Bacterial Magnetosomes Release Iron Ions and Induce Regulation of Iron Homeostasis in Endothelial Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Magnetosome Preparation
2.2. Examination of Purified MAGs and Amine Iron Oxide Nanoparticles
2.3. Cell Culture and Treatment
2.4. Cell Toxicity Assay
2.5. Flow Cytometry Analysis of the Cell Cycle
2.6. Determination of Lipid Peroxidation and Protein Oxidation
2.7. Intracellular Localization of MAGs and IONPs and Iron Quantification by Inductively Coupled PlasmaMass Spectrometry (ICP-MS)
2.8. Proteomic Analysis
2.9. Bioinformatics Analysis
2.10. Immunofluorescence and Western Blot Analysis
2.11. Statistical Methods
3. Results
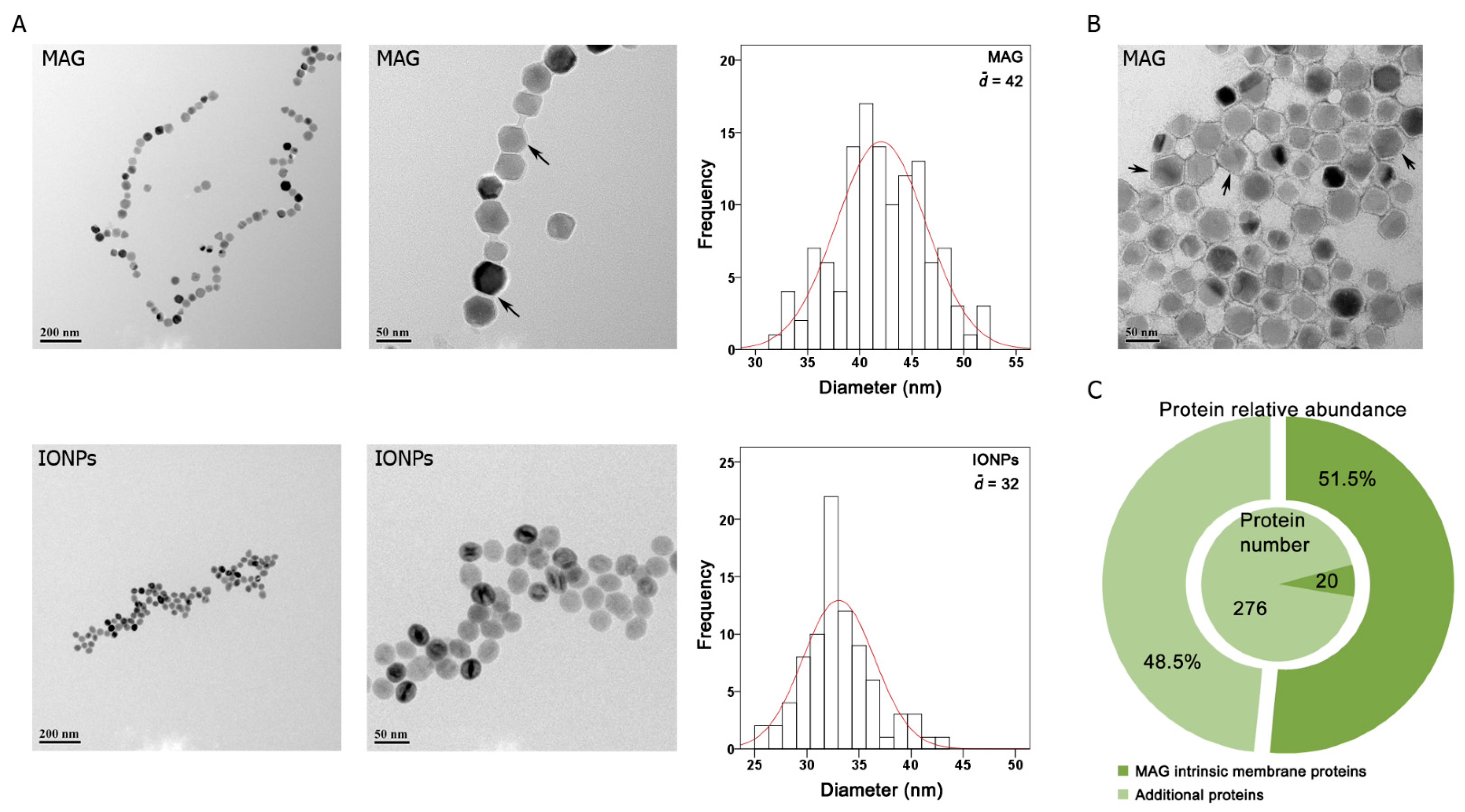
3.1. Particle Characterization
3.2. Cell Toxicity Analysis
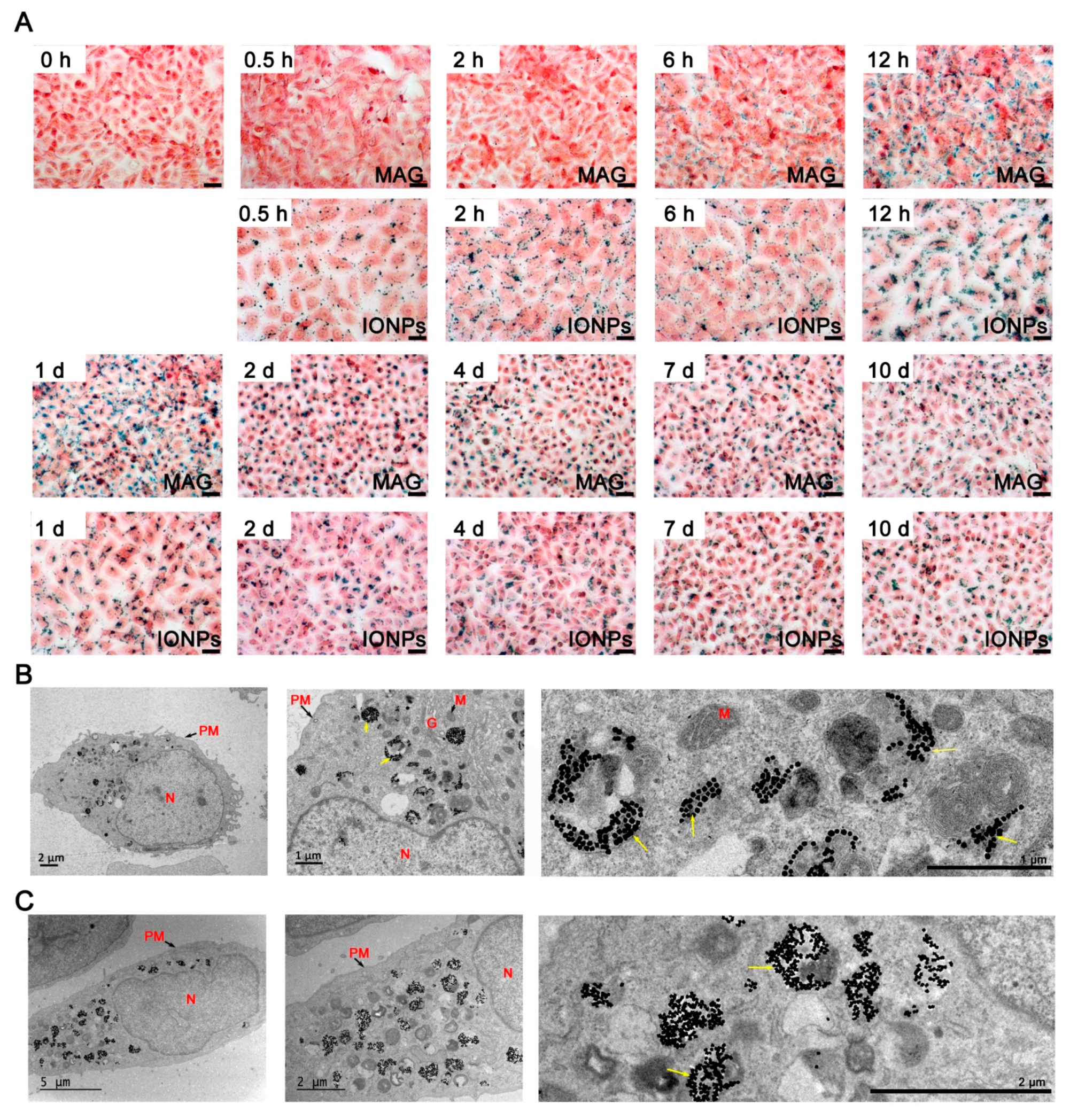
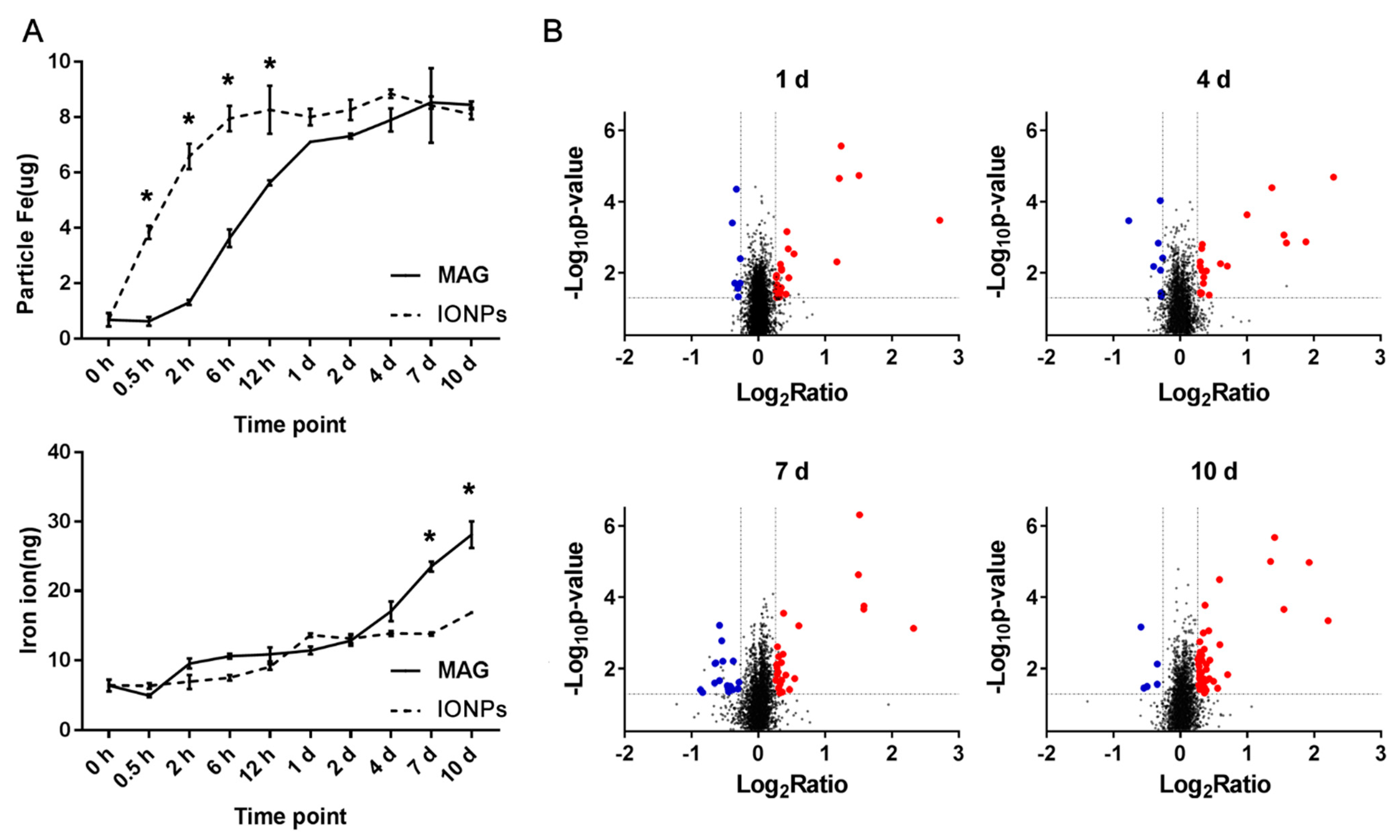
3.3. Cellular Internalization of MAGs and Iron Quantification
3.4. Overview of Differentially Expressed Proteins in MAG-Treated ECs
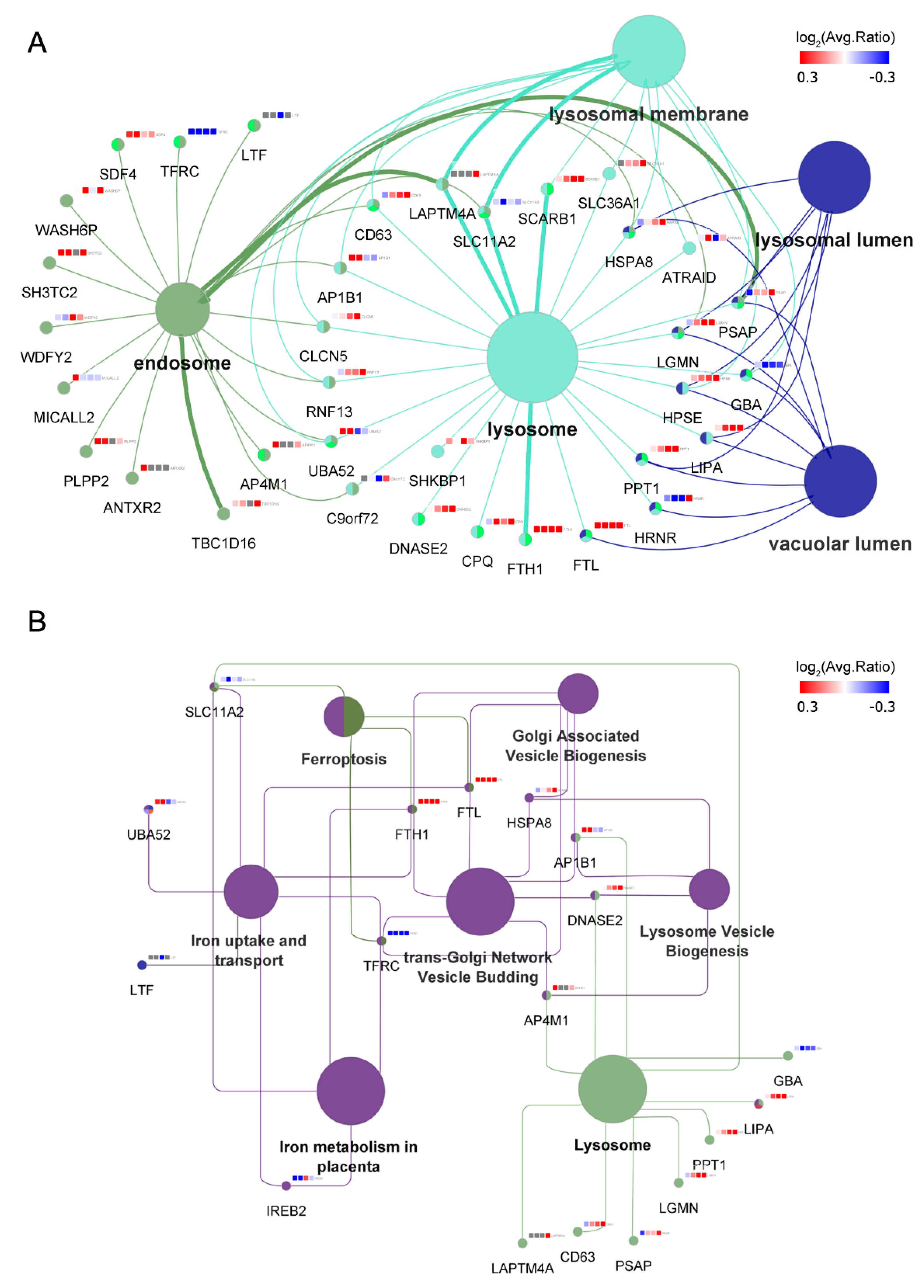
3.5. MAGs Induce Changes in Endosomal–Lysosomal Proteins
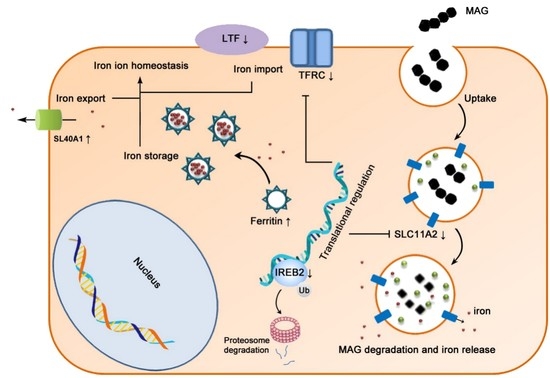
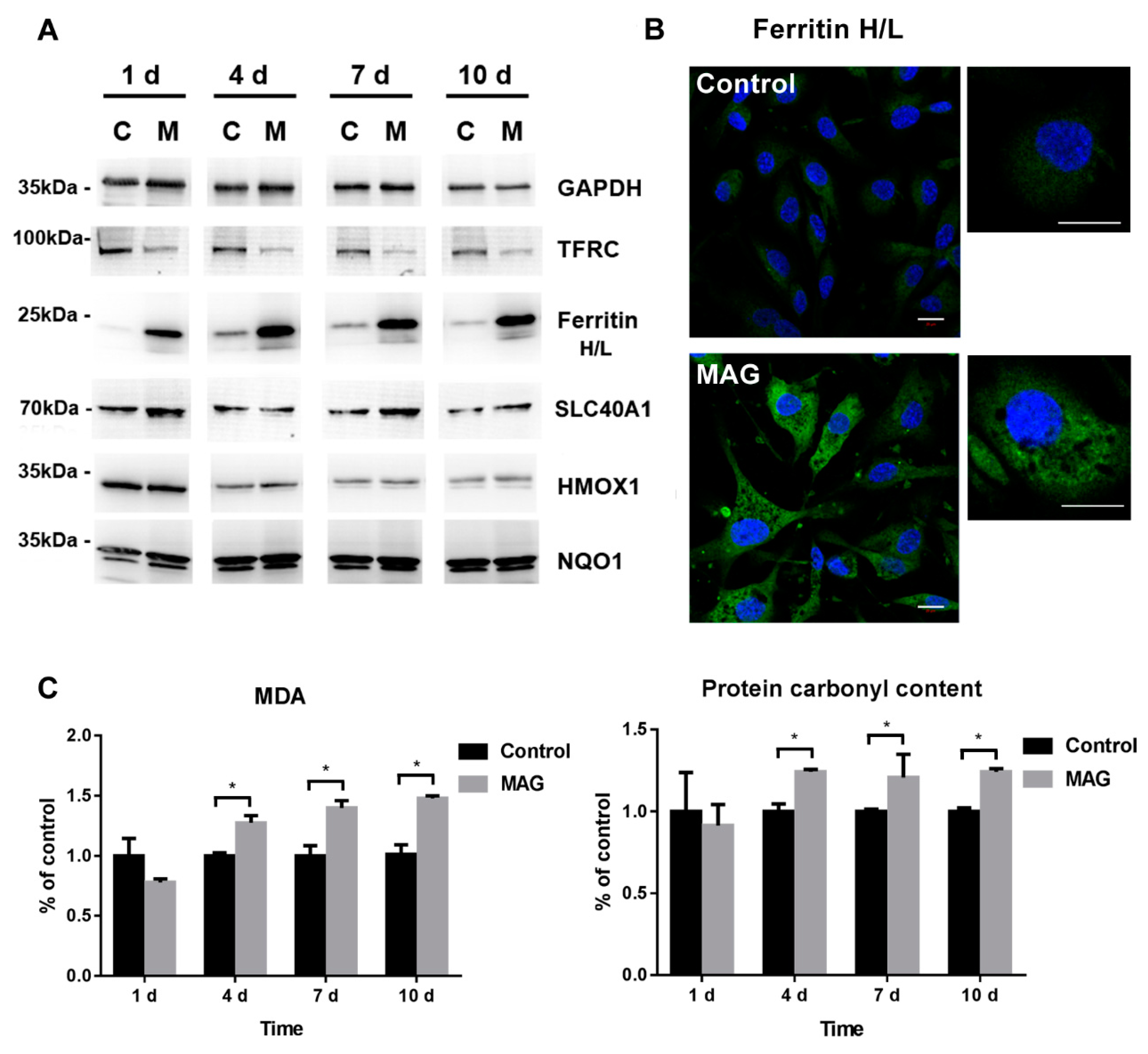
3.6. Iron Metabolism Regulation in ECs
3.7. Cell Oxidative Stress Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tong, S.; Zhu, H.; Bao, G. Magnetic iron oxide nanoparticles for disease detection and therapy. Mater. Today 2019, 31, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Baki, A.; Wiekhorst, F.; Bleul, R. Advances in Magnetic Nanoparticles Engineering for Biomedical Applications-A Review. Bioengineering 2021, 8, 134. [Google Scholar] [CrossRef] [PubMed]
- Gavilán, H.; Avugadda, S.K.; Fernández-Cabada, T.; Soni, N.; Cassani, M.; Mai, B.T.; Chantrell, R.; Pellegrino, T. Magnetic nanoparticles and clusters for magnetic hyperthermia: Optimizing their heat performance and developing combinatorial therapies to tackle cancer. Chem. Soc. Rev. 2021, 50, 11614–11667. [Google Scholar] [CrossRef]
- Chen, C.; Ge, J.; Gao, Y.; Chen, L.; Cui, J.; Zeng, J.; Gao, M. Ultrasmall superparamagnetic iron oxide nanoparticles: A next generation contrast agent for magnetic resonance imaging. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2021, 14, e1740. [Google Scholar] [CrossRef] [PubMed]
