MPTHub: An Open-Source Software for Characterizing the Transport of Particles in Biorelevant Media
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Software Development, Data Processing, and Performance
2.3. Application of MPTHub
2.3.1. Processing of Polystyrene Nanoparticles
2.3.2. Preparation of Mucus Surrogates
2.3.3. Microscope Configuration and Video Acquisition
3. Results and Discussion
3.1. MPTHub Development, Features, and Performance
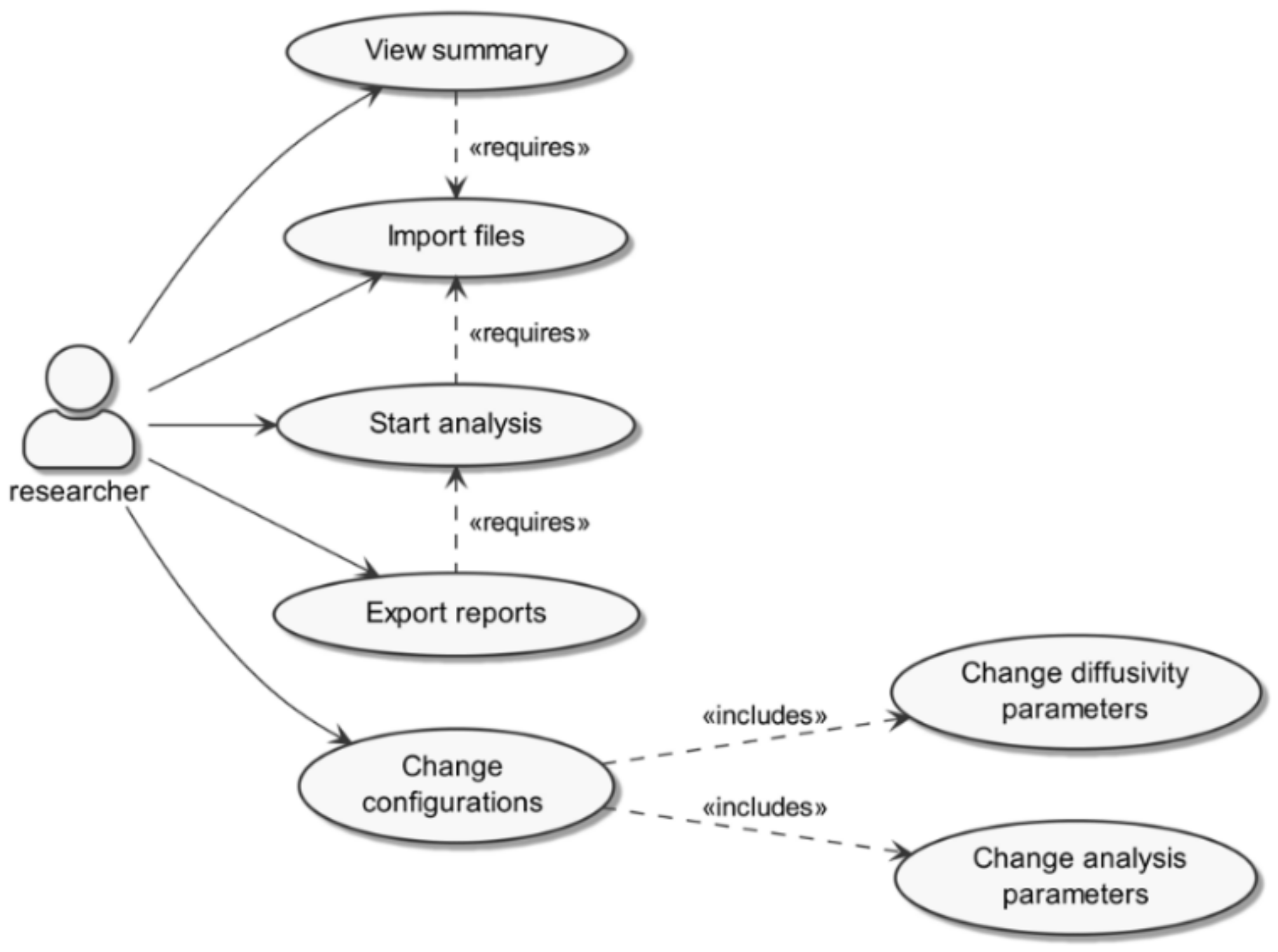
3.1.1. Software Programming and Workflow
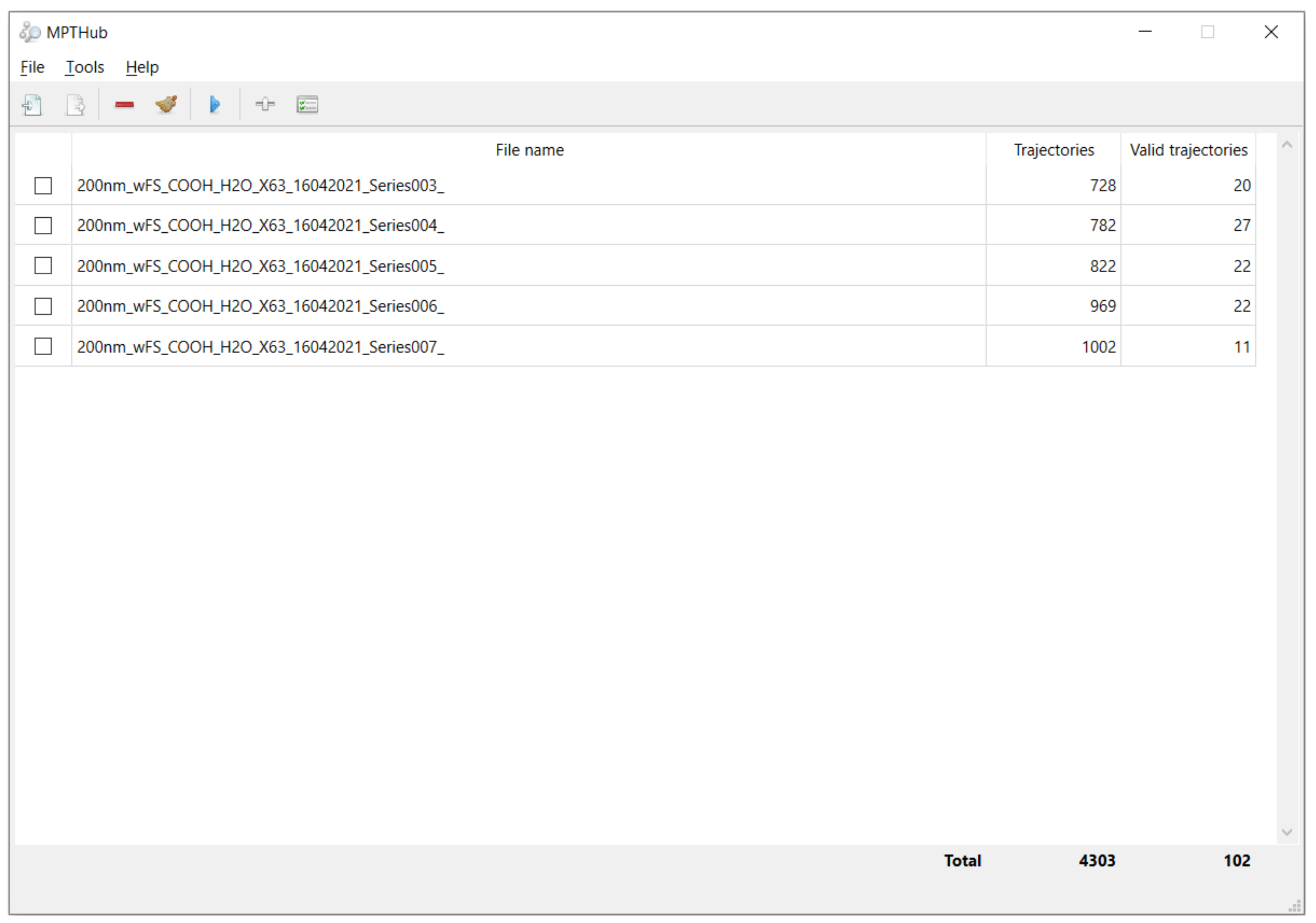
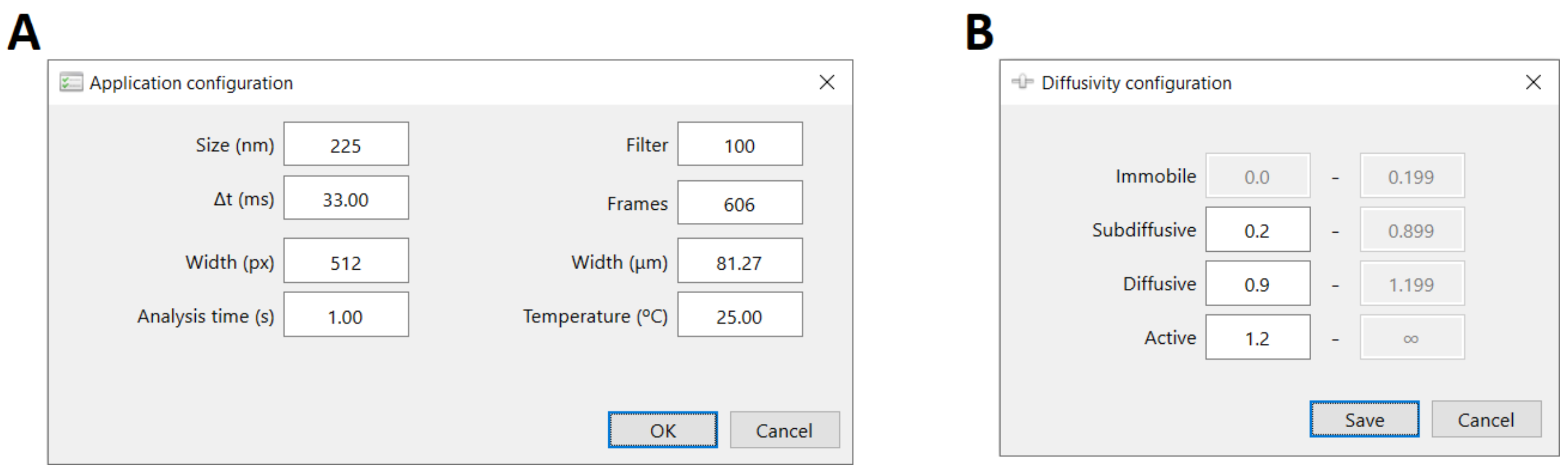
3.1.2. Graphical User Interface and Utilization
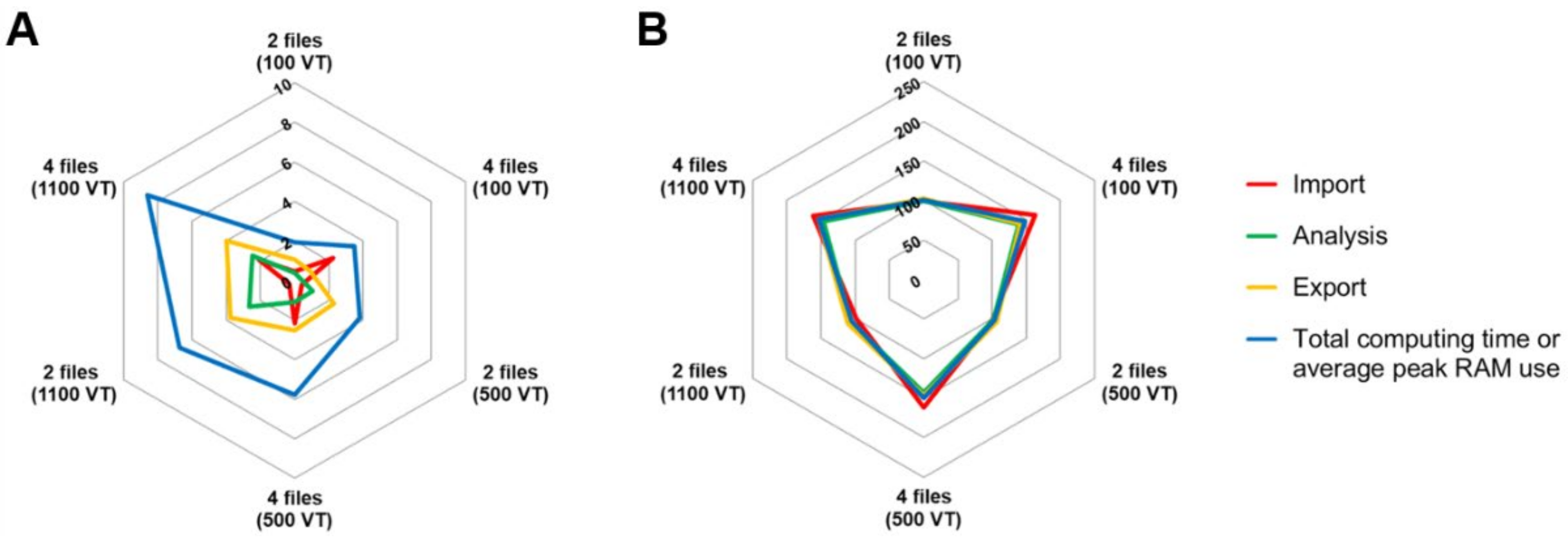
3.1.3. Software Performance
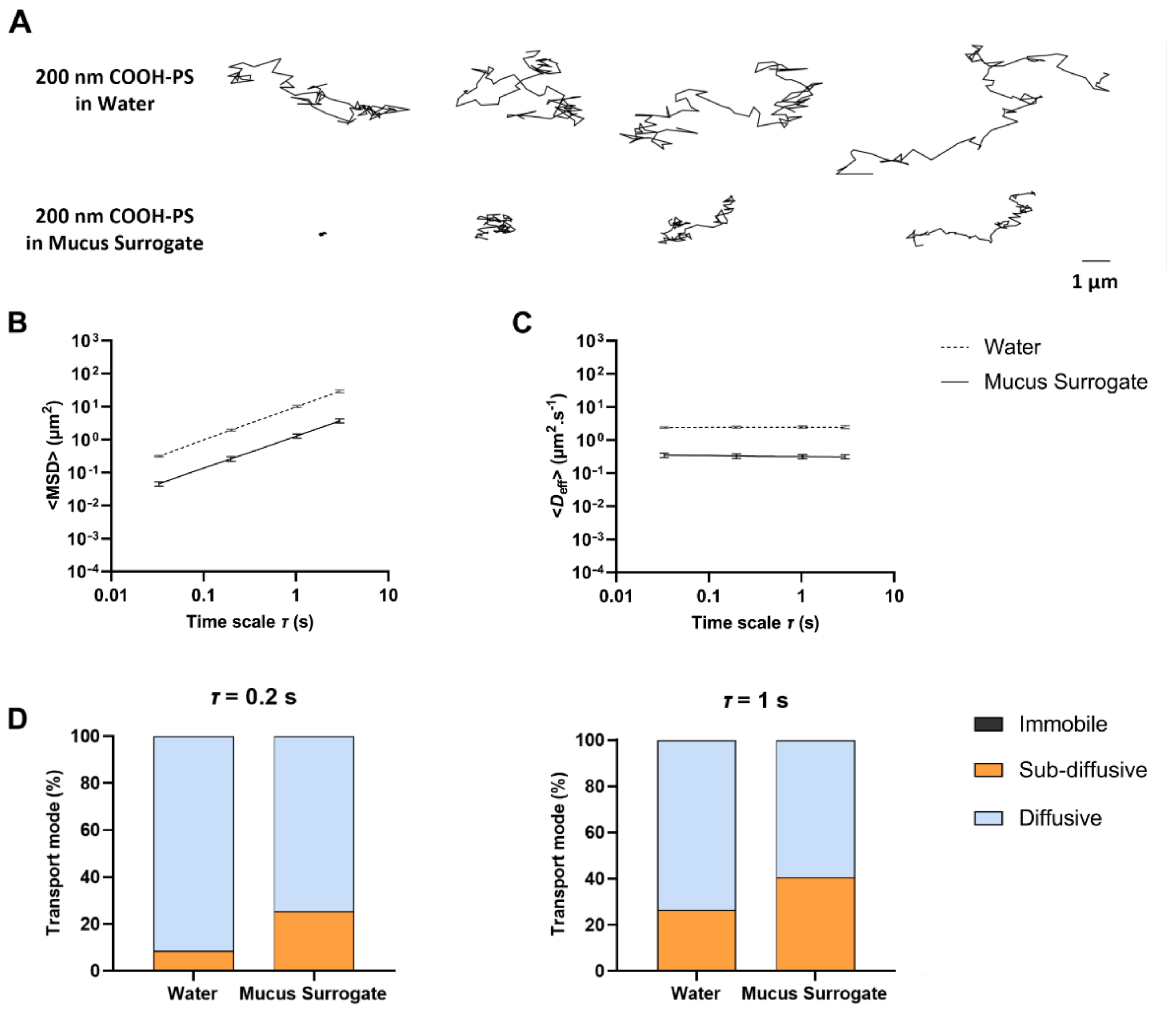
3.2. Application of MPTHub
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Talebian, S.; Rodrigues, T.; das Neves, J.; Sarmento, B.; Langer, R.; Conde, J. Facts and figures on materials science and nanotechnology progress and investment. ACS Nano 2021, 15, 15940–15952. [Google Scholar] [CrossRef] [PubMed]
- das Neves, J.; Sverdlov Arzi, R.; Sosnik, A. Molecular and cellular cues governing nanomaterial-mucosae interactions: From nanomedicine to nanotoxicology. Chem. Soc. Rev. 2020, 49, 5058–5100. [Google Scholar] [CrossRef] [PubMed]
- Olmsted, S.S.; Padgett, J.L.; Yudin, A.I.; Whaley, K.J.; Moench, T.R.; Cone, R.A. Diffusion of macromolecules and virus-like particles in human cervical mucus. Biophys. J. 2001, 81, 1930–1937. [Google Scholar] [CrossRef] [Green Version]
- Weiss, M.; Elsner, M.; Kartberg, F.; Nilsson, T. Anomalous subdiffusion is a measure for cytoplasmic crowding in living cells. Biophys. J. 2004, 87, 3518–3524. [Google Scholar] [CrossRef] [Green Version]
