From Residues to Added-Value Bacterial Biopolymers as Nanomaterials for Biomedical Applications
Abstract
1. Introduction
2. BC, PHAs, and PGA as Model Bacterial Biopolymers Produced by Upcycling Bioprocesses
2.1. Molecular Basis of the Biopolymer Synthesis
2.2. Main Properties of Natural Model Biopolymers
3. Sustainable Production of Bacterial Polymers
3.1. General Aspects of the Bioprocess
3.2. Bacterial Biopolymer Production from Renewable Sources
4. Major Biomedical Applications of Model Bacterial Biopolymers
4.1. Drug Delivery Systems
4.2. Tissue Engineering
4.3. Vascular Grafts, Cardiac Valves, and Vessel Stents
4.4. Wound Healing
4.5. Sutures and Biological Glues
5. Increasing Bacterial Polymers’ Structural Diversity by In Vivo and In Vitro Functionalization
5.1. In Vivo Functionalization Approaches
5.1.1. Metabolic Engineering
5.1.2. In Situ Modifications
5.2. In Vitro Functionalization Approaches
5.2.1. Peptide Affinity-Based Functionalization
5.2.2. Chemical Modifications
6. Regulatory Aspects and Transferability into Clinics
7. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, A.; Twardowski, T.; Wohlgemuth, R. Bioeconomy for Sustainable Development. Biotechnol. J. 2019, 14, 1800638. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.; Yadav, H.; Shah, V.G.; Shah, G.; Dhaka, G. Biomedical Biopolymers, Their Origin and Evolution in Biomedical Sciences: A Systematic Review. J. Clin. Diagn. Res. JCDR 2015, 9, ZE21–ZE25. [Google Scholar] [CrossRef]
- Dinjaski, N.; Kaplan, D.L. Recombinant Protein Blends: Silk beyond Natural Design. Curr. Opin. Biotechnol. 2016, 39, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Moradali, M.F.; Rehm, B.H.A. Bacterial Biopolymers: From Pathogenesis to Advanced Materials. Nat. Rev. Microbiol. 2020, 18, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Yau, A.; Lee, J.; Chen, Y. Nanomaterials for Protein Delivery in Anticancer Applications. Pharmaceutics 2021, 13, 155. [Google Scholar] [CrossRef] [PubMed]
- Grigoletto, A.; Tedeschini, T.; Canato, E.; Pasut, G. The Evolution of Polymer Conjugation and Drug Targeting for the Delivery of Proteins and Bioactive Molecules. WIREs Nanomed. Nanobiotechnol. 2020, e1689. [Google Scholar] [CrossRef]
- Kim, S.; Jung, U.T.; Kim, S.-K.; Lee, J.-H.; Choi, H.S.; Kim, C.-S.; Jeong, M.Y. Nanostructured Multifunctional Surface with Antireflective and Antimicrobial Characteristics. ACS Appl. Mater. Interfaces 2015, 7, 326–331. [Google Scholar] [CrossRef]
- Qiu, H.; Si, Z.; Luo, Y.; Feng, P.; Wu, X.; Hou, W.; Zhu, Y.; Chan-Park, M.B.; Xu, L.; Huang, D. The Mechanisms and the Applications of Antibacterial Polymers in Surface Modification on Medical Devices. Front. Bioeng. Biotechnol. 2020, 8, 910. [Google Scholar] [CrossRef]
- Elena, P.; Miri, K. Formation of Contact Active Antimicrobial Surfaces by Covalent Grafting of Quaternary Ammonium Compounds. Colloids Surf. B Biointerfaces 2018, 169, 195–205. [Google Scholar] [CrossRef]
- Blackman, L.D.; Qu, Y.; Cass, P.; Locock, K.E.S. Approaches for the Inhibition and Elimination of Microbial Biofilms Using Macromolecular Agents. Chem. Soc. Rev. 2021, 50, 1587–1616. [Google Scholar] [CrossRef]
- Tran, H.M.; Tran, H.; Booth, M.A.; Fox, K.E.; Nguyen, T.H.; Tran, N.; Tran, P.A. Nanomaterials for Treating Bacterial Biofilms on Implantable Medical Devices. Nanomaterials 2020, 10, 2253. [Google Scholar] [CrossRef] [PubMed]
- Sulaeva, I.; Henniges, U.; Rosenau, T.; Potthast, A. Bacterial Cellulose as a Material for Wound Treatment: Properties and Modifications. A Review. Biotechnol. Adv. 2015, 33, 1547–1571. [Google Scholar] [CrossRef]
- Castro, C.; Cleenwerck, I.; Trček, J.; Zuluaga, R.; De Vos, P.; Caro, G.; Aguirre, R.; Putaux, J.-L.; Gañán, P. Gluconacetobacter medellinensis Sp. Nov., Cellulose- and Non-Cellulose-Producing Acetic Acid Bacteria Isolated from Vinegar. Int. J. Syst. Evol. Microbiol. 2013, 63, 1119–1125. [Google Scholar] [CrossRef]
- Morikawa, M.; Kagihiro, S.; Haruki, M.; Takano, K.; Branda, S.; Kolter, R.; Kanaya, S. Biofilm Formation by a Bacillus Subtilis Strain That Produces γ-Polyglutamate. Microbiology 2006, 152, 2801–2807. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhu, C.; Yang, J.; Nie, Y.; Chen, C.; Sun, D. Recent Advances in Bacterial Cellulose. Cellulose 2014, 21, 1–30. [Google Scholar] [CrossRef]
- Yamada, Y.; Yukphan, P.; Lan Vu, H.T.; Muramatsu, Y.; Ochaikul, D.; Tanasupawat, S.; Nakagawa, Y. Description of Komagataeibacter Gen. Nov., with Proposals of New Combinations (Acetobacteraceae). J. Gen. Appl. Microbiol. 2012, 58, 397–404. [Google Scholar] [CrossRef]
- Romling, U.; Galperin, M.Y. Bacterial Cellulose Biosynthesis: Diversity of Operons, Subunits, Products, and Functions. Trends Microbiol. 2015, 23, 545–557. [Google Scholar] [CrossRef]
- Jacek, P.; Dourado, F.; Gama, M.; Bielecki, S. Molecular Aspects of Bacterial Nanocellulose Biosynthesis. Microb. Biotechnol. 2019, 12, 633–649. [Google Scholar] [CrossRef]
- Ross, P.; Weinhouse, H.; Aloni, Y.; Michaeli, D.; Weinberger-Ohana, P.; Mayer, R.; Braun, S.; de Vroom, E.; van der Marel, G.A.; van Boom, J.H.; et al. Regulation of Cellulose Synthesis in Acetobacter xylinum by Cyclic Diguanylic Acid. Nature 1987, 325, 279–281. [Google Scholar] [CrossRef]
- Hernández-Arriaga, A.M.; Del Cerro, C.; Urbina, L.; Eceiza, A.; Corcuera, M.A.; Retegi, A.; Auxiliadora Prieto, M. Genome Sequence and Characterization of the Bcs Clusters for the Production of Nanocellulose from the Low PH Resistant Strain Komagataeibacter medellinensis ID13488. Microb. Biotechnol. 2019, 12, 620–632. [Google Scholar] [CrossRef]
- Jedrzejczak-Krzepkowska, M.; Kubiak, K.; Ludwicka, K.; Bielecki, S. Chapter 2—Bacterial NanoCellulose Synthesis, Recent Findings. In Bacterial Nanocellulose; Gama, M., Dourado, F., Bielecki, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 19–46. ISBN 978-0-444-63458-0. [Google Scholar]
- Ryngajłło, M.; Kubiak, K.; Jędrzejczak-Krzepkowska, M.; Jacek, P.; Bielecki, S. Comparative Genomics of the Komagataeibacter Strains-Efficient Bionanocellulose Producers. Microbiologyopen 2019, 8, e00731. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Bedrán, H.; López-Isunza, F. The Unified Metabolism of Gluconacetobacter entanii in Continuous and Batch Processes. Process. Biochem. 2007, 42, 1180–1190. [Google Scholar] [CrossRef]
