PAMAM-Functionalized Cellulose Nanocrystals with Needle-Like Morphology for Effective Cancer Treatment
Abstract
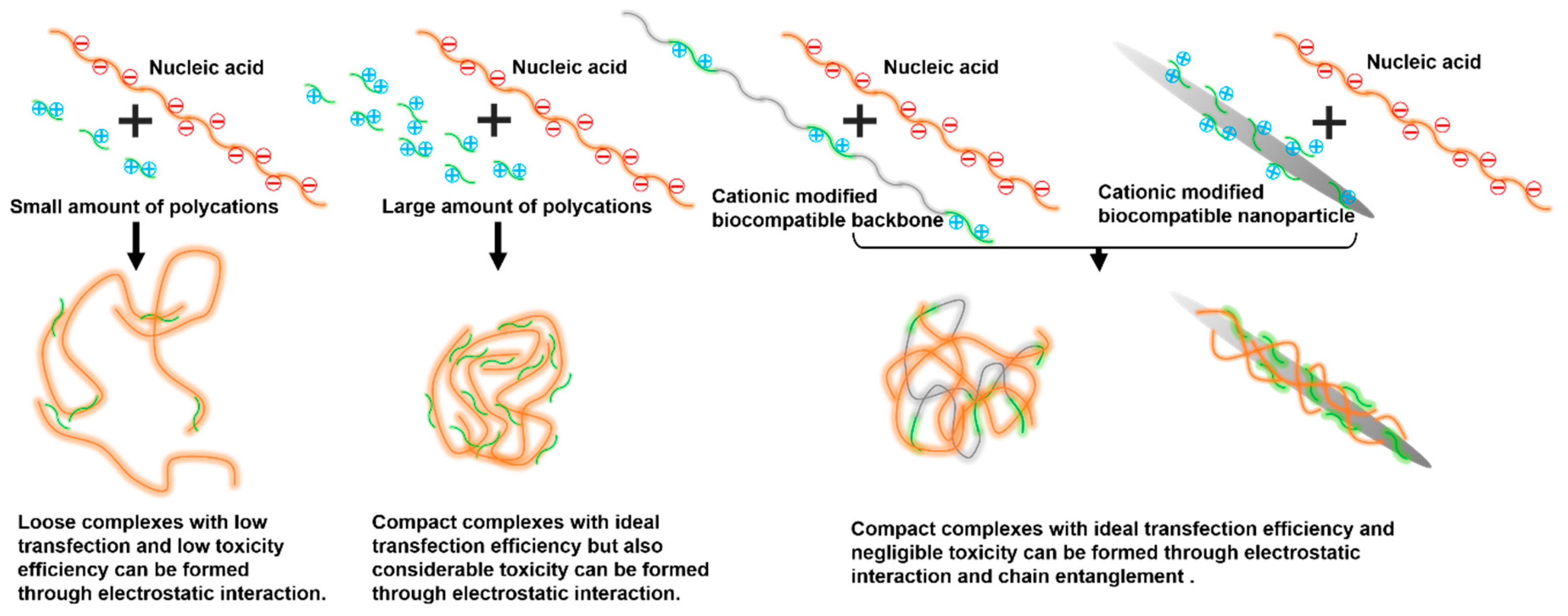
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Cellulose Nanocrystals (CNCs)
2.3. Preparation of PAMAM Dendrimer
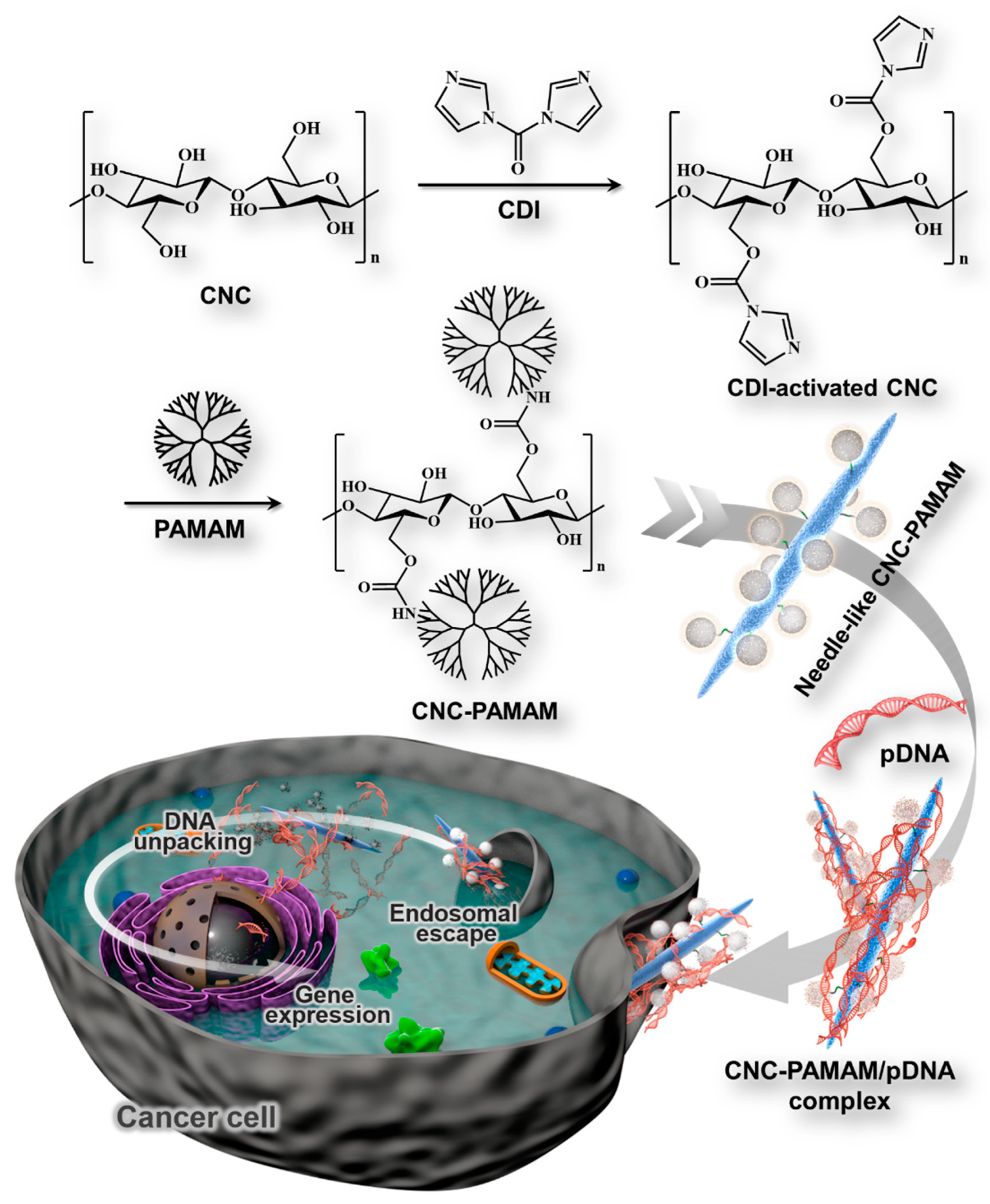
2.4. Synthesis of CNC-graft-PAMAM
2.5. Polymer Characterization
2.6. Preparation of CNC-PAMAM/pDNA Complexes
2.7. Characterization of CNC-PAMAM/pDNA Complexes
2.8. In Vitro Cytotoxicity Assay
2.9. In Vitro Transfection Assay
2.10. In Vitro Antitumor Activity
2.11. In Vivo Antitumor Activity
2.12. Statistical Analysis
3. Results and Discussion
3.1. Synthesis of the PAMAM Dendrimer
3.2. Preparation and Characterization of CNC-PAMAM
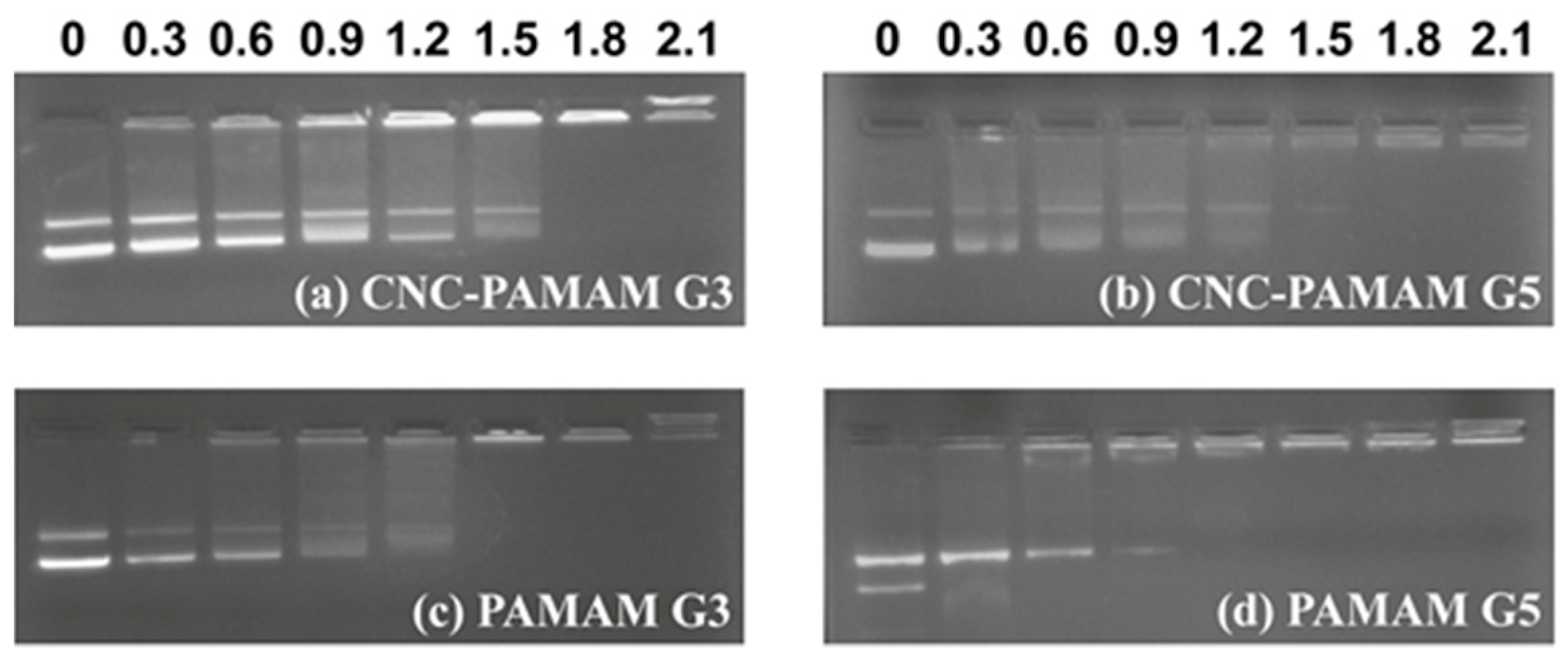
3.3. Characterization of CNC-PAMAM/pDNA Complexes
3.4. Cell Viability Assay
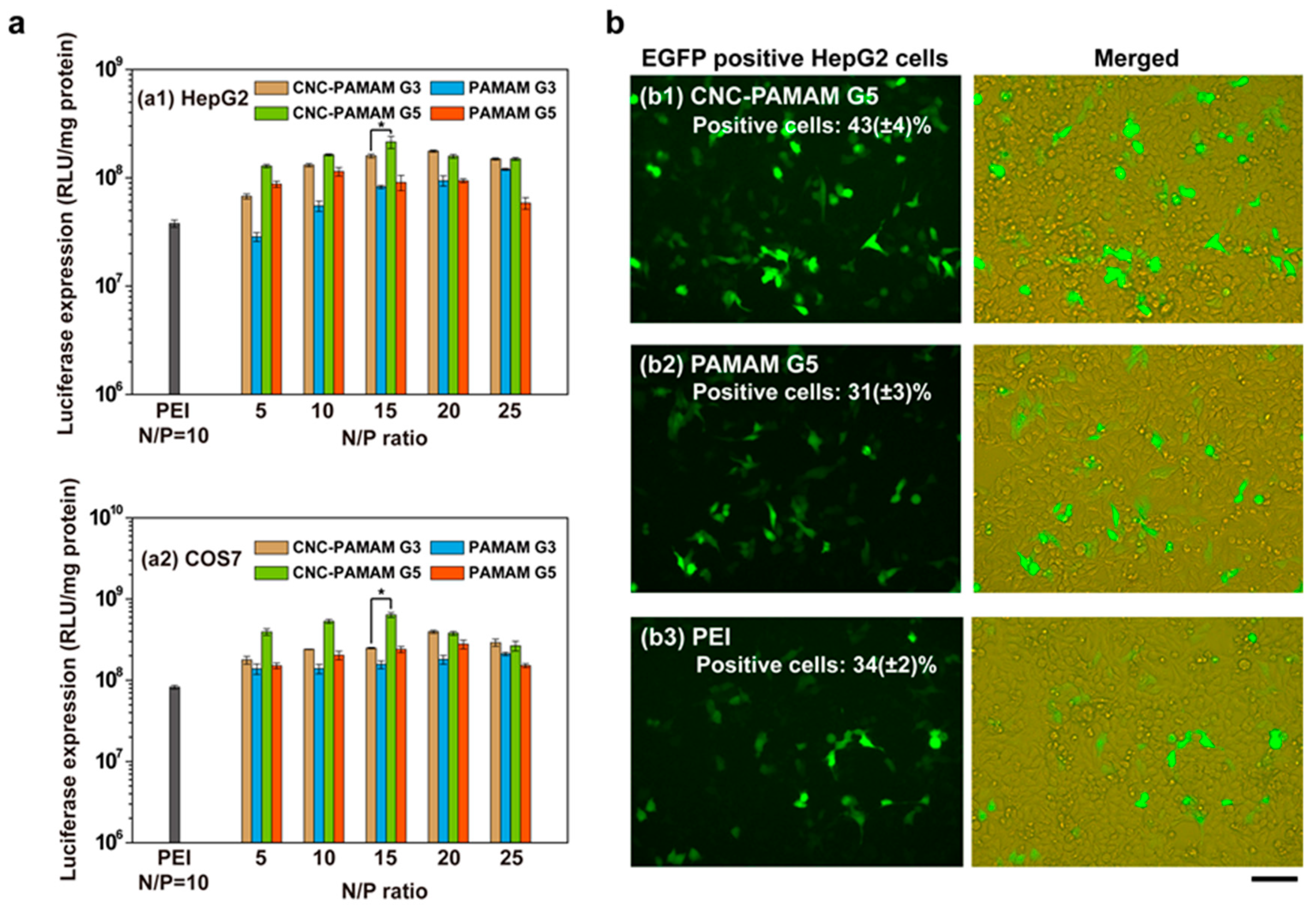
3.5. In Vitro Gene Transfection Assay
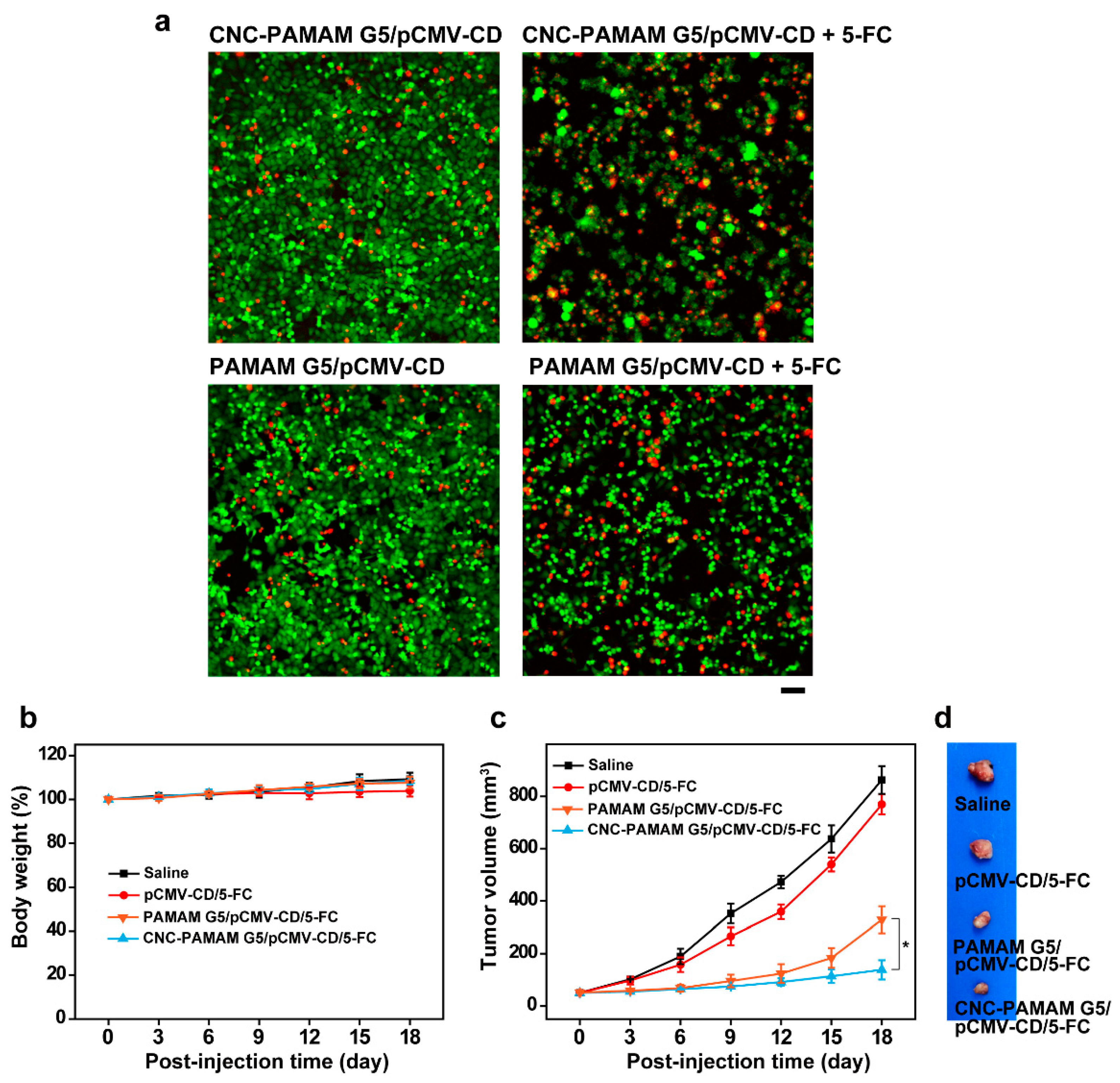
3.6. Antitumor Effects with a CD/5-FC Suicide Gene System
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Garafalo, A.V.; Cideciyan, A.V.; Heon, E.; Sheplock, R.; Pearson, A.; Yu, C.W.; Sumaroka, A.; Aguirre, G.D.; Jacobson, S.G. Progress in treating inherited retinal diseases: Early subretinal gene therapy clinical trials and candidates for future initiatives. Prog. Retin. Eye Res. 2020, 77, 100827. [Google Scholar] [CrossRef]