- Newby, J.M.; Seim, I.; Lysy, M.; Ling, Y.; Huckaby, J.; Lai, S.K.; Forest, M.G. Technological strategies to estimate and control diffusive passage times through the mucus barrier in mucosal drug delivery. Adv. Drug Deliv. Rev. 2018, 124, 64–81. [Google Scholar] [CrossRef] [Green Version]
- Murgia, X.; Loretz, B.; Hartwig, O.; Hittinger, M.; Lehr, C.M. The role of mucus on drug transport and its potential to affect therapeutic outcomes. Adv. Drug Deliv. Rev. 2018, 124, 82–97. [Google Scholar] [CrossRef]
- Rose, K.A.; Molaei, M.; Boyle, M.J.; Lee, D.; Crocker, J.C.; Composto, R.J. Particle tracking of nanoparticles in soft matter. J. Appl. Phys. 2020, 127, 191101. [Google Scholar] [CrossRef]
- Schuster, B.S.; Ensign, L.M.; Allan, D.B.; Suk, J.S.; Hanes, J. Particle tracking in drug and gene delivery research: State-of-the-art applications and methods. Adv. Drug Deliv. Rev. 2015, 91, 70–91. [Google Scholar] [CrossRef] [Green Version]
- Nance, E.A.; Woodworth, G.F.; Sailor, K.A.; Shih, T.-Y.; Xu, Q.; Swaminathan, G.; Xiang, D.; Eberhart, C.; Hanes, J. A dense poly(ethylene glycol) coating improves penetration of large polymeric nanoparticles within brain tissue. Sci. Transl. Med. 2012, 4, 149ra119. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Yu, T.; Wang, Y.-Y.; Lai, S.K.; Zeng, Q.; Miao, B.; Tang, B.C.; Simons, B.W.; Ensign, L.M.; Liu, G.; et al. Vaginal delivery of paclitaxel via nanoparticles with non-mucoadhesive surfaces suppresses cervical tumor growth. Adv. Healthc. Mater. 2014, 3, 1044–1052. [Google Scholar] [CrossRef] [Green Version]
- Siddique, S.; Chow, J.C.L. Gold nanoparticles for drug delivery and cancer therapy. Appl. Sci. 2020, 10, 3824. [Google Scholar] [CrossRef]