- Jang, W.D.; Kim, T.Y.; Kim, H.U.; Shim, W.Y.; Ryu, J.Y.; Park, J.H.; Lee, S.Y. Genomic and Metabolic Analysis of Komagataeibacter xylinus DSM 2325 Producing Bacterial Cellulose Nanofiber. Biotechnol. Bioeng. 2019. [Google Scholar] [CrossRef]
- Gao, L.; Wu, X.; Zhu, C.; Jin, Z.; Wang, W.; Xia, X. Metabolic Engineering to Improve the Biomanufacturing Efficiency of Acetic Acid Bacteria: Advances and Prospects. Crit. Rev. Biotechnol. 2020, 40, 522–538. [Google Scholar] [CrossRef]
- Ryngajłło, M.; Jacek, P.; Cielecka, I.; Kalinowska, H.; Bielecki, S. Effect of Ethanol Supplementation on the Transcriptional Landscape of Bionanocellulose Producer Komagataeibacter xylinus E25. Appl. Microbiol. Biotechnol. 2019, 103, 6673–6688. [Google Scholar] [CrossRef]
- Mezzina, M.P.; Pettinari, M.J. Phasins, Multifaceted Polyhydroxyalkanoate Granule-Associated Proteins. Appl. Environ. Microbiol. 2016, 82, 5060–5067. [Google Scholar] [CrossRef]
- Prieto, A.; Escapa, I.F.; Martinez, V.; Dinjaski, N.; Herencias, C.; de la Pena, F.; Tarazona, N.; Revelles, O. A Holistic View of Polyhydroxyalkanoate Metabolism in Pseudomonas putida. Environ. Microbiol. 2016, 18, 341–357. [Google Scholar] [CrossRef]
- Kniewel, R.; Lopez, O.R.; Prieto, M.A. Biogenesis of Medium-Chain-Length Polyhydroxyalkanoates. In Biogenesis of Fatty Acids, Lipids and Membranes; Geiger, O., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 457–481. ISBN 978-3-319-50430-8. [Google Scholar]
- Mezzina, M.P.; Manoli, M.T.; Prieto, M.A.; Nikel, P.I. Engineering Native and Synthetic Pathways in Pseudomonas putida for the Production of Tailored Polyhydroxyalkanoates. Biotechnol. J. 2020, 2000165. [Google Scholar] [CrossRef]
- Wang, Q.; Tappel, R.C.; Zhu, C.; Nomura, C.T. Development of a New Strategy for Production of Medium-Chain-Length Polyhydroxyalkanoates by Recombinant Escherichia coli via Inexpensive Non-Fatty Acid Feedstocks. Appl. Environ. Microbiol. 2012, 78, 519–527. [Google Scholar] [CrossRef]
- de Eugenio, L.I.; Escapa, I.F.; Morales, V.; Dinjaski, N.; Galan, B.; Garcia, J.L.; Prieto, M.A. The Turnover of Medium-Chain-Length Polyhydroxyalkanoates in Pseudomonas putida KT2442 and the Fundamental Role of PhaZ Depolymerase for the Metabolic Balance. Environ. Microbiol. 2010, 12, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Feng, J.; Sirisansaneeyakul, S.; Song, C.; Chisti, Y. Genetic and Metabolic Engineering for Microbial Production of Poly-γ-Glutamic Acid. Biotechnol. Adv. 2018, 36, 1424–1433. [Google Scholar] [CrossRef] [PubMed]
- Ashiuchi, M. Microbial Production and Chemical Transformation of Poly-γ-Glutamate. Microb. Biotechnol. 2013, 6, 664–674. [Google Scholar] [CrossRef] [PubMed]
- Ogunleye, A.; Bhat, A.; Irorere, V.U.; Hill, D.; Williams, C.; Radecka, I. Poly-γ-Glutamic Acid: Production, Properties and Applications. Microbiology 2015, 161, 1–17. [Google Scholar] [CrossRef]
- Scoffone, V.; Dondi, D.; Biino, G.; Borghese, G.; Pasini, D.; Galizzi, A.; Calvio, C. Knockout of PgdS and Ggt Genes Improves γ-PGA Yield in B. Subtilis. Biotechnol. Bioeng. 2013, 110, 2006–2012. [Google Scholar] [CrossRef]
- Feng, J.; Quan, Y.; Gu, Y.; Liu, F.; Huang, X.; Shen, H.; Dang, Y.; Cao, M.; Gao, W.; Lu, X.; et al. Enhancing Poly-γ-Glutamic Acid Production in Bacillus amyloliquefaciens by Introducing the Glutamate Synthesis Features from Corynebacterium glutamicum. Microb. Cell Fact. 2017, 16, 88. [Google Scholar] [CrossRef]
- Cao, M.; Geng, W.; Zhang, W.; Sun, J.; Wang, S.; Feng, J.; Zheng, P.; Jiang, A.; Song, C. Engineering of Recombinant Escherichia coli Cells Co-Expressing Poly-γ-Glutamic Acid (γ-PGA) Synthetase and Glutamate Racemase for Differential Yielding of γ-PGA. Microb. Biotechnol. 2013, 6, 675–684. [Google Scholar] [CrossRef]
- Xu, G.; Zha, J.; Cheng, H.; Ibrahim, M.H.A.; Yang, F.; Dalton, H.; Cao, R.; Zhu, Y.; Fang, J.; Chi, K.; et al. Engineering Corynebacterium glutamicum for the de novo Biosynthesis of Tailored Poly-γ-Glutamic Acid. Metab Eng. 2019, 56, 39–49. [Google Scholar] [CrossRef]
- Urushibata, Y.; Tokuyama, S.; Tahara, Y. Characterization of the Bacillus Subtilis YwsC Gene, Involved in Gamma-Polyglutamic Acid Production. J. Bacteriol. 2002, 184, 337–343. [Google Scholar] [CrossRef]
- Ohsawa, T.; Tsukahara, K.; Ogura, M. Bacillus subtilis Response Regulator DegU Is a Direct Activator of PgsB Transcription Involved in γ-Poly-Glutamic Acid Synthesis. Biosci. Biotechnol. Biochem. 2009, 73, 2096–2102. [Google Scholar] [CrossRef]
- Feng, J.; Gu, Y.; Quan, Y.; Cao, M.; Gao, W.; Zhang, W.; Wang, S.; Yang, C.; Song, C. Improved Poly-γ-Glutamic Acid Production in Bacillus amyloliquefaciens by Modular Pathway Engineering. Metab Eng. 2015, 32, 106–115. [Google Scholar] [CrossRef]
- Yadav, V.; Paniliatis, B.J.; Shi, H.; Lee, K.; Cebe, P.; Kaplan, D.L. Novel in Vivo-Degradable Cellulose-Chitin Copolymer from Metabolically Engineered Gluconacetobacter xylinus. Appl. Environ. Microbiol. 2010, 76, 6257–6265. [Google Scholar] [CrossRef]
- Shrivastav, A.; Kim, H.-Y.; Kim, Y.-R. Advances in the Applications of Polyhydroxyalkanoate Nanoparticles for Novel Drug Delivery System. BioMed Res. Int. 2013, 2013, 581684. [Google Scholar] [CrossRef]
- Murakami, S.; Aoki, N.; Matsumura, S. Bio-Based Biodegradable Hydrogels Prepared by Crosslinking of Microbial Poly(γ-Glutamic Acid) with L-Lysine in Aqueous Solution. Polym. J. 2011, 43, 414–420. [Google Scholar] [CrossRef]
- Fan, K.; Gonzales, D.; Sevoian, M. Hydrolytic and Enzymatic Degradation of Poly(γ-Glutamic Acid) Hydrogels and Their Application in Slow-Release Systems for Proteins. J. Environ. Polym. Degrad. 1996, 4, 253–260. [Google Scholar] [CrossRef]
- Xu, T.; Zhan, S.; Yi, M.; Chi, B.; Xu, H.; Mao, C. Degradation Performance of Polyglutamic Acid and Its Application of Calcium Supplement. Polym. Adv. Technol. 2018, 29, 1966–1973. [Google Scholar] [CrossRef]
- Kumbhar, J.V.; Jadhav, S.H.; Bodas, D.S.; Barhanpurkar-Naik, A.; Wani, M.R.; Paknikar, K.M.; Rajwade, J.M. In Vitro and in Vivo Studies of a Novel Bacterial Cellulose-Based Acellular Bilayer Nanocomposite Scaffold for the Repair of Osteochondral Defects. Int. J. Nanomed. 2017, 12, 6437–6459. [Google Scholar] [CrossRef]