- Belen Cerda, M.; Batalla, M.; Anton, M.; Cafferata, E.; Podhajcer, O.; Plank, C.; Mykhaylyk, O.; Policastro, L. Enhancement of nucleic acid delivery to hard-to-transfect human colorectal cancer cells by magnetofection at laminin coated substrates and promotion of the endosomal/lysosomal escape. RSC Adv. 2015, 5, 58345–58354. [Google Scholar] [CrossRef]
- Sun, Y.; Ma, X.; Hu, H. Application of nano-drug delivery system based on cascade technology in cancer treatment. Int. J. Mol. Sci. 2021, 22, 5698. [Google Scholar] [CrossRef]
- Ibraheem, D.; Elaissari, A.; Fessi, H. Gene therapy and DNA delivery systems. Int. J. Pharm. 2014, 459, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Vu, M.N.; Kelly, H.G.; Wheatley, A.K.; Peng, S.; Pilkington, E.H.; Veldhuis, N.A.; Davis, T.P.; Kent, S.J.; Truong, N.P. Cellular interactions of liposomes and PISA nanoparticles during human blood flow in a microvascular network. Small 2020, 16, 2002861. [Google Scholar] [CrossRef]
- Nitta, S.K.; Numata, K. Biopolymer-based nanoparticles for drug/gene delivery and tissue engineering. Int. J. Mol. Sci. 2013, 14, 1629–1654. [Google Scholar] [CrossRef] [Green Version]