- Zierden, H.C.; Ortiz, J.I.; DeLong, K.; Yu, J.; Li, G.; Dimitrion, P.; Bensouda, S.; Laney, V.; Bailey, A.; Anders, N.M.; et al. Enhanced drug delivery to the reproductive tract using nanomedicine reveals therapeutic options for prevention of preterm birth. Sci. Transl. Med. 2021, 13, eabc6245. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, A.; Forkey, J.N.; McKinney, S.A.; Ha, T.; Goldman, Y.E.; Selvin, P.R. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science 2003, 300, 2061–2065. [Google Scholar] [CrossRef] [Green Version]
- Lill, Y.; Lill, M.A.; Fahrenkrog, B.; Schwarz-Herion, K.; Paulillo, S.; Aebi, U.; Hecht, B. Single hepatitis-B virus core capsid binding to individual nuclear pore complexes in Hela cells. Biophy. J. 2006, 91, 3123–3130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoang, T.; Toler, E.; DeLong, K.; Mafunda, N.A.; Bloom, S.M.; Zierden, H.C.; Moench, T.R.; Coleman, J.S.; Hanes, J.; Kwon, D.S.; et al. The cervicovaginal mucus barrier to HIV-1 is diminished in bacterial vaginosis. PLoS Pathog. 2020, 16, e1008236. [Google Scholar] [CrossRef] [Green Version]
- Herraez-Aguilar, D.; Madrazo, E.; Lopez-Menendez, H.; Ramirez, M.; Monroy, F.; Redondo-Munoz, J. Multiple particle tracking analysis in isolated nuclei reveals the mechanical phenotype of leukemia cells. Sci. Rep. 2020, 10, 6707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, S.K.; Wang, Y.-Y.; Hida, K.; Cone, R.; Hanes, J. Nanoparticles reveal that human cervicovaginal mucus is riddled with pores larger than viruses. Proc. Natl. Acad. Sci. USA 2010, 107, 598–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chisholm, J.F.; Shenoy, S.K.; Shade, J.K.; Kim, V.; Putcha, N.; Carson, K.A.; Wise, R.; Hansel, N.N.; Hanes, J.S.; Suk, J.S.; et al. Nanoparticle diffusion in spontaneously expectorated sputum as a biophysical tool to probe disease severity in COPD. Eur. Respir. J. 2019, 54, 1900088. [Google Scholar] [CrossRef] [PubMed]