- Dieudonné, A.; Pignol, D.; Prévéral, S. Magnetosomes: Biogenic iron nanoparticles produced by environmental bacteria. Appl. Microbiol. Biotechnol. 2019, 103, 3637–3649. [Google Scholar] [CrossRef]
- Prozorov, T.; Bazylinski, D.A.; Mallapragada, S.K.; Prozorov, R. Novel magnetic nanomaterials inspired by magnetotactic bacteria: Topical review. Mater. Sci. Eng. R Rep. 2013, 74, 133–172. [Google Scholar] [CrossRef]
- Kralj, S.; Marchesan, S. Bioinspired Magnetic Nanochains for Medicine. Pharmaceutics 2021, 13, 1262. [Google Scholar] [CrossRef]
- Le Fèvre, R.; Durand-Dubief, M.; Chebbi, I.; Mandawala, C.; Lagroix, F.; Valet, J.-P.; Idbaih, A.; Adam, C.; Delattre, J.-Y.; Schmitt, C.; et al. Enhanced antitumor efficacy of biocompatible magnetosomes for the magnetic hyperthermia treatment of glioblastoma. Theranostics 2017, 7, 4618–4631. [Google Scholar] [CrossRef]
- Jacob, J.J.; Suthindhiran, K. Magnetotactic bacteria and magnetosomes—Scope and challenges. Mater. Sci. Eng. C 2016, 68, 919–928. [Google Scholar] [CrossRef]
- Oltolina, F.; Peigneux, A.; Colangelo, D.; Clemente, N.; D’Urso, A.; Valente, G.; Iglesias, G.R.; Jiménez-Lopez, C.; Prat, M. Biomimetic Magnetite Nanoparticles as Targeted Drug Nanocarriers and Mediators of Hyperthermia in an Experimental Cancer Model. Cancers 2020, 12, 2564. [Google Scholar] [CrossRef]
- Nan, X.; Teng, Y.; Tian, J.; Hu, Z.; Fang, Q. A comprehensive assessment of the biocompatibility of Magnetospirillum gryphiswaldense MSR-1 bacterial magnetosomes in vitro and in vivo. Toxicology 2021, 462, 152949. [Google Scholar] [CrossRef] [PubMed]
- Mickoleit, F.; Jörke, C.; Geimer, S.; Maier, D.S.; Müller, J.P.; Demut, J.; Gräfe, C.; Schüler, D.; Clement, J.H. Biocompatibility, uptake and subcellular localization of bacterial magnetosomes in mammalian cells. Nanoscale Adv. 2021, 3, 3799–3815. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Lv, X.; Zhang, T.; Jia, P.; Yan, R.; Li, S.; Zou, R.; Xue, Y.; Dai, L. Cytotoxicity and genotoxicity of bacterial magnetosomes against human retinal pigment epithelium cells. Sci. Rep. 2016, 6, 26961. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Tang, T.; Duan, J.; Xu, P.-x.; Wang, Z.; Zhang, Y.; Wu, L.; Li, Y. Biocompatibility of bacterial magnetosomes: Acute toxicity, immunotoxicity and cytotoxicity. Nanotoxicology 2010, 4, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Chen, C.; Chen, C.; Li, Y.; Pan, W.; Song, T. The interaction of bacterial magnetosomes and human liver cancer cells in vitro. J. Magn. Magn. Mater. 2017, 427, 105–110. [Google Scholar] [CrossRef]
