Enterococcus spp. Cell-Free Extract: An Abiotic Route for Synthesis of Selenium Nanoparticles (SeNPs), Their Characterisation and Inhibition of Escherichia coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culturing, Growing and Concentrating the Selenium-Reducing Bacteria
2.2. Chemicals, Culture Media and Solutions
2.3. Abiotic Synthesis of SeNPs
2.4. Cell-Free Extract Preparation
2.5. Characterisation of Selenium Nanoparticles (SeNPs)
2.6. Antibacterial Properties
3. Results and Discussion
3.1. SeO32− Reduction and SeNP Formation
3.2. XRD Analysis
3.3. SeNP Morphology and Particle-Size Distribution
3.4. FTIR Analysis
3.5. Antibacterial Efficiency of the SeNPs against E. coli
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Filipović, N.; Ušjak, D.; Milenković, M.T.; Zheng, K.; Liverani, L.; Boccaccini, A.R.; Stevanović, M.M. Comparative Study of the Antimicrobial Activity of Selenium Nanoparticles With Different Surface Chemistry and Structure. Front. Bioeng. Biotechnol. 2021, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Nayak, V.; Singh, K.R.; Singh, A.K.; Singh, R.P. Potentialities of selenium nanoparticles in biomedical science. New J. Chem. 2021, 45, 2849–2878. [Google Scholar] [CrossRef]
- Rezaei, H.; Jouyban, A.; Rahimpour, E. Development of a new method based on gold nanoparticles for determination of uric acid in urine samples. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 272, 120995. [Google Scholar] [CrossRef] [PubMed]
- Nancharaiah, Y.V.; Lens, P.N. Selenium biomineralization for biotechnological applications. Trends Biotechnol. 2015, 33, 323–330. [Google Scholar] [CrossRef]
- Brozmanová, J.; Mániková, D.; Vlčková, V.; Chovanec, M. Selenium: A double-edged sword for defense and offence in cancer. Arch. Toxicol. 2010, 84, 919–938. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, G.; Tang, Y. Towards selenium recovery: Biocathode induced selenate reduction to extracellular elemental selenium nanoparticles. Chem. Eng. J. 2018, 351, 1095–1103. [Google Scholar] [CrossRef]
- Gore, F.; Fawell, J.; Bartram, J. Too much or too little? A review of the conundrum of selenium. J. Water Heal. 2009, 8, 405–416. [Google Scholar] [CrossRef]
- Nakamaru, Y.; Tagami, K.; Uchida, S. Distribution coefficient of selenium in Japanese agricultural soils. Chemosphere 2005, 58, 1347–1354. [Google Scholar] [CrossRef]
- Fernandez-Martinez, A.; Charlet, L. Selenium environmental cycling and bioavailability: A structural chemist point of view. Rev. Environ. Sci. Bio. Technol. 2009, 8, 81–110. [Google Scholar] [CrossRef]
- Kuroda, M.; Notaguchi, E.; Sato, A.; Yoshioka, M.; Hasegawa, A.; Kagami, T.; Narita, T.; Yamashita, M.; Sei, K.; Soda, S.; et al. Characterization of Pseudomonas stutzeri NT-I capable of removing soluble selenium from the aqueous phase under aerobic conditions. J. Biosci. Bioeng. 2011, 112, 259–264. [Google Scholar] [CrossRef]