- Singh, A.K.; Srivastava, J.K.; Chandel, A.K.; Sharma, L.; Mallick, N.; Singh, S.P. Biomedical Applications of Microbially Engineered Polyhydroxyalkanoates: An Insight into Recent Advances, Bottlenecks, and Solutions. Appl. Microbiol. Biotechnol. 2019, 103, 2007–2032. [Google Scholar] [CrossRef]
- Karabasz, A.; Szczepanowicz, K.; Cierniak, A.; Bereta, J.; Bzowska, M. In Vitro Toxicity Studies of Biodegradable, Polyelectrolyte Nanocapsules. Int. J. Nanomed. 2018, 13, 5159–5172. [Google Scholar] [CrossRef]
- Karabasz, A.; Szczepanowicz, K.; Cierniak, A.; Mezyk-Kopec, R.; Dyduch, G.; Szczech, M.; Bereta, J.; Bzowska, M. In Vivo Studies on Pharmacokinetics, Toxicity and Immunogenicity of Polyelectrolyte Nanocapsules Functionalized with Two Different Polymers: Poly-L-Glutamic Acid or PEG. Int. J. Nanomed. 2019, 14, 9587–9602. [Google Scholar] [CrossRef]
- Discher, D.E.; Janmey, P.; Wang, Y.L. Tissue Cells Feel and Respond to the Stiffness of Their Substrate. Science 2005, 310, 1139–1143. [Google Scholar] [CrossRef] [PubMed]
- Gorgieva, S.; Trček, J. Bacterial Cellulose: Production, Modification and Perspectives in Biomedical Applications. Nanomaterials 2019, 9, 1352. [Google Scholar] [CrossRef] [PubMed]
- Klemm, D.; Heublein, B.; Fink, H.-P.; Bohn, A. Cellulose: Fascinating Biopolymer and Sustainable Raw Material. Angew. Chem. Int. Ed. 2005, 44, 3358–3393. [Google Scholar] [CrossRef] [PubMed]
- Czaja, W.; Krystynowicz, A.; Bielecki, S.; Brown, R.M. Microbial Cellulose—the Natural Power to Heal Wounds. Biomaterials 2006, 27, 145–151. [Google Scholar] [CrossRef]
- Pogorelova, N.; Rogachev, E.; Digel, I.; Chernigova, S.; Nardin, D. Bacterial Cellulose Nanocomposites: Morphology and Mechanical Properties. Materials 2020, 13, 2849. [Google Scholar] [CrossRef]
- Volova, T.G.; Prudnikova, S.V.; Sukovatyi, A.G.; Shishatskaya, E.I. Production and Properties of Bacterial Cellulose by the Strain Komagataeibacter xylinus B-12068. Appl. Microbiol. Biotechnol. 2018, 102, 7417–7428. [Google Scholar] [CrossRef]
- Chen, S.-Q.; Lopez-Sanchez, P.; Wang, D.; Mikkelsen, D.; Gidley, M.J. Mechanical Properties of Bacterial Cellulose Synthesised by Diverse Strains of the Genus Komagataeibacter. Food Hydrocoll. 2018, 81, 87–95. [Google Scholar] [CrossRef]
- McKenna, B.A.; Mikkelsen, D.; Wehr, J.B.; Gidley, M.J.; Menzies, N.W. Mechanical and Structural Properties of Native and Alkali-Treated Bacterial Cellulose Produced by Gluconacetobacter xylinus Strain ATCC 53524. Cellulose 2009, 16, 1047–1055. [Google Scholar] [CrossRef]
- Machado, R.T.A.; Gutierrez, J.; Tercjak, A.; Trovatti, E.; Uahib, F.G.M.; Moreno, G.P.; Nascimento, A.P.; Berreta, A.A.; Ribeiro, S.J.L.; Barud, H.S. Komagataeibacter rhaeticus as an Alternative Bacteria for Cellulose Production. Carbohydr. Polym. 2016, 152, 841–849. [Google Scholar] [CrossRef]
- Urbina, L.; Hernández-Arriaga, A.M.; Eceiza, A.; Gabilondo, N.; Corcuera, M.A.; Prieto, M.A.; Retegi, A. By-Products of the Cider Production: An Alternative Source of Nutrients to Produce Bacterial Cellulose. Cellulose 2017, 24, 2071–2082. [Google Scholar] [CrossRef]
- Andritsou, V.; de Melo, E.M.; Tsouko, E.; Ladakis, D.; Maragkoudaki, S.; Koutinas, A.A.; Matharu, A.S. Synthesis and Characterization of Bacterial Cellulose from Citrus-Based Sustainable Resources. ACS Omega 2018, 3, 10365–10373. [Google Scholar] [CrossRef]
- Güzel, M.; Akpınar, Ö. Preparation and Characterization of Bacterial Cellulose Produced from Fruit and Vegetable Peels by Komagataeibacter Hansenii GA2016. Int. J. Biol. Macromol. 2020, 162, 1597–1604. [Google Scholar] [CrossRef] [PubMed]
- Rai, R.; Keshavarz, T.; Roether, J.A.; Boccaccini, A.R.; Roy, I. Medium Chain Length Polyhydroxyalkanoates, Promising New Biomedical Materials for the Future. Mater. Sci. Eng. R Rep. 2011, 72, 29–47. [Google Scholar] [CrossRef]
- Tortajada, M.; da Silva, L.F.; Prieto, M.A. Second-Generation Functionalized Medium-Chain-Length Polyhydroxyalkanoates: The Gateway to High-Value Bioplastic Applications. Int. Microbiol. 2013, 16, 1–15. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, G.; Zhou, X.R.; Chen, G.-Q. Biosynthesis of Poly(3-Hydroxydecanoate) and 3-Hydroxydodecanoate Dominating Polyhydroxyalkanoates by β-Oxidation Pathway Inhibited Pseudomonas Putida. Metab. Eng. 2011, 13, 11–17. [Google Scholar] [CrossRef]
- Sudesh, K.; Abe, H.; Doi, Y. Synthesis, Structure and Properties of Polyhydroxyalkanoates: Biological Polyesters. Prog. Polym. Sci. 2000, 25, 1503–1555. [Google Scholar] [CrossRef]
- Martin, D.P.; Williams, S.F. Medical Applications of Poly-4-Hydroxybutyrate: A Strong Flexible Absorbable Biomaterial. Biochem. Eng. J. 2003, 16, 97–105. [Google Scholar] [CrossRef]
- Anjum, A.; Zuber, M.; Zia, K.M.; Noreen, A.; Anjum, M.N.; Tabasum, S. Microbial Production of Polyhydroxyalkanoates (PHAs) and Its Copolymers: A Review of Recent Advancements. Int. J. Biol. Macromol. 2016, 89, 161–174. [Google Scholar] [CrossRef]
- Saito, Y.; Doi, Y. Microbial Synthesis and Properties of Poly(3-Hydroxybutyrate-Co-4-Hydroxybutyrate) in Comamonas Acidovorans. Int. J. Biol. Macromol. 1994, 16, 99–104. [Google Scholar] [CrossRef]
- Volova, T.G.; Syrvacheva, D.A.; Zhila, N.O.; Sukovatiy, A.G. Synthesis of P(3HB-Co-3HHx) Copolymers Containing High Molar Fraction of 3-Hydroxyhexanoate Monomer by Cupriavidus eutrophus B10646. J. Chem. Technol. Biotechnol. 2016, 91, 416–425. [Google Scholar] [CrossRef]
- Abraham, G.A.; Gallardo, A.; San Roman, J.; Olivera, E.R.; Jodra, R.; García, B.; Miñambres, B.; García, J.L.; Luengo, J.M. Microbial Synthesis of Poly(β-Hydroxyalkanoates) Bearing Phenyl Groups from Pseudomonas putida: Chemical Structure and Characterization. Biomacromolecules 2001, 2, 562–567. [Google Scholar] [CrossRef]
- Mizuno, S.; Katsumata, S.; Hiroe, A.; Tsuge, T. Biosynthesis and Thermal Characterization of Polyhydroxyalkanoates Bearing Phenyl and Phenylalkyl Side Groups. Polym. Degrad. Stab. 2014, 109, 379–384. [Google Scholar] [CrossRef]
- Shen, R.; Cai, L.; Meng, D.; Wu, L.; Guo, K.; Dong, G.; Liu, L.; Chen, J.; Wu, Q.; Chen, G. Benzene Containing Polyhydroxyalkanoates Homo- and Copolymers Synthesized by Genome Edited Pseudomonas entomophila. Sci. China Life Sci. 2014, 57, 4–10. [Google Scholar] [CrossRef]
