. Int. J. Mol. Sci. 2020, 22, 26. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaiswal, S.; Manhas, A.; Pandey, A.K.; Priya, S.; Sharma, S.K. Engineered Nanoparticle-Protein Interactions Influence Protein Structural Integrity and Biological Significance. Nanomaterials 2022, 12, 1214. https://doi.org/10.3390/nano12071214
Jaiswal S, Manhas A, Pandey AK, Priya S, Sharma SK. Engineered Nanoparticle-Protein Interactions Influence Protein Structural Integrity and Biological Significance. Nanomaterials. 2022; 12(7):1214. https://doi.org/10.3390/nano12071214
Chicago/Turabian StyleJaiswal, Surabhi, Amit Manhas, Alok Kumar Pandey, Smriti Priya, and Sandeep K. Sharma. 2022. "Engineered Nanoparticle-Protein Interactions Influence Protein Structural Integrity and Biological Significance" Nanomaterials 12, no. 7: 1214. https://doi.org/10.3390/nano12071214
APA StyleJaiswal, S., Manhas, A., Pandey, A. K., Priya, S., & Sharma, S. K. (2022). Engineered Nanoparticle-Protein Interactions Influence Protein Structural Integrity and Biological Significance. Nanomaterials, 12(7), 1214. https://doi.org/10.3390/nano12071214