- Cypriano, J.; Werckmann, J.; Vargas, G.; dos Santos, A.L.; Silva, K.T.; Leao, P.; Almeida, F.P.; Bazylinski, D.A.; Farina, M.; Lins, U.; et al. Uptake and persistence of bacterial magnetite magnetosomes in a mammalian cell line: Implications for medical and biotechnological applications. PLoS ONE 2019, 14, e0215657. [Google Scholar] [CrossRef]
- Tsoi, K.M.; MacParland, S.A.; Ma, X.-Z.; Spetzler, V.N.; Echeverri, J.; Ouyang, B.; Fadel, S.M.; Sykes, E.A.; Goldaracena, N.; Kaths, J.M.; et al. Mechanism of hard-nanomaterial clearance by the liver. Nat. Mater. 2016, 15, 1212–1221. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Syed, A.M.; MacMillan, P.; Rocheleau, J.V.; Chan, W.C.W. Flow Rate Affects Nanoparticle Uptake into Endothelial Cells. Adv. Mater. 2020, 32, 1906274. [Google Scholar] [CrossRef]
- Liu, L.Y.; Ma, X.Z.; Ouyang, B.; Ings, D.P.; Marwah, S.; Liu, J.; Chen, A.Y.; Gupta, R.; Manuel, J.; Chen, X.C.; et al. Nanoparticle Uptake in a Spontaneous and Immunocompetent Woodchuck Liver Cancer Model. ACS Nano 2020, 14, 4698–4715. [Google Scholar] [CrossRef]
- Bosseboeuf, E.; Raimondi, C. Signalling, Metabolic Pathways and Iron Homeostasis in Endothelial Cells in Health, Atherosclerosis and Alzheimer’s Disease. Cells 2020, 9, 2055. [Google Scholar] [CrossRef]
- Sindhwani, S.; Syed, A.M.; Ngai, J.; Kingston, B.R.; Maiorino, L.; Rothschild, J.; MacMillan, P.; Zhang, Y.; Rajesh, N.U.; Hoang, T.; et al. The entry of nanoparticles into solid tumours. Nat. Mater. 2020, 19, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.; Li, D.; Wang, Q.; Nan, X.; Xiang, Z.; Ma, Y.; Liu, Y.; Chen, J.; Tian, J.; Fang, Q. A Protein Corona Adsorbed to a Bacterial Magnetosome Affects Its Cellular Uptake. Int. J Nanomed. 2020, 15, 1481–1498. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Wei, J.; Jianbo, S.; Guili, W.; Feng, G.; Ying, L. Purified and sterilized magnetosomes from Magnetospirillum gryphiswaldense MSR-1 were not toxic to mouse fibroblasts in vitro. Lett. Appl. Microbiol. 2007, 45, 75–81. [Google Scholar] [CrossRef]