- Aróstegui, S.M.; Aponte, M.A.; Díaz, E.; Schröder, E. Bacterial Polyesters Produced by Pseudomonas oleovorans Containing Nitrophenyl Groups. Macromolecules 1999, 32, 2889–2895. [Google Scholar] [CrossRef]
- Escapa, I.F.; Morales, V.; Martino, V.P.; Pollet, E.; Avérous, L.; García, J.L.; Prieto, M.A. Disruption of β-Oxidation Pathway in Pseudomonas putida KT2442 to Produce New Functionalized PHAs with Thioester Groups. Appl. Microbiol. Biotechnol. 2011, 89, 1583–1598. [Google Scholar] [CrossRef]
- Arkin, A.H.; Hazer, B. Chemical Modification of Chlorinated Microbial Polyesters. Biomacromolecules 2002, 3, 1327–1335. [Google Scholar] [CrossRef]
- Thuronyi, B.W.; Privalsky, T.M.; Chang, M.C.Y. Engineered Fluorine Metabolism and Fluoropolymer Production in Living Cells. Angew. Chem. Int. Ed. 2017, 56, 13637–13640. [Google Scholar] [CrossRef]
- Wang, L.-L.; Chen, J.-T.; Wang, L.-F.; Wu, S.; Zhang, G.; Yu, H.-Q.; Ye, X.; Shi, Q.-S. Conformations and Molecular Interactions of Poly-γ-Glutamic Acid as a Soluble Microbial Product in Aqueous Solutions. Sci. Rep. 2017, 7, 12787. [Google Scholar] [CrossRef]
- Matsutani, M.; Ito, K.; Azuma, Y.; Ogino, H.; Shirai, M.; Yakushi, T.; Matsushita, K. Adaptive Mutation Related to Cellulose Producibility in Komagataeibacter medellinensis (Gluconacetobacter xylinus) NBRC 3288. Appl. Microbiol. Biotechnol. 2015, 99, 7229–7240. [Google Scholar] [CrossRef]
- Beekmann, U.; Schmölz, L.; Lorkowski, S.; Werz, O.; Thamm, J.; Fischer, D.; Kralisch, D. Process Control and Scale-up of Modified Bacterial Cellulose Production for Tailor-Made Anti-Inflammatory Drug Delivery Systems. Carbohydr. Polym. 2020, 236, 116062. [Google Scholar] [CrossRef]
- Campano, C.; Balea, A.; Blanco, A.; Negro, C. Enhancement of the Fermentation Process and Properties of Bacterial Cellulose: A Review. Cellulose 2016, 23, 57–91. [Google Scholar] [CrossRef]
- Cakar, F.; Ozer, I.; Aytekin, A.O.; Sahin, F. Improvement Production of Bacterial Cellulose by Semi-Continuous Process in Molasses Medium. Carbohydr. Polym. 2014, 106, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Salari, M.; Sowti Khiabani, M.; Rezaei Mokarram, R.; Ghanbarzadeh, B.; Samadi Kafil, H. Preparation and Characterization of Cellulose Nanocrystals from Bacterial Cellulose Produced in Sugar Beet Molasses and Cheese Whey Media. Int. J. Biol. Macromol. 2019, 122, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Karmann, S.; Panke, S.; Zinn, M. Fed-Batch Cultivations of Rhodospirillum rubrum Under Multiple Nutrient-Limited Growth Conditions on Syngas as a Novel Option to Produce Poly(3-Hydroxybutyrate) (PHB). Front. Bioeng. Biotechnol. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Kourmentza, C.; Plácido, J.; Venetsaneas, N.; Burniol-Figols, A.; Varrone, C.; Gavala, H.N.; Reis, M.A.M. Recent Advances and Challenges towards Sustainable Polyhydroxyalkanoate (PHA) Production. Bioengineering 2017, 4, 55. [Google Scholar] [CrossRef]
- Koller, M.; Marsalek, L.; de Sousa Dias, M.M.; Braunegg, G. Producing Microbial Polyhydroxyalkanoate (PHA) Biopolyesters in a Sustainable Manner. New Biotechnol. 2017, 37, 24–38. [Google Scholar] [CrossRef]
- Davis, R.; Duane, G.; Kenny, S.T.; Cerrone, F.; Guzik, M.W.; Babu, R.P.; Casey, E.; O’Connor, K.E. High Cell Density Cultivation of Pseudomonas putida KT2440 Using Glucose without the Need for Oxygen Enriched Air Supply. Biotechnol. Bioeng. 2015, 112, 725–733. [Google Scholar] [CrossRef]
- Richard, A.; Margaritis, A. Rheology, Oxygen Transfer, and Molecular Weight Characteristics of Poly(Glutamic Acid) Fermentation by Bacillus subtilis. Biotechnol. Bioeng. 2003, 82, 299–305. [Google Scholar] [CrossRef]
- Fang, J.; Huan, C.; Liu, Y.; Xu, L.; Yan, Z. Bioconversion of Agricultural Waste into Poly-Gamma-Glutamic Acid in Solid-State Bioreactors at Different Scales. Waste Manag. 2020, 102, 939–948. [Google Scholar] [CrossRef]
- Tang, B.; Lei, P.; Xu, Z.; Jiang, Y.; Xu, Z.; Liang, J.; Feng, X.; Xu, H. Highly Efficient Rice Straw Utilization for Poly-(Gamma-Glutamic Acid) Production by Bacillus subtilis NX-2. Bioresour. Technol. 2015, 193, 370–376. [Google Scholar] [CrossRef]
- Buescher, J.M.; Margaritis, A. Microbial Biosynthesis of Polyglutamic Acid Biopolymer and Applications in the Biopharmaceutical, Biomedical and Food Industries. Crit. Rev. Biotechnol. 2007, 27, 1–19. [Google Scholar] [CrossRef]
- Brigham, C. Perspectives for the Biotechnological Production of Biofuels from CO2 and H2 Using Ralstonia eutropha and Other ‘Knallgas’ Bacteria. Appl. Microbiol. Biotechnol. 2019, 103, 2113–2120. [Google Scholar] [CrossRef]
- Papademas, P.; Kotsaki, P. Technological Utilization of Whey towards Sustainable Exploitation. Adv. Dairy Res. 2019, 7, 1–10. [Google Scholar]
- Koller, M.; Salerno, A.; Muhr, A.; Reiterer, A.; Chiellini, E.; Sergio, C.; Horvat, P.; Braunegg, G. Whey Lactose as a Raw Material for Microbial Production of Biodegradable Polyesters; TU Graz: Graz, Austria, 2012; pp. 51–92. ISBN 978-953-51-0770-5. [Google Scholar]
- Bustamante, D.; Segarra, S.; Tortajada, M.; Ramon, D.; Del Cerro, C.; Auxiliadora Prieto, M.; Iglesias, J.R.; Rojas, A. In Silico Prospection of Microorganisms to Produce Polyhydroxyalkanoate from Whey: Caulobacter segnis DSM 29236 as a Suitable Industrial Strain. Microb. Biotechnol. 2019, 12, 487–501. [Google Scholar] [CrossRef]
- Gustavsson, J.; Cederberg, C.; Sonesson, U.; Otterdijk, R.; McYbeck, A. Global Food Losses and Food Waste: Extent, Causes and Prevention; Food and Agriculture Organization of the United Nations: Rome, Italy, 2011. [Google Scholar]
- Heng, K.-S.; Hatti-Kaul, R.; Adam, F.; Fukui, T.; Sudesh, K. Conversion of Rice Husks to Polyhydroxyalkanoates (PHA) via a Three-Step Process: Optimized Alkaline Pretreatment, Enzymatic Hydrolysis, and Biosynthesis by Burkholderia Cepacia USM (JCM 15050). J. Chem. Technol. Biotechnol. 2017, 92, 100–108. [Google Scholar] [CrossRef]
- Abdelraof, M.; Hasanin, M.S.; El-Saied, H. Ecofriendly Green Conversion of Potato Peel Wastes to High Productivity Bacterial Cellulose. Carbohydr. Polym. 2019, 211, 75–83. [Google Scholar] [CrossRef]