- Ečimović, S.; Velki, M.; Vuković, R.; Čamagajevac, I.; Petek, A.; Bošnjaković, R.; Grgić, M.; Engelmann, P.; Bodó, K.; Filipović-Marijić, V.; et al. Acute toxicity of selenate and selenite and their impacts on oxidative status, efflux pump activity, cellular and genetic parameters in earthworm Eisenia andrei. Chemosphere 2018, 212, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Chasteen, T.G.; Bentley, R. Biomethylation of Selenium and Tellurium: Microorganisms and Plants. Chem. Rev. 2002, 103, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.; Grosell, M.; Buchwalter, D.; Fisher, N.; Luoma, S.; Mathews, T.; Orr, P.; Wang, W.-X. Bioaccumulation and Trophic Transfer of Selenium. In Ecological Assessment of Selenium in the Aquatic Environment; Chapman, P.M., Adams, W.J., Brooks, M.L., Delos, C.G., Luoma, S.N., Maher, W.A., Ohlendorf, H.M., Presser, T.S., Shaw, D.P., Eds.; SETAC Press: Pensacola, FL, USA, 2010; pp. 93–139. [Google Scholar] [CrossRef]
- Schrauzer, G.N. Nutritional Selenium Supplements: Product Types, Quality, and Safety. J. Am. Coll. Nutr. 2001, 20, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Kabir, M.Z.; Emelianova, E.V.; Arkhipov, V.I.; Yunus, M.; Kasap, S.O.; Adriaenssens, G. The effects of large signals on charge collection in radiation detectors: Application to amorphous selenium detectors. J. Appl. Phys. 2006, 99, 124501. [Google Scholar] [CrossRef]
- Tan, L.C.; Nancharaiah, Y.V.; van Hullebusch, E.D.; Lens, P.N.L. Selenium: Environmental significance, pollution, and biological treatment technologies. Biotechnol. Adv. 2016, 34, 886–907. [Google Scholar] [CrossRef]
- Lenz, M.; Lens, P.N. The essential toxin: The changing perception of selenium in environmental sciences. Sci. Total Environ. 2009, 407, 3620–3633. [Google Scholar] [CrossRef]
- Golder-Associates-Inc. Literature Review of Treatment Technologies to Remove Selenium from Mining Influenced Water; Golder Associates Inc.: Lakewood, CA, USA, 2009; Available online: http://www.namc.org/docs/00057713.PDF (accessed on 1 February 2022).
- Zhang, W.; Chen, Z.; Liu, H.; Zhang, L.; Gao, P.; Li, D. Biosynthesis and structural characteristics of selenium nanoparticles by Pseudomonas alcaliphila. Colloids Surf. B Biointerfaces 2011, 88, 196–201. [Google Scholar] [CrossRef]
- Shakibaie, M.; Khorramizadeh, M.; Faramarzi, M.A.; Sabzevari, O.; Shahverdi, A.R. Biosynthesis and recovery of selenium nanoparticles and the effects on matrix metalloproteinase-2 expression. Biotechnol. Appl. Biochem. 2010, 56, 7–15. [Google Scholar] [CrossRef]
- Sonkusre, P. Improved Extraction of Intracellular Biogenic Selenium Nanoparticles and their Specificity for Cancer Chemoprevention. J. Nanomed. Nanotechnol. 2014, 5, 7–15. [Google Scholar] [CrossRef] [Green Version]