- Meng, Q.Y.; Hu, H.; Zhou, L.P.; Zhang, Y.X.; Yu, B.; Shen, Y.Q.; Cong, H.L. Logical design and application of prodrug platforms. Polym. Chem. 2019, 10, 306–324. [Google Scholar] [CrossRef]
- Truong, N.P.; Gu, W.; Prasadam, I.; Jia, Z.; Crawford, R.; Xiao, Y.; Monteiro, M.J. An influenza virus-inspired polymer system for the timed release of siRNA. Nat. Commun. 2013, 4, 1902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, S.; Gao, Y.G.; Lin, X.; Li, Y.; Dang, K.; Tian, Y.; Zhang, W.J.; Jiang, S.F.; Qadir, A.; Qian, A.R. The development of functional non-viral vectors for gene delivery. Int. J. Mol. Sci. 2019, 20, 5491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, H.; Yuan, W.; Liu, F.S.; Cheng, G.; Xu, F.J.; Ma, J. Redox-responsive polycation-functionalized cotton cellulose nanocrystals for effective cancer treatment. ACS Appl. Mater. Interfaces 2015, 7, 8942–8951. [Google Scholar] [CrossRef]
- Zukancic, D.; Suys, E.J.; Pilkington, E.H.; Algarni, A.; Al-Wassiti, H.; Truong, N.P. The importance of poly(ethylene glycol) and lipid structure in targeted gene delivery to lymph nodes by lipid nanoparticles. Pharmaceutics 2020, 12, 1068. [Google Scholar] [CrossRef]
- Bravo-Anaya, L.M.; Rosselgong, J.; Fernandez-Solis, K.G.; Xiao, Y.; Vax, A.; Ibarboure, E.; Ruban, A.; Lebleu, C.; Joucla, G.; Garbay, B.; et al. Coupling of RAFT polymerization and chemoselective post-modifications of elastin-like polypeptides for the synthesis of gene delivery hybrid vectors. Polym. Chem. 2021, 12, 226–241. [Google Scholar] [CrossRef]