- Skiba, E.A.; Budaeva, V.V.; Ovchinnikova, E.V.; Gladysheva, E.K.; Kashcheyeva, E.I.; Pavlov, I.N.; Sakovich, G. V A Technology for Pilot Production of Bacterial Cellulose from Oat Hulls. Chem. Eng. J. 2020, 383, 123128. [Google Scholar] [CrossRef]
- Kulpreecha, S.; Boonruangthavorn, A.; Meksiriporn, B.; Thongchul, N. Inexpensive Fed-Batch Cultivation for High Poly(3-Hydroxybutyrate) Production by a New Isolate of Bacillus megaterium. J. Biosci. Bioeng. 2009, 107, 240–245. [Google Scholar] [CrossRef]
- Bae, S.O.; Shoda, M. Production of Bacterial Cellulose by Acetobacter xylinum BPR2001 Using Molasses Medium in a Jar Fermentor. Appl. Microbiol. Biotechnol. 2005, 67, 45–51. [Google Scholar] [CrossRef]
- Zhang, D.; Feng, X.; Zhou, Z.; Zhang, Y.; Xu, H. Economical Production of Poly(γ-Glutamic Acid) Using Untreated Cane Molasses and Monosodium Glutamate Waste Liquor by Bacillus subtilis NX-2. Bioresour. Technol. 2012, 114, 583–588. [Google Scholar] [CrossRef]
- Feng, X.; Tang, B.; Jiang, Y.; Xu, Z.; Lei, P.; Liang, J.; Xu, H. Efficient Production of Poly-γ-Glutamic Acid from Cane Molasses by Bacillus subtilis NX-2 Immobilized on Chemically Modified Sugarcane Bagasse. J. Chem. Technol. Biotechnol. 2016, 91, 2085–2093. [Google Scholar] [CrossRef]
- Nielsen, C.; Rahman, A.; Rehman, A.U.; Walsh, M.K.; Miller, C.D. Food Waste Conversion to Microbial Polyhydroxyalkanoates. Microb. Biotechnol. 2017, 10, 1338–1352. [Google Scholar] [CrossRef] [PubMed]
- Obruca, S.; Petrik, S.; Benesova, P.; Svoboda, Z.; Eremka, L.; Marova, I. Utilization of Oil Extracted from Spent Coffee Grounds for Sustainable Production of Polyhydroxyalkanoates. Appl. Microbiol. Biotechnol. 2014, 98, 5883–5890. [Google Scholar] [CrossRef] [PubMed]
- Kamilah, H.; Al-Gheethi, A.; Yang, T.; Sudesh, K. Correction to: The Use of Palm Oil-Based Waste Cooking Oil to Enhance the Production of Polyhydroxybutyrate [P(3HB)] by Cupriavidus necator H16 Strain. Arab. J. Sci. Eng. 2018, 44. [Google Scholar] [CrossRef]
- Taniguchi, I.; Kagotani, K.; Kimura, Y. Microbial Production of Poly(Hydroxyalkanoate)s from Waste Edible Oils. Green Chem. 2003, 5, 545–548. [Google Scholar] [CrossRef]
- Fernández, D.; Rodríguez, E.; Bassas, M.; Viñas, M.; Solanas, A.M.; Llorens, J.; Marqués, A.M.; Manresa, A. Agro-Industrial Oily Wastes as Substrates for PHA Production by the New Strain Pseudomonas aeruginosa NCIB 40045: Effect of Culture Conditions. Biochem. Eng. J. 2005, 26, 159–167. [Google Scholar] [CrossRef]
- Żywicka, A.; Junka, A.; Szymczyk, P.; Chodaczek, G.; Grzesiak, J.; Sedghizadeh, P.; Fijałkowski, K. Bacterial Cellulose Yield Increased over 500% by Supplementation of Medium with Vegetable Oil. Carbohydr. Polym. 2018, 199. [Google Scholar] [CrossRef]
- Berwig, K.H.; Baldasso, C.; Dettmer, A. Production and Characterization of Poly(3-Hydroxybutyrate) Generated by Alcaligenes Latus Using Lactose and Whey after Acid Protein Precipitation Process. Bioresour. Technol. 2016, 218, 31–37. [Google Scholar] [CrossRef]
- Pais, J.; Serafim, L.S.; Freitas, F.; Reis, M.A.M. Conversion of Cheese Whey into Poly(3-Hydroxybutyrate-Co-3-Hydroxyvalerate) by Haloferax mediterranei. New Biotechnol. 2016, 33, 224–230. [Google Scholar] [CrossRef]
- Povolo, S.; Toffano, P.; Basaglia, M.; Casella, S. Polyhydroxyalkanoates Production by Engineered Cupriavidus necator from Waste Material Containing Lactose. Bioresour. Technol. 2010, 101, 7902–7907. [Google Scholar] [CrossRef]
- Bekatorou, A.; Plioni, I.; Sparou, K.; Maroutsiou, R.; Tsafrakidou, P.; Petsi, T.; Kordouli, E. Bacterial Cellulose Production Using the Corinthian Currant Finishing Side-Stream and Cheese Whey: Process Optimization and Textural Characterization. Foods 2019, 8. [Google Scholar] [CrossRef]
- Battad-Bernardo, E.; McCrindle, S.L.; Couperwhite, I.; Neilan, B.A. Insertion of an E. Coli LacZ Gene in Acetobacter xylinus for the Production of Cellulose in Whey. FEMS Microbiol. Lett. 2004, 231, 253–260. [Google Scholar] [CrossRef]
- Chaudhry, W.N.; Jamil, N.; Ali, I.; Ayaz, M.H.; Hasnain, S. Screening for Polyhydroxyalkanoate (PHA)-Producing Bacterial Strains and Comparison of PHA Production from Various Inexpensive Carbon Sources. Ann. Microbiol. 2011, 61, 623–629. [Google Scholar] [CrossRef]
- Albuquerque, M.; Torres, C.A.V.; Reis, M. Polyhydroxyalkanoate (PHA) Production by a Mixed Microbial Culture Using Sugar Molasses: Effect of the Influent Substrate Concentration on Culture Selection. Water Res. 2010, 44, 3419–3433. [Google Scholar] [CrossRef]
- Kumar, V.; Sharma, D.K.; Bansal, V.; Mehta, D.; Sangwan, R.S.; Yadav, S.K. Efficient and Economic Process for the Production of Bacterial Cellulose from Isolated Strain of Acetobacter pasteurianus of RSV-4 Bacterium. Bioresour. Technol. 2019, 275, 430–433. [Google Scholar] [CrossRef]
- Pramanik, A.; Mitra, A.; Arumugam, M.; Bhattacharyya, A.; Sadhukhan, S.; Ray, A.; Haldar, S.; Mukhopadhyay, U.K.; Mukherjee, J. Utilization of Vinasse for the Production of Polyhydroxybutyrate by Haloarcula marismortui. Folia Microbiol. Praha 2012, 57, 71–79. [Google Scholar] [CrossRef]
- Yu, J.; Stahl, H. Microbial Utilization and Biopolyester Synthesis of Bagasse Hydrolysates. Bioresour. Technol. 2008, 99, 8042–8048. [Google Scholar] [CrossRef]
- Silva, L.F.; Taciro, M.K.; Michelin Ramos, M.E.; Carter, J.M.; Pradella, J.G.C.; Gomez, J.G.C. Poly-3-Hydroxybutyrate (P3HB) Production by Bacteria from Xylose, Glucose and Sugarcane Bagasse Hydrolysate. J. Ind. Microbiol. Biotechnol. 2004, 31, 245–254. [Google Scholar] [CrossRef]
- Follonier, S.; Riesen, R.; Zinn, M. Pilot-Scale Production of Functionalized Mcl-PHA from Grape Pomace Supplemented with Fatty Acids. Chem. Biochem. Eng. Q. 2015, 29, 113–121. [Google Scholar] [CrossRef]
- Lin, D.; Lopez-Sanchez, P.; Li, R.; Li, Z. Production of Bacterial Cellulose by Gluconacetobacter hansenii CGMCC 3917 Using Only Waste Beer Yeast as Nutrient Source. Bioresour. Technol. 2014, 151, 113–119. [Google Scholar] [CrossRef]
- Fan, X.; Gao, Y.; He, W.; Hu, H.; Tian, M.; Wang, K.; Pan, S. Production of Nano Bacterial Cellulose from Beverage Industrial Waste of Citrus Peel and Pomace Using Komagataeibacter xylinus. Carbohydr. Polym. 2016, 151, 1068–1072. [Google Scholar] [CrossRef] [PubMed]
- Kurosumi, A.; Sasaki, C.; Yamashita, Y.; Nakamura, Y. Utilization of Various Fruit Juices as Carbon Source for Production of Bacterial Cellulose by Acetobacter xylinum NBRC 13693. Carbohydr. Polym. 2009, 76, 333–335. [Google Scholar] [CrossRef]