- Jain, R.O.; Gonzalez-Gil, G.R.; Singh, V.I.; Van Hullebusch, E.D.; Farges, F.; Lens, P.N. Biogenic Selenium Nanoparticles: Production, Characterization and Challenges; Studium Press LLC: Houston, TX, USA, 2014; pp. 365–394. [Google Scholar] [CrossRef]
- Tendenedzai, J.T.; Chirwa, E.M.N.; Brink, H.G. Performance Evaluation of Selenite (SeO32−) Reduction by Enterococcus spp. Catalysts 2021, 11, 1024. [Google Scholar] [CrossRef]
- Roestorff, M.M.; Chirwa, E.M.N. Correction to: Cr(VI) mediated hydrolysis of algae cell walls to release TOC for enhanced biotransformation of Cr(VI) by a culture of Cr(VI) reducing bacteria. J. Appl. Phycol. 2019, 31, 3651. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Cheng, H.; Wang, F.; Wei, D.; Wang, X. An improved 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) reduction assay for evaluating the viability of Escherichia coli cells. J. Microbiol. Methods 2010, 82, 330–333. [Google Scholar] [CrossRef]
- Kagami, T.; Narita, T.; Kuroda, M.; Notaguchi, E.; Yamashita, M.; Sei, K.; Soda, S.; Ike, M. Effective selenium volatilization under aerobic conditions and recovery from the aqueous phase by Pseudomonas stutzeri NT-I. Water Res. 2012, 47, 1361–1368. [Google Scholar] [CrossRef] [PubMed]
- Dungan, R.S.; Frankenberger, W.T. Reduction of Selenite to Elemental Selenium by Enterobacter cloacae SLD1a-1. J. Environ. Qual. 1998, 27, 1301–1306. [Google Scholar] [CrossRef]
- Tendenedzai, J.; Chirwa, E.M.; Brink, H.G. Reduction of Selenite by Use of Pseudomonas Stutzeri NT-I Cell Free Extract. Chem. Eng. Trans. 2020, 79, 373–378. [Google Scholar] [CrossRef]
- Tendenedzai, J.T.; Brink, H.G. The Effect of Nitrogen on the Reduction of Selenite to Elemental Selenium by Pseudomonas stutzeri NT-I. Chem. Eng. Transanctions 2019, 74, 529–534. [Google Scholar] [CrossRef]
- Javed, S.; Sarwar, A.; Tassawar, M.; Faisal, M. Conversion of selenite to elemental selenium by indigenous bacteria isolated from polluted areas. Chem. Speciat. Bioavailab. 2015, 27, 162–168. [Google Scholar] [CrossRef] [Green Version]
- Hageman, S.P.; van der Weijden, R.D.; Stams, A.J.; Buisman, C.J. Bio-production of selenium nanoparticles with diverse physical properties for recovery from water. Int. J. Miner. Process. 2017, 169, 7–15. [Google Scholar] [CrossRef]
- Cruz, L.Y.; Wang, D.; Liu, J. Biosynthesis of selenium nanoparticles, characterization and X-ray induced radiotherapy for the treatment of lung cancer with interstitial lung disease. J. Photochem. Photobiol. B Biol. 2018, 191, 123–127. [Google Scholar] [CrossRef]