- Lai, W.; Wang, Q.; Li, L.; Hu, Z.; Chen, J.; Fang, Q. Interaction of gold and silver nanoparticles with human plasma: Analysis of protein corona reveals specific binding patterns. Colloids Surf. B 2017, 152, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Chevion, M.; Berenshtein, E.; Stadtman, E.R. Human studies related to protein oxidation: Protein carbonyl content as a marker of damage. Free Radic. Re.s 2000, 33, S99–S108. [Google Scholar]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2010. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Orue, I.; Marcano, L.; Bender, P.; García-Prieto, A.; Valencia, S.; Mawass, M.A.; Gil-Cartón, D.; Alba Venero, D.; Honecker, D.; García-Arribas, A.; et al. Configuration of the magnetosome chain: A natural magnetic nanoarchitecture. Nanoscale 2018, 10, 7407–7419. [Google Scholar] [CrossRef]
- Bergeron, J.R.C.; Hutto, R.; Ozyamak, E.; Hom, N.; Hansen, J.; Draper, O.; Byrne, M.E.; Keyhani, S.; Komeili, A.; Kollman, J.M. Structure of the magnetosome-associated actin-like MamK filament at subnanometer resolution. Protein Sci. 2017, 26, 93–102. [Google Scholar] [CrossRef]
- Scheffel, A.; Gruska, M.; Faivre, D.; Linaroudis, A.; Plitzko, J.M.; Schüler, D. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 2006, 440, 110–114. [Google Scholar] [CrossRef]
- Arvizo, R.R.; Miranda, O.R.; Thompson, M.A.; Pabelick, C.M.; Bhattacharya, R.; Robertson, J.D.; Rotello, V.M.; Prakash, Y.S.; Mukherjee, P. Effect of Nanoparticle Surface Charge at the Plasma Membrane and Beyond. Nano Lett. 2010, 10, 2543–2548. [Google Scholar] [CrossRef] [PubMed]
- Escamilla-Rivera, V.; Uribe-Ramirez, M.; Gonzalez-Pozos, S.; Lozano, O.; Lucas, S.; De Vizcaya-Ruiz, A. Protein corona acts as a protective shield against Fe3O4-PEG inflammation and ROS-induced toxicity in human macrophages. Toxicol. Lett. 2016, 240, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Lartigue, L.; Alloyeau, D.; Kolosnjaj-Tabi, J.; Javed, Y.; Guardia, P.; Riedinger, A.; Pechoux, C.; Pellegrino, T.; Wilhelm, C.; Gazeaut, F. Biodegradation of Iron Oxide Nanocubes: High-Resolution In Situ Monitoring. ACS Nano 2013, 7, 3939–3952. [Google Scholar] [CrossRef]
- Liu, R.-t.; Liu, J.; Tong, J.-q.; Tang, T.; Kong, W.-C.; Wang, X.-w.; Li, Y.; Tang, J.-t. Heating effect and biocompatibility of bacterial magnetosomes as potential materials used in magnetic fluid hyperthermia. Prog. Nat. Sci. Mater. Int. 2012, 22, 31–39. [Google Scholar] [CrossRef]