- Huck, B.C.; Hartwig, O.; Biehl, A.; Schwarzkopf, K.; Wagner, C.; Loretz, B.; Murgia, X.; Lehr, C.M. Macro- and microrheological properties of mucus surrogates in comparison to native intestinal and pulmonary mucus. Biomacromolecules 2019, 20, 3504–3512. [Google Scholar] [CrossRef]
- Chenouard, N.; Smal, I.; de Chaumont, F.; Maska, M.; Sbalzarini, I.F.; Gong, Y.; Cardinale, J.; Carthel, C.; Coraluppi, S.; Winter, M.; et al. Objective comparison of particle tracking methods. Nat. Methods 2014, 11, 281–289. [Google Scholar] [CrossRef] [Green Version]
- Sbalzarini, I.F.; Koumoutsakos, P. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 2005, 151, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.; Lipinski, H.G.; Wiemann, M. Dark field nanoparticle tracking analysis for size characterization of plasmonic and non-plasmonic particles. J. Nanoparticle Res. 2014, 16, 2419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tinevez, J.Y.; Perry, N.; Schindelin, J.; Hoopes, G.M.; Reynolds, G.D.; Laplantine, E.; Bednarek, S.Y.; Shorte, S.L.; Eliceiri, K.W. TrackMate: An open and extensible platform for single-particle tracking. Methods 2017, 115, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Arts, M.; Smal, I.; Paul, M.W.; Wyman, C.; Meijering, E. Particle mobility analysis using deep learning and the moment scaling spectrum. Sci. Rep. 2019, 9, 17160. [Google Scholar] [CrossRef]