- Gonzalez-Gil, G.; Lens, P.N.L.; Saikaly, P. Selenite Reduction by Anaerobic Microbial Aggregates: Microbial Community Structure, and Proteins Associated to the Produced Selenium Spheres. Front. Microbiol. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Wang, H.; Bao, Y.; Zhang, L. Nano red elemental selenium has no size effect in the induction of seleno-enzymes in both cultured cells and mice. Life Sci. 2004, 75, 237–244. [Google Scholar] [CrossRef]
- Nguyen, T.-C.; Rajeswari, V.D.; Al-Kheraif, A.A.; Brindhadevi, K. Study of antimicrobial properties of Piper betel coated nanozirconium on cotton gauze. Appl. Nanosci. 2021, 11, 1–7. [Google Scholar] [CrossRef]
- Tugarova, A.; Mamchenkova, P.; Dyatlova, Y.A.; Kamnev, A.A. FTIR and Raman spectroscopic studies of selenium nanoparticles synthesised by the bacterium Azospirillum thiophilum. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 192, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Piacenza, E.; Presentato, A.; Ferrante, F.; Cavallaro, G.; Alduina, R.; Martino, D.C. Biogenic Selenium Nanoparticles: A Fine Characterization to Unveil Their Thermodynamic Stability. Nanomaterials 2021, 11, 1195. [Google Scholar] [CrossRef]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [Green Version]
- Kamnev, A.A.; Sadovnikova, J.N.; Tarantilis, P.A.; Polissiou, M.G.; Antonyuk, L.P. Responses of Azospirillum brasilense to Nitrogen Deficiency and to Wheat Lectin: A Diffuse Reflectance Infrared Fourier Transform (DRIFT) Spectroscopic Study. Microb. Ecol. 2008, 56, 615–624. [Google Scholar] [CrossRef]
- Faghihzadeh, F.; Anaya, N.M.; Schifman, L.A.; Oyanedel-Craver, V. Fourier transform infrared spectroscopy to assess molecular-level changes in microorganisms exposed to nanoparticles. Nanotechnol. Environ. Eng. 2016, 1, 1–16. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Fourier Transform Infrared (FTIR) Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Guisbiers, G.; Wang, Q.; Khachatryan, E.; Mimun, L.; Mendoza-Cruz, R.; Larese-Casanova, P.; Webster, T.; Nash, K. Inhibition of E. coli and S. aureus with selenium nanoparticles synthesized by pulsed laser ablation in deionized water. Int. J. Nanomed. 2016, 11, 3731–3736. [Google Scholar] [CrossRef] [Green Version]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Muchová, J.; Hearnden, V.; Michlovská, L.; Vištejnová, L.; Zavaďáková, A.; Šmerková, K.; Kočiová, S.; Adam, V.; Kopel, P.; Vojtová, L. Mutual influence of selenium nanoparticles and FGF2-STAB® on biocompatible properties of collagen/chitosan 3D scaffolds: In vitro and ex ovo evaluation. J. Nanobiotechnol. 2021, 19, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Vahdati, M.; Moghadam, T.T. Synthesis and Characterization of Selenium Nanoparticles-Lysozyme Nanohybrid System with Synergistic Antibacterial Properties. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Radovic-Moreno, A.F.; Wu, J.; Langer, R.; Shi, J. Nanomedicine in the management of microbial infection—Overview and perspectives. Nano Today 2014, 9, 478–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Concentration (mM) | Average SeO32− Remaining (mM) | Average Se0 Formation (mM) | Total Average Se (mM) |