- Gutierrez, L.; Romero, S.; da Silva, G.B.; Costo, R.; Vargas, M.D.; Ronconi, C.M.; Serna, C.J.; Veintemillas-Verdaguer, S.; Morales, M.D. Degradation of magnetic nanoparticles mimicking lysosomal conditions followed by AC susceptibility. Biomed. Eng. Biomed. Tech. 2015, 60, 417–425. [Google Scholar] [CrossRef]
- Van de Walle, A.; Kolosnjaj-Tabi, J.; Lalatonne, Y.; Wilhelm, C. Ever-Evolving Identity of Magnetic Nanoparticles within Human Cells: The Interplay of Endosomal Confinement, Degradation, Storage, and Neocrystallization. Acc. Chem. Res. 2020, 53, 2212–2224. [Google Scholar] [CrossRef]
- Arbab, A.S.; Wilson, L.B.; Ashari, P.; Jordan, E.K.; Lewis, B.K.; Frank, J.A. A model of lysosomal metabolism of dextran coated superparamagnetic iron oxide (SPIO) nanoparticles: Implications for cellular magnetic resonance imaging. NMR Biomed. 2005, 18, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Stepien, G.; Moros, M.; Perez-Hernandez, M.; Monge, M.; Gutierrez, L.; Fratila, R.M.; Las Heras, M.; Menao Guillen, S.; Puente Lanzarote, J.J.; Solans, C.; et al. Effect of Surface Chemistry and Associated Protein Corona on the Long-Term Biodegradation of Iron Oxide Nanoparticles In Vivo. ACS Appl. Mater. Interfaces 2018, 10, 4548–4560. [Google Scholar] [CrossRef]
- Nan, X.; Lai, W.; Li, D.; Tian, J.; Hu, Z.; Fang, Q. Biocompatibility of Bacterial Magnetosomes as MRI Contrast Agent: A Long-Term In Vivo Follow-Up Study. Nanomaterials 2021, 11, 1235. [Google Scholar] [CrossRef]
- Lippiat, J.; Smith, A. The CLC-5 2Cl−/H+ exchange transporter in endosomal function and Dent’s disease. Front. Physiol. 2012, 3, 449. [Google Scholar] [CrossRef]
- Fölsch, H. Role of the epithelial cell-specific clathrin adaptor complex AP-1B in cell polarity. Cell. Logist. 2015, 5, e1074331. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Kang, R.; Kroemer, G.; Tang, D. Broadening horizons: The role of ferroptosis in cancer. Nat. Rev. Clin. Oncol. 2021, 18, 280–296. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Kang, R.; Tang, D.L. Signaling pathways and defense mechanisms of ferroptosis. FEBS J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Montalbetti, N.; Simonin, A.; Kovacs, G.; Hediger, M.A. Mammalian iron transporters: Families SLC11 and SLC40. Mol. Aspects Med. 2013, 34, 270–287. [Google Scholar] [CrossRef]
- Chen, X.; Yu, C.; Kang, R.; Tang, D. Iron Metabolism in Ferroptosis. Front. Cell Dev. Biol. 2020, 8, 590226. [Google Scholar] [CrossRef]