- Yang, X.Y.; Huang, C.; Guo, H.J.; Xiong, L.; Luo, J.; Wang, B.; Lin, X.Q.; Chen, X.F.; Chen, X.D. Bacterial Cellulose Production from the Litchi Extract by Gluconacetobacter xylinus. Prep. Biochem. Biotechnol. 2016, 46, 39–43. [Google Scholar] [CrossRef]
- Jozala, A.F.; Pertile, R.A.; dos Santos, C.A.; de Carvalho Santos-Ebinuma, V.; Seckler, M.M.; Gama, F.M.; Pessoa Jr., A. Bacterial Cellulose Production by Gluconacetobacter xylinus by Employing Alternative Culture Media. Appl. Microbiol. Biotechnol. 2015, 99, 1181–1190. [Google Scholar] [CrossRef]
- Rani, M.U.; Appaiah, K.A. Production of Bacterial Cellulose by Gluconacetobacter hansenii UAC09 Using Coffee Cherry Husk. J. Food Sci. Technol. 2013, 50, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Gomes, F.P.; Silva, N.H.C.S.; Trovatti, E.; Serafim, L.S.; Duarte, M.F.; Silvestre, A.J.D.; Neto, C.P.; Freire, C.S.R. Production of Bacterial Cellulose by Gluconacetobacter sacchari Using Dry Olive Mill Residue. Biomass Bioenergy 2013, 55, 205–211. [Google Scholar] [CrossRef]
- Jiang, K.; Tang, B.; Wang, Q.; Xu, Z.; Sun, L.; Ma, J.; Li, S.; Xu, H.; Lei, P. The Bio-Processing of Soybean Dregs by Solid State Fermentation Using a Poly Gamma-Glutamic Acid Producing Strain and Its Effect as Feed Additive. Bioresour Technol 2019, 291, 121841. [Google Scholar] [CrossRef]
- López-Cuellar, M.R.; Alba-Flores, J.; Rodríguez, J.N.G.; Pérez-Guevara, F. Production of Polyhydroxyalkanoates (PHAs) with Canola Oil as Carbon Source. Int. J. Biol. Macromol. 2011, 48, 74–80. [Google Scholar] [CrossRef]
- Anne, E.; Le Grand, A.; Yves-Marie, C.; le fellic, M.; Hachet, N.; Le Tilly, V.; Loulergue, P.; Audic, J.-L.; Bruzaud, S. Valorisation of Local Agro-Industrial Processing Waters as Growth Media for Polyhydroxyalkanoates (PHA) Production. Ind. Crop. Prod. 2016, 80, 1–5. [Google Scholar] [CrossRef]
- Gayathri, G.; Srinikethan, G. Bacterial Cellulose Production by K. saccharivorans BC1 Strain Using Crude Distillery Effluent as Cheap and Cost Effective Nutrient Medium. Int. J. Biol. Macromol. 2019, 138, 950–957. [Google Scholar] [CrossRef]
- Wu, J.M.; Liu, R.H. Thin Stillage Supplementation Greatly Enhances Bacterial Cellulose Production by Gluconacetobacter Xylinus. Carbohydr. Polym. 2012, 90, 116–121. [Google Scholar] [CrossRef]
- Huang, C.; Guo, H.J.; Xiong, L.; Wang, B.; Shi, S.L.; Chen, X.F.; Lin, X.Q.; Wang, C.; Luo, J.; Chen, X.D. Using Wastewater after Lipid Fermentation as Substrate for Bacterial Cellulose Production by Gluconacetobacter xylinus. Carbohydr. Polym. 2016, 136, 198–202. [Google Scholar] [CrossRef]
- Erbas Kiziltas, E.; Kiziltas, A.; Gardner, D.J. Synthesis of Bacterial Cellulose Using Hot Water Extracted Wood Sugars. Carbohydr. Polym. 2015, 124, 131–138. [Google Scholar] [CrossRef]
- Huang, C.; Yang, X.Y.; Xiong, L.; Guo, H.J.; Luo, J.; Wang, B.; Zhang, H.R.; Lin, X.Q.; Chen, X.D. Evaluating the Possibility of Using Acetone-Butanol-Ethanol (ABE) Fermentation Wastewater for Bacterial Cellulose Production by Gluconacetobacter xylinus. Lett. Appl. Microbiol. 2015, 60, 491–496. [Google Scholar] [CrossRef]
- Li, Z.; Wang, L.; Hua, J.; Jia, S.; Zhang, J.; Liu, H. Production of Nano Bacterial Cellulose from Waste Water of Candied Jujube-Processing Industry Using Acetobacter xylinum. Carbohydr. Polym. 2015, 120, 115–119. [Google Scholar] [CrossRef]
- Campanari, S.; e Silva, F.A.; Bertin, L.; Villano, M.; Majone, M. Effect of the Organic Loading Rate on the Production of Polyhydroxyalkanoates in a Multi-Stage Process Aimed at the Valorization of Olive Oil Mill Wastewater. Int. J. Biol. Macromol. 2014, 71, 34–41. [Google Scholar] [CrossRef]
- Farghaly, A.; Enitan, A.M.; Kumari, S.; Bux, F.; Tawfik, A. Polyhydroxyalkanoates Production from Fermented Paperboard Mill Wastewater Using Acetate-Enriched Bacteria. Clean Technol. Environ. Policy 2017, 19, 935–947. [Google Scholar] [CrossRef]
- Tamisa, J.; Luzkov, K.; Jiang, Y.; van Loosdrecht, M.C.; Kleerebezem, R. Enrichment of Plasticicumulans acidivorans at Pilot-Scale for PHA Production on Industrial Wastewater. J. Biotechnol. 2014, 192 Pt A, 161–169. [Google Scholar] [CrossRef]
- Moretto, G.; Russo, I.; Bolzonella, D.; Pavan, P.; Majone, M.; Valentino, F. An Urban Biorefinery for Food Waste and Biological Sludge Conversion into Polyhydroxyalkanoates and Biogas. Water Res. 2020, 170, 115371. [Google Scholar] [CrossRef]
- Valentino, F.; Moretto, G.; Lorini, L.; Bolzonella, D.; Pavan, P.; Majone, M. Pilot-Scale Polyhydroxyalkanoate Production from Combined Treatment of Organic Fraction of Municipal Solid Waste and Sewage Sludge. Ind. Eng. Chem. Res. 2019, 58, 12149–12158. [Google Scholar] [CrossRef]
- Elmowafy, E.; Abdal-Hay, A.; Skouras, A.; Tiboni, M.; Casettari, L.; Guarino, V. Polyhydroxyalkanoate (PHA): Applications in Drug Delivery and Tissue Engineering. Expert Rev. Med. Devices 2019, 16, 467–482. [Google Scholar] [CrossRef]
- Salmaso, S.; Caliceti, P. Stealth Properties to Improve Therapeutic Efficacy of Drug Nanocarriers. J. Drug Deliv. 2013, 2013, 374252. [Google Scholar] [CrossRef]
- Amoozgar, Z.; Yeo, Y. Recent Advances in Stealth Coating of Nanoparticle Drug Delivery Systems. WIREs Nanomed. Nanobiotechnology 2012, 4, 219–233. [Google Scholar] [CrossRef]
- Hu, J.; Wang, M.; Xiao, X.; Zhang, B.; Xie, Q.; Xu, X.; Li, S.; Zheng, Z.; Wei, D.; Zhang, X. A Novel Long-Acting Azathioprine Polyhydroxyalkanoate Nanoparticle Enhances Treatment Efficacy for Systemic Lupus Erythematosus with Reduced Side Effects. Nanoscale 2020, 12, 10799–10808. [Google Scholar] [CrossRef]
- Urimi, D.; Agrawal, A.K.; Kushwah, V.; Jain, S. Polyglutamic Acid Functionalization of Chitosan Nanoparticles Enhances the Therapeutic Efficacy of Insulin Following Oral Administration. AAPS PharmSciTech 2019, 20, 131. [Google Scholar] [CrossRef]
- Shao, W.; Liu, H.; Wang, S.; Wu, J.; Huang, M.; Min, H.; Liu, X. Controlled Release and Antibacterial Activity of Tetracycline Hydrochloride-Loaded Bacterial Cellulose Composite Membranes. Carbohydr. Polym. 2016, 145, 114–120. [Google Scholar] [CrossRef]