- Li, J.; Liang, H.M.; Liu, J.; Wang, Z.Y. Poly (amidoamine) (PAMAM) dendrimer mediated delivery of drug and pDNA/siRNA for cancer therapy. Int. J. Pharm. 2018, 546, 215–225. [Google Scholar] [CrossRef]
- Abedi-Gaballu, F.; Dehghan, G.; Ghaffari, M.; Yekta, R.; Abbaspour-Ravasjani, S.; Baradaran, B.; Dolatabadi, J.E.N.; Hamblin, M.R. PAMAM dendrimers as efficient drug and gene delivery nanosystems for cancer therapy. Appl. Mater. Today 2018, 12, 177–190. [Google Scholar] [CrossRef]
- Luong, D.; Kesharwani, P.; Deshmukh, R.; Amin, M.; Gupta, U.; Greish, K.; Iyer, A.K. PEGylated PAMAM dendrimers: Enhancing efficacy and mitigating toxicity for effective anticancer drug and gene delivery. Acta Biomater. 2016, 43, 14–29. [Google Scholar] [CrossRef]
- Hu, H.; Xu, F.J. Rational design and latest advances of polysaccharide-based hydrogels for wound healing. Biomater. Sci. 2020, 8, 2084–2101. [Google Scholar] [CrossRef]
- Sun, Y.; Jing, X.; Ma, X.; Feng, Y.; Hu, H. Versatile types of polysaccharide-based drug delivery systems: From strategic design to cancer therapy. Int. J. Mol. Sci. 2020, 21, 9159. [Google Scholar] [CrossRef]