- Brown, C.W.; Amante, J.J.; Chhoy, P.; Elaimy, A.L.; Liu, H.; Zhu, L.J.; Baer, C.E.; Dixon, S.J.; Mercurio, A.M. Prominin2 Drives Ferroptosis Resistance by Stimulating Iron Export. Dev. Cell 2019, 51, 575–586.e4. [Google Scholar] [CrossRef]
- Iwai, K.; Drake, S.K.; Wehr, N.B.; Weissman, A.M.; LaVaute, T.; Minato, N.; Klausner, R.D.; Levine, R.L.; Rouault, T.A. Iron-dependent oxidation, ubiquitination, and degradation of iron regulatory protein 2: Implications for degradation of oxidized proteins. Proc. Natl. Acad. Sci. USA 1998, 95, 4924–4928. [Google Scholar] [CrossRef]
- Lieu, P.T.; Heiskala, M.; Peterson, P.A.; Yang, Y. The roles of iron in health and disease. Mol. Aspects Med. 2001, 22, 1–87. [Google Scholar] [CrossRef]
- Anandhan, A.; Dodson, M.; Schmidlin, C.J.; Liu, P.F.; Zhang, D.D. Breakdown of an Ironclad Defense System: The Critical Role of NRF2 in Mediating Ferroptosis. Cell Chem. Biol. 2020, 27, 436–447. [Google Scholar] [CrossRef]
- Liu, L.; Bai, X.; Martikainen, M.V.; Kårlund, A.; Roponen, M.; Xu, W.; Hu, G.; Tasciotti, E.; Lehto, V.P. Cell membrane coating integrity affects the internalization mechanism of biomimetic nanoparticles. Nat. Commun. 2021, 12, 5726. [Google Scholar] [CrossRef]
- Zelepukin, I.V.; Yaremenko, A.V.; Ivanov, I.N.; Yuryev, M.V.; Cherkasov, V.R.; Deyev, S.M.; Nikitin, P.I.; Nikitin, M.P. Long-Term Fate of Magnetic Particles in Mice: A Comprehensive Study. ACS Nano 2021, 15, 11341–11357. [Google Scholar] [CrossRef] [PubMed]
- Kotakadi, S.M.; Borelli, D.P.R.; Nannepaga, J.S. Therapeutic Applications of Magnetotactic Bacteria and Magnetosomes: A Review Emphasizing on the Cancer Treatment. Front. Bioeng. Biotechnol. 2022, 10, 789016. [Google Scholar] [CrossRef] [PubMed]
- Felfoul, O.; Mohammadi, M.; Taherkhani, S.; de Lanauze, D.; Zhong Xu, Y.; Loghin, D.; Essa, S.; Jancik, S.; Houle, D.; Lafleur, M.; et al. Magneto-aerotactic bacteria deliver drug-containing nanoliposomes to tumour hypoxic regions. Nat. Nanotechnol. 2016, 11, 941–947. [Google Scholar] [CrossRef] [PubMed]
- Raschdorf, O.; Bonn, F.; Zeytuni, N.; Zarivach, R.; Becher, D.; Schüler, D. A quantitative assessment of the membrane-integral sub-proteome of a bacterial magnetic organelle. J. Proteomics 2018, 172, 89–99. [Google Scholar] [CrossRef]