- Matysik, A.; Kraut, R.S. TrackArt: The user friendly interface for single molecule tracking data analysis and simulation applied to complex diffusion in mica supported lipid bilayers. BMC Res. Notes 2014, 7, 274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallotton, P.; van Oijen, A.M.; Whitchurch, C.B.; Gelfand, V.; Yeo, L.; Tsiavaliaris, G.; Heinrich, S.; Dultz, E.; Weis, K.; Grünwald, D. Diatrack particle tracking software: Review of applications and performance evaluation. Traffic 2017, 18, 840–852. [Google Scholar] [CrossRef]
- Lee, B.H.; Park, H.Y. HybTrack: A hybrid single particle tracking software using manual and automatic detection of dim signals. Sci. Rep. 2018, 8, 212. [Google Scholar] [CrossRef]
- Rösch, T.C.; Oviedo-Bocanegra, L.M.; Fritz, G.; Graumann, P.L. SMTracker: A tool for quantitative analysis, exploration and visualization of single-molecule tracking data reveals highly dynamic binding of B. subtilis global repressor AbrB throughout the genome. Sci. Rep. 2018, 8, 15747. [Google Scholar] [CrossRef] [Green Version]
- Rahm, J.; Malkusch, S.; Endesfelder, U.; Dietz, M.; Heilemann, M. Diffusion state transitions in single-particle trajectories of MET receptor tyrosine kinase measured in live cells. Front. Comput. Sci. 2021, 3, 757653. [Google Scholar] [CrossRef]
- Shepherd, J.W.; Higgins, E.J.; Wollman, A.J.M.; Leake, M.C. PySTACHIO: Python Single-molecule TrAcking stoiCHiometry Intensity and simulatiOn, a flexible, extensible, beginner-friendly and optimized program for analysis of single-molecule microscopy data. Comput. Struct. Biotechnol. J. 2021, 19, 4049–4058. [Google Scholar] [CrossRef]
- Malloy, A.; Carr, B. NanoParticle tracking analysis–The halo™ system. Part. Part. Syst. Charact. 2006, 23, 197–204. [Google Scholar] [CrossRef]
- Plaza-Oliver, M.; Cano, E.L.; Arroyo-Jimenez, M.M.; Gamez, M.; Lozano-Lopez, M.V.; Santander-Ortega, M.J. Taking particle tracking into practice by novel software and screening approach: Case-study of oral lipid nanocarriers. Pharmaceutics 2021, 13, 370. [Google Scholar] [CrossRef]