- Jing, X.; Sun, Y.D.; Ma, X.; Hu, H. Marine polysaccharides: Green and recyclable resources as wound dressings. Mater. Chem. Front. 2021. [Google Scholar] [CrossRef]
- Habibi, Y.; Lucia, L.A.; Rojas, O.J. Cellulose nanocrystals: Chemistry, self-assembly, and applications. Chem. Rev. 2010, 110, 3479–3500. [Google Scholar] [CrossRef]
- Huang, X.; Teng, X.; Chen, D.; Tang, F.; He, J. The effect of the shape of mesoporous silica nanoparticles on cellular uptake and cell function. Biomaterials 2010, 31, 438–448. [Google Scholar] [CrossRef]
- Hu, Y.; Li, Y.; Xu, F.J. Versatile functionalization of polysaccharides via polymer grafts: From design to biomedical applications. Acc. Chem. Res. 2017, 50, 281–292. [Google Scholar] [CrossRef]
- Cong, H.L.; Zhou, L.P.; Meng, Q.Y.; Zhang, Y.X.; Yu, B.; Shen, Y.Q.; Hu, H. Preparation and evaluation of PAMAM dendrimer-based polymer gels physically cross-linked by hydrogen bonding. Biomater. Sci. 2019, 7, 3918–3925. [Google Scholar] [CrossRef]
- Hu, H.; Hou, X.J.; Wang, X.C.; Nie, J.J.; Cai, Q.; Xu, F.J. Gold nanoparticle-conjugated heterogeneous polymer brush-wrapped cellulose nanocrystals prepared by combining different controllable polymerization techniques for theranostic applications. Polym. Chem. 2016, 7, 3107–3116. [Google Scholar] [CrossRef]