- Dobrovolskaia, M.A.; McNeil, S.E. Understanding the correlation between in vitro and in vivo immunotoxicity tests for nanomedicines. J. Control. Release 2013, 172, 456–466. [Google Scholar] [CrossRef]
- Wang, Y.X.J.; Hussain, S.M.; Krestin, G.P. Superparamagnetic iron oxide contrast agents: Physicochemical characteristics and applications in MR imaging. Eur. Radiol. 2001, 11, 2319–2331. [Google Scholar] [CrossRef]
- Weinstein, J.S.; Varallyay, C.G.; Dosa, E.; Gahramanov, S.; Hamilton, B.; Rooney, W.D.; Muldoon, L.L.; Neuwelt, E.A. Superparamagnetic iron oxide nanoparticles: Diagnostic magnetic resonance imaging and potential therapeutic applications in neurooncology and central nervous system inflammatory pathologies, a review. J. Cerebr. Blood F Met. 2010, 30, 15–35. [Google Scholar] [CrossRef]
- Periasamy, V.S.; Athinarayanan, J.; Alhazmi, M.; Alatiah, K.A.; Alshatwi, A.A. Fe3O4 nanoparticle redox system modulation via cell-cycle progression and gene expression in human mesenchymal stem cells. Environ. Toxicol. 2016, 31, 901–912. [Google Scholar] [CrossRef]
- Laffon, B.; Fernández-Bertólez, N.; Costa, C.; Brandão, F.; Teixeira, J.P.; Pásaro, E.; Valdiglesias, V. Cellular and Molecular Toxicity of Iron Oxide Nanoparticles. In Cellular and Molecular Toxicology of Nanoparticles; Saquib, Q., Faisal, M., Al-Khedhairy, A.A., Alatar, A.A., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 199–213. [Google Scholar]
- Sun, J.-B.; Wang, Z.-L.; Duan, J.-H.; Ren, J.; Yang, X.-D.; Dai, S.-L.; Li, Y. Targeted Distribution of Bacterial Magnetosomes Isolated from Magnetospirillum gryphiswaldense MSR-1 in Healthy Sprague-Dawley Rats. J. Nanosci. Nanotechnol. 2009, 9, 1881–1885. [Google Scholar] [CrossRef]
- Frtús, A.; Smolková, B.; Uzhytchak, M.; Lunova, M.; Jirsa, M.; Kubinová, Š.; Dejneka, A.; Lunov, O. Analyzing the mechanisms of iron oxide nanoparticles interactions with cells: A road from failure to success in clinical applications. J. Control. Release 2020, 328, 59–77. [Google Scholar] [CrossRef]
- Kolosnjaj-Tabi, J.; Lartigue, L.; Javed, Y.; Luciani, N.; Pellegrino, T.; Wilhelm, C.; Alloyeau, D.; Gazeau, F. Biotransformations of magnetic nanoparticles in the body. Nano Today 2016, 11, 280–284. [Google Scholar] [CrossRef]
- Levy, M.; Luciani, N.; Alloyeau, D.; Elgrabli, D.; Deveaux, V.; Pechoux, C.; Chat, S.; Wang, G.; Vats, N.; Gendron, F.; et al. Long term in vivo biotransformation of iron oxide nanoparticles. Biomaterials 2011, 32, 3988–3999. [Google Scholar] [CrossRef] [PubMed]