- Faez, S.; Carattino, A.; Mosk, A. PyNTA: An open source software application for live particle tracking. Preprints 2019, 2019060251. [Google Scholar] [CrossRef]
- AlTarawneh, G.; Thorne, S. A pilot study exploring spreadsheet risk in scientific research. arXiv 2017, arXiv:1703.09785. [Google Scholar]
- Allan, D.B.; Caswell, T.; Keim, N.C.; van der Wel, C.M.; Verweij, R.W. Soft Matter/Trackpy: Trackpy v0.5.0; Zenodo: Geneva, Switzerland, 2021. [Google Scholar] [CrossRef]
- Suh, J.; Dawson, M.; Hanes, J. Real-time multiple-particle tracking: Applications to drug and gene delivery. Adv. Drug Deliv. Rev. 2005, 57, 63–78. [Google Scholar] [CrossRef]
- Lanoiselée, Y.; Sikora, G.; Grzesiek, A.; Grebenkov, D.S.; Wyłomańska, A. Optimal parameters for anomalous-diffusion-exponent estimation from noisy data. Phys. Rev. E 2018, 98, 062139. [Google Scholar] [CrossRef] [Green Version]
- Crater, J.S.; Carrier, R.L. Barrier properties of gastrointestinal mucus to nanoparticle transport. Macromol. Biosci. 2010, 10, 1473–1483. [Google Scholar] [CrossRef]
- das Neves, J.; Rocha, C.M.; Gonçalves, M.P.; Carrier, R.L.; Amiji, M.; Bahia, M.F.; Sarmento, B. Interactions of microbicide nanoparticles with a simulated vaginal fluid. Mol. Pharm. 2012, 9, 3347–3356. [Google Scholar] [CrossRef]
- Prechelt, L. Are scripting languages any good? A validation of Perl, Python, Rexx, and Tcl against C, C++, and Java. Adv. Comput. 2003, 57, 205–270. [Google Scholar]
- Crocker, J.C.; Grier, D.G. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 1996, 179, 298–310. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Lai, S.K.; Wang, Y.-Y.; Zhong, W.; Happe, C.; Zhang, M.; Fu, J.; Hanes, J. Biodegradable nanoparticles composed entirely of safe materials that rapidly penetrate human mucus. Angew. Chem. Int. Ed. Engl. 2011, 50, 2597–2600. [Google Scholar] [CrossRef]