- Wang, Z.H.; Li, W.B.; Ma, J.; Tang, G.P.; Yang, W.T.; Xu, F.J. Functionalized nonionic dextran backbones by atom transfer radical polymerization for efficient gene delivery. Macromolecules 2011, 44, 230–239. [Google Scholar] [CrossRef]
- Xu, F.J.; Ping, Y.; Ma, J.; Tang, G.P.; Yang, W.T.; Li, J.; Kang, E.T.; Neoh, K.G. Comb-sbaped copolymers composed of hydroxypropyl cellulose backbones and cationic poly((2-dimethyl amino)ethyl methacrylate) side chains for gene delivery. Bioconjug. Chem. 2009, 20, 1449–1458. [Google Scholar] [CrossRef]
- Zhou, Z.X.; Liu, X.R.; Zhu, D.C.; Wang, Y.; Zhang, Z.; Zhou, X.F.; Qiu, N.S.; Chen, X.S.; Shen, Y.Q. Nonviral cancer gene therapy: Delivery cascade and vector nanoproperty integration. Adv. Drug Deliv. Rev. 2017, 115, 115–154. [Google Scholar] [CrossRef]
- Sun, Y.; Hu, H.; Jing, X.; Meng, Q.; Yu, B.; Cong, H.; Shen, Y. Co-delivery of chemotherapeutic drugs and cell cycle regulatory agents using nanocarriers for cancer therapy. Sci. China Mater. 2021. [Google Scholar] [CrossRef]
- Kopec, W.; Zak, A.; Jamroz, D.; Nakahata, R.; Yusa, S.I.; Gapsys, V.; Kepczynski, M. Polycation-anionic lipid membrane interactions. Langmuir 2020, 36, 12435–12450. [Google Scholar] [CrossRef]