- Silva, N.H.C.S.; Rodrigues, A.F.; Almeida, I.F.; Costa, P.C.; Rosado, C.; Neto, C.P.; Silvestre, A.J.D.; Freire, C.S.R. Bacterial Cellulose Membranes as Transdermal Delivery Systems for Diclofenac: In Vitro Dissolution and Permeation Studies. Carbohydr. Polym. 2014, 106, 264–269. [Google Scholar] [CrossRef]
- Dutta, S.D.; Patel, D.K.; Lim, K.T. Functional Cellulose-Based Hydrogels as Extracellular Matrices for Tissue Engineering. J. Biol Eng. 2019, 13, 55. [Google Scholar] [CrossRef]
- Lim, J.; You, M.; Li, J.; Li, Z. Emerging Bone Tissue Engineering via Polyhydroxyalkanoate (PHA)-Based Scaffolds. Mater. Sci Eng. C Mater. Biol. Appl. 2017, 79, 917–929. [Google Scholar] [CrossRef]
- Codreanu, A.; Balta, C.; Herman, H.; Cotoraci, C.; Mihali, C.V.; Zurbau, N.; Zaharia, C.; Rapa, M.; Stanescu, P.; Radu, I.C.; et al. Bacterial Cellulose-Modified Polyhydroxyalkanoates Scaffolds Promotes Bone Formation in Critical Size Calvarial Defects in Mice. Mater. Basel 2020, 13. [Google Scholar] [CrossRef]
- Nemati Hayati, A.; Hosseinalipour, S.M.; Rezaie, H.R.; Shokrgozar, M.A. Characterization of Poly(3-Hydroxybutyrate)/Nano-Hydroxyapatite Composite Scaffolds Fabricated without the Use of Organic Solvents for Bone Tissue Engineering Applications. Mater. Sci. Eng. C 2012, 32, 416–422. [Google Scholar] [CrossRef]
- Khan, S.; Ul-Islam, M.; Ikram, M.; Islam, S.U.; Ullah, M.W.; Israr, M.; Jang, J.H.; Yoon, S.; Park, J.K. Preparation and Structural Characterization of Surface Modified Microporous Bacterial Cellulose Scaffolds: A Potential Material for Skin Regeneration Applications in Vitro and in Vivo. Int. J. Biol. Macromol. 2018, 117, 1200–1210. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Wang, X.; Liu, S.; Zhang, W.; Wang, P.; Liu, X.; Ren, Y.; Tan, X.; Chi, B. Bioinspired Poly (Gamma-Glutamic Acid) Hydrogels for Enhanced Chondrogenesis of Bone Marrow-Derived Mesenchymal Stem Cells. Int J. Biol Macromol 2020, 142, 332–344. [Google Scholar] [CrossRef] [PubMed]
- Constantinides, C.; Basnett, P.; Lukasiewicz, B.; Carnicer, R.; Swider, E.; Majid, Q.A.; Srinivas, M.; Carr, C.A.; Roy, I. In Vivo Tracking and 1H/19F Magnetic Resonance Imaging of Biodegradable Polyhydroxyalkanoate/Polycaprolactone Blend Scaffolds Seeded with Labeled Cardiac Stem Cells. ACS Appl. Mater. Interfaces 2018, 10, 25056–25068. [Google Scholar] [CrossRef]
- Milleret, V.; Hefti, T.; Hall, H.; Vogel, V.; Eberli, D. Influence of the Fiber Diameter and Surface Roughness of Electrospun Vascular Grafts on Blood Activation. Acta Biomater. 2012, 8, 4349–4356. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Yang, S.; Peng, M.; Gama, M.; Yang, Z.; Deng, X.; Zhou, J.; Ouyang, C.; Luo, H. Controllable Synthesis of Biomimetic Nano/Submicro-Fibrous Tubes for Potential Small-Diameter Vascular Grafts. J. Mater. Chem. B 2020, 8, 5694–5706. [Google Scholar] [CrossRef]
- Sodian, R.; Sperling, J.S.; Martin, D.P.; Egozy, A.; Stock, U.; Mayer, J.E.; Vacanti, J.P. Technical Report: Fabrication of a Trileaflet Heart Valve Scaffold from a Polyhydroxyalkanoate Biopolyester for Use in Tissue Engineering. Tissue Eng. 2000, 6, 183–188. [Google Scholar] [CrossRef]
- Wu, S.; Liu, Y.-L.; Cui, B.; Qu, X.-H.; Chen, G.-Q. Study on Decellularized Porcine Aortic Valve/Poly (3-Hydroxybutyrate-Co-3-Hydroxyhexanoate) Hybrid Heart Valve in Sheep Model. Artif. Organs 2007, 31, 689–697. [Google Scholar] [CrossRef]
- Protopopov, A.V.; Kochkina, T.A.; Konstantinov, E.P.; Shishatskaya, E.I.; Efremov, S.N.; Volova, T.G.; Gitelson, I.I. Investigation of Application of PHA Coating to Enhance Biocompatibility of Vascular Stents. Dokl. Biol. Sci. 2005, 401, 85–87. [Google Scholar] [CrossRef]
- Aboelnaga, A.; Elmasry, M.; Adly, O.A.; Elbadawy, M.A.; Abbas, A.H.; Abdelrahman, I.; Salah, O.; Steinvall, I. Microbial Cellulose Dressing Compared with Silver Sulphadiazine for the Treatment of Partial Thickness Burns: A Prospective, Randomised, Clinical Trial. Burns 2018, 44, 1982–1988. [Google Scholar] [CrossRef]
- Portela, R.; Leal, C.R.; Almeida, P.L.; Sobral, R.G. Bacterial Cellulose: A Versatile Biopolymer for Wound Dressing Applications. Microb. Biotechnol. 2019, 12, 586–610. [Google Scholar] [CrossRef]
- Shishatskaya, E.I.; Nikolaeva, E.D.; Vinogradova, O.N.; Volova, T.G. Experimental Wound Dressings of Degradable PHA for Skin Defect Repair. J. Mater. Sci. Mater. Med. 2016, 27, 165. [Google Scholar] [CrossRef]
- Liu, W.-C.; Wang, H.-Y.; Lee, T.-H.; Chung, R.-J. Gamma-Poly Glutamate/Gelatin Composite Hydrogels Crosslinked by Proanthocyanidins for Wound Healing. Mater. Sci. Eng. C 2019, 101, 630–639. [Google Scholar] [CrossRef]
- Dennis, C.; Sethu, S.; Nayak, S.; Mohan, L.; Morsi, Y.Y.; Manivasagam, G. Suture Materials—Current and Emerging Trends. J. Biomed. Mater. Res. A 2016, 104, 1544–1559. [Google Scholar] [CrossRef]
- Matz, D.; Teuteberg, S.; Wiencierz, A.; Soysal, S.D.; Heizmann, O. Do Antibacterial Skin Sutures Reduce Surgical Site Infections after Elective Open Abdominal Surgery?—Study Protocol of a Prospective, Randomized Controlled Single Center Trial. Trials 2019, 20, 390. [Google Scholar] [CrossRef]
- Hsu, S.H.; Lin, C.H. The Properties of Gelatin-Poly (Gamma-Glutamic Acid) Hydrogels as Biological Glues. Biorheology 2007, 44, 17–28. [Google Scholar]
- Chen, G.-Q.; Jiang, X.-R.; Guo, Y. Synthetic Biology of Microbes Synthesizing Polyhydroxyalkanoates (PHA). Synth. Syst. Biotechnol. 2016, 1, 236–242. [Google Scholar] [CrossRef]
- Chen, G.-Q.; Hajnal, I. The ‘PHAome’. Trends Biotechnol. 2015, 33, 559–564. [Google Scholar] [CrossRef]
- Ouyang, S.-P.; Luo, R.C.; Chen, S.-S.; Liu, Q.; Chung, A.; Wu, Q.; Chen, G.-Q. Production of Polyhydroxyalkanoates with High 3-Hydroxydodecanoate Monomer Content by FadB and FadA Knockout Mutant of Pseudomonas Putida KT2442. Biomacromolecules 2007, 8, 2504–2511. [Google Scholar] [CrossRef]
- Tripathi, L.; Wu, L.-P.; Dechuan, M.; Chen, J.; Wu, Q.; Chen, G.-Q. Pseudomonas Putida KT2442 as a Platform for the Biosynthesis of Polyhydroxyalkanoates with Adjustable Monomer Contents and Compositions. Bioresour. Technol. 2013, 142, 225–231. [Google Scholar] [CrossRef]