- Bansil, R.; Turner, B.S. The biology of mucus: Composition, synthesis and organization. Adv. Drug Deliv. Rev. 2018, 124, 3–15. [Google Scholar] [CrossRef]
- Hsieh, C.L.; Spindler, S.; Ehrig, J.; Sandoghdar, V. Tracking single particles on supported lipid membranes: Multimobility diffusion and nanoscopic confinement. J. Phys. Chem. B 2014, 118, 1545–1554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster, B.S.; Suk, J.S.; Woodworth, G.F.; Hanes, J. Nanoparticle diffusion in respiratory mucus from humans without lung disease. Biomaterials 2013, 34, 3439–3446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.-Y.; Nunn, K.L.; Harit, D.; McKinley, S.A.; Lai, S.K. Minimizing biases associated with tracking analysis of submicron particles in heterogeneous biological fluids. J. Control. Release 2015, 220, 37–43. [Google Scholar] [CrossRef] [Green Version]
- Lieleg, O.; Vladescu, I.; Ribbeck, K. Characterization of particle translocation through mucin hydrogels. Biophys. J. 2010, 98, 1782–1789. [Google Scholar] [CrossRef] [Green Version]
- Lai, S.K.; O’Hanlon, D.E.; Harrold, S.; Man, S.T.; Wang, Y.-Y.; Cone, R.; Hanes, J. Rapid transport of large polymeric nanoparticles in fresh undiluted human mucus. Proc. Natl. Acad. Sci. USA 2007, 104, 1482–1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cobarrubia, A.; Tall, J.; Crispin-Smith, A.; Luque, A. Unifying framework for the diffusion of microscopic particles in mucus. bioRxiv 2020. [Google Scholar] [CrossRef]
- Huckaby, J.T.; Lai, S.K. PEGylation for enhancing nanoparticle diffusion in mucus. Adv. Drug Deliv. Rev. 2018, 124, 125–139. [Google Scholar] [CrossRef]