- Zhang, L.M.; Lu, Z.X.; Zhao, Q.H.; Huang, J.; Shen, H.; Zhang, Z.J. Enhanced chemotherapy efficacy by sequential delivery of siRNA and anticancer drugs using PEI-grafted graphene oxide. Small 2011, 7, 460–464. [Google Scholar] [CrossRef]
- Liu, C.J.; Zhang, P.; Zhai, X.Y.; Tian, F.; Li, W.C.; Yang, J.H.; Liu, Y.; Wang, H.B.; Wang, W.; Liu, W.G. Nano-carrier for gene delivery and bioimaging based on carbon dots with PEI-passivation enhanced fluorescence. Biomaterials 2012, 33, 3604–3613. [Google Scholar] [CrossRef]
- Mahmoud, K.A.; Mena, J.A.; Male, K.B.; Hrapovic, S.; Kamen, A.; Luong, J.H.T. Effect of surface charge on the cellular uptake and cytotoxicity of fluorescent labeled cellulose nanocrystals. ACS Appl. Mater. Interfaces 2010, 2, 2924–2932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van de Wetering, P.; Moret, E.E.; Schuurmans-Nieuwenbroek, N.M.; van Steenbergen, M.J.; Hennink, W.E. Structure-activity relationships of water-soluble cationic methacrylate/methacrylamide polymers for nonviral gene delivery. Bioconjug. Chem. 1999, 10, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Ping, Y.A.; Liu, C.D.; Tang, G.P.; Li, J.S.; Li, J.; Yang, W.T.; Xu, F.J. Functionalization of chitosan via atom transfer radical polymerization for gene delivery. Adv. Funct. Mater. 2010, 20, 3106–3116. [Google Scholar] [CrossRef]