- Volatron, J.; Carn, F.; Kolosnjaj-Tabi, J.; Javed, Y.; Vuong, Q.L.; Gossuin, Y.; Menager, C.; Luciani, N.; Charron, G.; Hemadi, M.; et al. Ferritin Protein Regulates the Degradation of Iron Oxide Nanoparticles. Small 2017, 13, 1602030. [Google Scholar] [CrossRef] [PubMed]
- Vashisht, A.A.; Zumbrennen, K.B.; Huang, X.; Powers, D.N.; Durazo, A.; Sun, D.; Bhaskaran, N.; Persson, A.; Uhlen, M.; Sangfelt, O.; et al. Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 2009, 326, 718–721. [Google Scholar] [CrossRef] [PubMed]
- Herborn, C.U.; Papanikolaou, N.; Reszka, R.; Grunberg, K.; Schuler, D.; Debatin, J.F. Magnetosomes as biological model for iron binding: Relaxivity determination with MRI. RoeFo-Fortschr. Geb. Roentgenstrahlen Neuen Bildgeb. 2003, 175, 830–834. [Google Scholar]
- Lisy, M.R.; Hartung, A.; Lang, C.; Schuler, D.; Richter, W.; Reichenbach, J.R.; Kaiser, W.A.; Hilger, I. Fluorescent bacterial magnetic nanoparticles as bimodal contrast agents. Invest. Radiol. 2007, 42, 235–241. [Google Scholar] [CrossRef]
- Xiong, L.; Zhang, L.; Yang, Y.; Li, N.; Lai, W.; Wang, F.; Zhu, X.; Wang, T. ER complex proteins are required for rhodopsin biosynthesis and photoreceptor survival in Drosophila and mice. Cell Death Differ. 2020, 27, 646–661. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, W.; Li, D.; Wang, Q.; Ma, Y.; Tian, J.; Fang, Q. Bacterial Magnetosomes Release Iron Ions and Induce Regulation of Iron Homeostasis in Endothelial Cells. Nanomaterials 2022, 12, 3995. https://doi.org/10.3390/nano12223995
Lai W, Li D, Wang Q, Ma Y, Tian J, Fang Q. Bacterial Magnetosomes Release Iron Ions and Induce Regulation of Iron Homeostasis in Endothelial Cells. Nanomaterials. 2022; 12(22):3995. https://doi.org/10.3390/nano12223995
Chicago/Turabian StyleLai, Wenjia, Dan Li, Qingsong Wang, Yan Ma, Jiesheng Tian, and Qiaojun Fang. 2022. "Bacterial Magnetosomes Release Iron Ions and Induce Regulation of Iron Homeostasis in Endothelial Cells" Nanomaterials 12, no. 22: 3995. https://doi.org/10.3390/nano12223995
APA StyleLai, W., Li, D., Wang, Q., Ma, Y., Tian, J., & Fang, Q. (2022). Bacterial Magnetosomes Release Iron Ions and Induce Regulation of Iron Homeostasis in Endothelial Cells. Nanomaterials, 12(22), 3995. https://doi.org/10.3390/nano12223995