- Chen, P.; Yue, H.; Zhai, X.; Huang, Z.; Ma, G.-H.; Wei, W.; Yan, L.-T. Transport of a graphene nanosheet sandwiched inside cell membranes. Sci. Adv. 2019, 5, eaaw3192. [Google Scholar] [CrossRef] [Green Version]
| NPs | Coating | Hydrodynamic Diameter (nm) | PdI | Zeta Potential (mV) |
|---|---|---|---|---|
| 200 nm | – | 225 ± 2 | 0.019 ± 0.011 | −51.4 ± 1.8 |
| 100 nm | – | 120 ± 2 | 0.102 ± 0.064 | −42.6 ± 0.2 |
| 100 nm | Poloxamer 407 | 127 ± 4 | 0.036 ± 0.023 | −4.6 ± 0.8 |
| Coating | Mucin Content (w/w%) | Dw/Deff | α |
|---|---|---|---|
| – | 3% | 6.0 | 0.91 |
| – | 5% | 250.1 | 0.39 |
| Poloxamer 407 | 3% | 1.7 | 0.98 |
| Poloxamer 407 | 5% | 4.9 | 0.88 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gabriel, L.; Almeida, H.; Avelar, M.; Sarmento, B.; das Neves, J. MPTHub: An Open-Source Software for Characterizing the Transport of Particles in Biorelevant Media. Nanomaterials 2022, 12, 1899. https://doi.org/10.3390/nano12111899
Gabriel L, Almeida H, Avelar M, Sarmento B, das Neves J. MPTHub: An Open-Source Software for Characterizing the Transport of Particles in Biorelevant Media. Nanomaterials. 2022; 12(11):1899. https://doi.org/10.3390/nano12111899
Chicago/Turabian StyleGabriel, Leandro, Helena Almeida, Marta Avelar, Bruno Sarmento, and José das Neves. 2022. "MPTHub: An Open-Source Software for Characterizing the Transport of Particles in Biorelevant Media" Nanomaterials 12, no. 11: 1899. https://doi.org/10.3390/nano12111899
APA StyleGabriel, L., Almeida, H., Avelar, M., Sarmento, B., & das Neves, J. (2022). MPTHub: An Open-Source Software for Characterizing the Transport of Particles in Biorelevant Media. Nanomaterials, 12(11), 1899. https://doi.org/10.3390/nano12111899