- Lin, X.; Zhao, N.; Yan, P.; Hu, H.; Xu, F.J. The shape and size effects of polycation functionalized silica nanoparticles on gene transfection. Acta Biomater. 2015, 11, 381–392. [Google Scholar] [CrossRef]
- Liang, B.; He, M.L.; Chan, C.Y.; Chen, Y.C.; Li, X.P.; Li, Y.; Zheng, D.X.; Lin, M.C.; Kung, H.F.; Shuai, X.T.; et al. The use of folate-PEG-grafted-hybranched-PEI nonviral vector for the inhibition of glioma growth in the rat. Biomaterials 2009, 30, 4014–4020. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Ma, X.; Jing, X.; Hu, H. PAMAM-Functionalized Cellulose Nanocrystals with Needle-Like Morphology for Effective Cancer Treatment. Nanomaterials 2021, 11, 1640. https://doi.org/10.3390/nano11071640
Sun Y, Ma X, Jing X, Hu H. PAMAM-Functionalized Cellulose Nanocrystals with Needle-Like Morphology for Effective Cancer Treatment. Nanomaterials. 2021; 11(7):1640. https://doi.org/10.3390/nano11071640
Chicago/Turabian StyleSun, Yanzhen, Xiaoli Ma, Xiaodong Jing, and Hao Hu. 2021. "PAMAM-Functionalized Cellulose Nanocrystals with Needle-Like Morphology for Effective Cancer Treatment" Nanomaterials 11, no. 7: 1640. https://doi.org/10.3390/nano11071640
APA StyleSun, Y., Ma, X., Jing, X., & Hu, H. (2021). PAMAM-Functionalized Cellulose Nanocrystals with Needle-Like Morphology for Effective Cancer Treatment. Nanomaterials, 11(7), 1640. https://doi.org/10.3390/nano11071640