|---|---|---|---|
| 1 | 0.742 | 0.348 | 1.09 |
| 3 | 2.535 | 0.558 | 3.093 |
| 5 | 3.372 | 1.501 | 4.873 |
| Selenium Concentration | Distribution 1 (Red Peak) | Distribution 2 (Green Peak) | Distribution 3 (Blue Peak) | Total Distribution (Dashed Line/Bars) | ||||
|---|---|---|---|---|---|---|---|---|
| % of Total | d50 Predicted1 | % of Total | d50 Predicted1 | % of Total | d50 Predicted1 | d50 Predicted 1 | d50 Measured 2 | |
| 1 mM | 52.5 | 178.4 | 19.7 | 323.6 | 29.5 | 1940.1 | 267.2 | 278.8 |
| 3 mM | 43.3 | 166.6 | 48.7 | 329.4 | 10.9 | 3892.5 | 247.3 | 244.9 |
| 5 mM | 43.8 | 163.2 | 54.8 | 318.1 | 1.5 | 4846.5 | 248.5 | 246.5 |
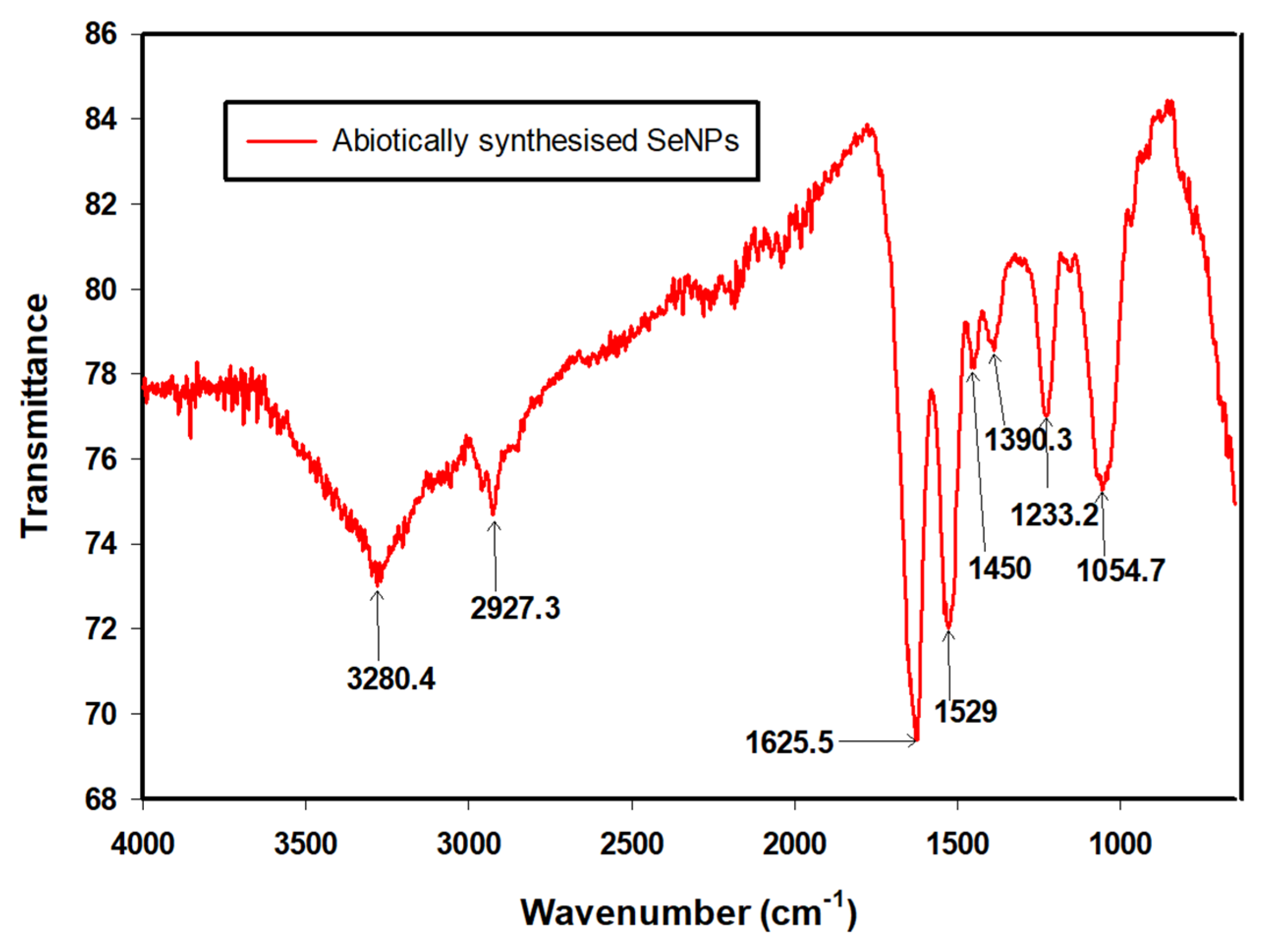
| Wavenumber (cm−1) | Functional Groups | References |
|---|---|---|
| 3280.4 | N–H2, aminoacidic group | [36] |
| 2927.3 | C–H, C–H2 stretch, Alkanes, aliphatic groups, fatty acid aliphatic chains | [37] |
| 1625.5 | N–H stretch, Secondary amine, amide I | [38] |
| 1529 | C–N stretch, amide II band, alkanes | [36] |
| 1450 | –CH2/–CH3 (in proteins, lipids, polyesters, etc.) | [36,39] |
| 1390.3 | Carboxyl (–COO−) stretching vibration | [37,40] |
| 1233.2 | C–N stretch, amide III band, O–P–O | [36] |
| 1054.7 | Cyclohexane ring vibrations/Aromatic C–H in-plane bend/Aliphatic fluoro compounds, C–F stretch/Primary amine C–N stretch C–O, C–C, (In polysaccharides, proteins and polyesters) | [36,41] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tendenedzai, J.T.; Chirwa, E.M.N.; Brink, H.G. Enterococcus spp. Cell-Free Extract: An Abiotic Route for Synthesis of Selenium Nanoparticles (SeNPs), Their Characterisation and Inhibition of Escherichia coli. Nanomaterials 2022, 12, 658. https://doi.org/10.3390/nano12040658
Tendenedzai JT, Chirwa EMN, Brink HG. Enterococcus spp. Cell-Free Extract: An Abiotic Route for Synthesis of Selenium Nanoparticles (SeNPs), Their Characterisation and Inhibition of Escherichia coli. Nanomaterials. 2022; 12(4):658. https://doi.org/10.3390/nano12040658
Chicago/Turabian StyleTendenedzai, Job T., Evans M. N. Chirwa, and Hendrik G. Brink. 2022. "Enterococcus spp. Cell-Free Extract: An Abiotic Route for Synthesis of Selenium Nanoparticles (SeNPs), Their Characterisation and Inhibition of Escherichia coli" Nanomaterials 12, no. 4: 658. https://doi.org/10.3390/nano12040658
APA StyleTendenedzai, J. T., Chirwa, E. M. N., & Brink, H. G. (2022). Enterococcus spp. Cell-Free Extract: An Abiotic Route for Synthesis of Selenium Nanoparticles (SeNPs), Their Characterisation and Inhibition of Escherichia coli. Nanomaterials, 12(4), 658. https://doi.org/10.3390/nano12040658