- Vigneswari, S.; Vijaya, S.; Majid, M.I.A.; Sudesh, K.; Sipaut, C.S.; Azizan, M.N.M.; Amirul, A.A. Enhanced Production of Poly(3-Hydroxybutyrate-Co-4-Hydroxybutyrate) Copolymer with Manipulated Variables and Its Properties. J. Ind. Microbiol. Biotechnol. 2009, 36, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Ballistreri, A.; Giuffrida, M.; Guglielmino, S.P.P.; Carnazza, S.; Ferreri, A.; Impallomeni, G. Biosynthesis and Structural Characterization of Medium-Chain-Length Poly(3-Hydroxyalkanoates) Produced by Pseudomonas aeruginosa from Fatty Acids. Int. J. Biol. Macromol. 2001, 29, 107–114. [Google Scholar] [CrossRef]
- Ewering, C.; Lütke-Eversloh, T.; Luftmann, H.; Steinbüchel, A. Identification of Novel Sulfur-Containing Bacterial Polyesters: Biosynthesis of Poly(3-Hydroxy-S-Propyl-ω-Thioalkanoates) Containing Thioether Linkages in the Side Chains. Microbiology 2002, 148, 1397–1406. [Google Scholar] [CrossRef][Green Version]
- Dinjaski, N.; Fernández-Gutiérrez, M.; Selvam, S.; Parra-Ruiz, F.J.; Lehman, S.M.; San Román, J.; García, E.; García, J.L.; García, A.J.; Prieto, M.A. PHACOS, a Functionalized Bacterial Polyester with Bactericidal Activity against Methicillin-Resistant Staphylococcus aureus. Biomaterials 2014, 35, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Florea, M.; Hagemann, H.; Santosa, G.; Abbott, J.; Micklem, C.N.; Spencer-Milnes, X.; de Arroyo Garcia, L.; Paschou, D.; Lazenbatt, C.; Kong, D.; et al. Engineering Control of Bacterial Cellulose Production Using a Genetic Toolkit and a New Cellulose-Producing Strain. Proc. Natl. Acad. Sci. USA 2016, 113, E3431–E3440. [Google Scholar] [CrossRef] [PubMed]
- Teh, M.Y.; Ooi, K.H.; Danny Teo, S.X.; Bin Mansoor, M.E.; Shaun Lim, W.Z.; Tan, M.H. An Expanded Synthetic Biology Toolkit for Gene Expression Control in Acetobacteraceae. ACS Synth. Biol. 2019, 8, 708–723. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Kawano, S.; Tajima, K.; Kondo, T. In Vivo Curdlan/Cellulose Bionanocomposite Synthesis by Genetically Modified Gluconacetobacter Xylinus. Biomacromolecules 2015, 16, 3154–3160. [Google Scholar] [CrossRef]
- Gao, M.; Li, J.; Bao, Z.; Hu, M.; Nian, R.; Feng, D.; An, D.; Li, X.; Xian, M.; Zhang, H. A Natural in Situ Fabrication Method of Functional Bacterial Cellulose Using a Microorganism. Nat. Commun. 2019, 10, 437. [Google Scholar] [CrossRef]
- Yadav, V.; Sun, L.; Panilaitis, B.; Kaplan, D.L. In Vitro Chondrogenesis with Lysozyme Susceptible Bacterial Cellulose as a Scaffold. J. Tissue Eng. Regen. Med. 2015, 9, E276–E288. [Google Scholar] [CrossRef]
- Halmschlag, B.; Steurer, X.; Putri, S.P.; Fukusaki, E.; Blank, L.M. Tailor-Made Poly-γ-Glutamic Acid Production. Metab. Eng. 2019, 55, 239–248. [Google Scholar] [CrossRef]
- Poo, H.; Park, C.; Kwak, M.-S.; Choi, D.-Y.; Hong, S.-P.; Lee, I.-H.; Lim, Y.T.; Choi, Y.K.; Bae, S.-R.; Uyama, H.; et al. New Biological Functions and Applications of High-Molecular-Mass Poly-γ-Glutamic Acid. Chem. Biodivers. 2010, 7, 1555–1562. [Google Scholar] [CrossRef]
- Khalil, I.R.; Burns, A.T.; Radecka, I.; Kowalczuk, M.; Khalaf, T.; Adamus, G.; Johnston, B.; Khechara, M.P. Bacterial-Derived Polymer Poly-y-Glutamic Acid (y-PGA)-Based Micro/Nanoparticles as a Delivery System for Antimicrobials and Other Biomedical Applications. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef]
- Stumpf, T.R.; Yang, X.; Zhang, J.; Cao, X. In Situ and Ex Situ Modifications of Bacterial Cellulose for Applications in Tissue Engineering. Mater. Sci. Eng. C 2018, 82, 372–383. [Google Scholar] [CrossRef]
- Butchosa, N.; Brown, C.; Larsson, P.T.; Berglund, L.A.; Bulone, V.; Zhou, Q. Nanocomposites of Bacterial Cellulose Nanofibers and Chitin Nanocrystals: Fabrication, Characterization and Bactericidal Activity. Green Chem. 2013, 15, 3404–3413. [Google Scholar] [CrossRef]
- Zhang, P.; Chen, L.; Zhang, Q.; Hong, F.F. Using In Situ Dynamic Cultures to Rapidly Biofabricate Fabric-Reinforced Composites of Chitosan/Bacterial Nanocellulose for Antibacterial Wound Dressings. Front. Microbiol. 2016, 7, 260. [Google Scholar] [CrossRef]
- Abdelraof, M.; Hasanin, M.S.; Farag, M.M.; Ahmed, H.Y. Green Synthesis of Bacterial Cellulose/Bioactive Glass Nanocomposites: Effect of Glass Nanoparticles on Cellulose Yield, Biocompatibility and Antimicrobial Activity. Int. J. Biol. Macromol. 2019, 138, 975–985. [Google Scholar] [CrossRef]
- Chen, J.; Chen, C.; Liang, G.; Xu, X.; Hao, Q.; Sun, D. In Situ Preparation of Bacterial Cellulose with Antimicrobial Properties from Bioconversion of Mulberry Leaves. Carbohydr. Polym. 2019, 220, 170–175. [Google Scholar] [CrossRef]
- Castro-Mayorga, J.L.; Freitas, F.; Reis, M.A.M.; Prieto, M.A.; Lagaron, J.M. Biosynthesis of Silver Nanoparticles and Polyhydroxybutyrate Nanocomposites of Interest in Antimicrobial Applications. Int. J. Biol. Macromol. 2018, 108, 426–435. [Google Scholar] [CrossRef]
- Geng, Y.; Wang, S.; Qi, Q. Expression of Active Recombinant Human Tissue-Type Plasminogen Activator by Using in Vivo Polyhydroxybutyrate Granule Display. Appl. Environ. Microbiol. 2010, 76, 7226–7230. [Google Scholar] [CrossRef]
- Yao, Y.C.; Zhan, X.Y.; Zhang, J.; Zou, X.H.; Wang, Z.H.; Xiong, Y.C.; Chen, J.; Chen, G.Q. A Specific Drug Targeting System Based on Polyhydroxyalkanoate Granule Binding Protein PhaP Fused with Targeted Cell Ligands. Biomaterials 2008, 29, 4823–4830. [Google Scholar] [CrossRef]
- Fan, F.; Wang, L.; Ouyang, Z.; Wen, Y.; Lu, X. Development and Optimization of a Tumor Targeting System Based on Microbial Synthesized PHA Biopolymers and PhaP Mediated Functional Modification. Appl. Microbiol. Biotechnol. 2018, 102, 3229–3241. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jung, S.-G.; Park, C.-S.; Kim, H.-Y.; Batt, C.; Kim, Y.-R. Tumor-Specific Hybrid Polyhydroxybutyrate Nanoparticle: Surface Modification of Nanoparticle by Enzymatically Synthesized Functional Block Copolymer. Bioorg. Med. Chem. Lett. 2011, 21, 2941–2944. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Park, J.P.; Park, T.J.; Lee, S.Y.; Lee, S.; Park, J.K. Selective Immobilization of Fusion Proteins on Poly(Hydroxyalkanoate) Microbeads. Anal. Chem. 2005, 77, 5755–5759. [Google Scholar] [CrossRef] [PubMed]
- Park, T.J.; Yoo, S.; Keum, K.; Lee, S.Y. Microarray of DNA–Protein Complexes on Poly-3-Hydroxybutyrate Surface for Pathogen Detection. Anal. Bioanal. Chem. 2009, 393, 1639–1647. [Google Scholar] [CrossRef] [PubMed]
- Backstrom, B.T.; Brockelbank, J.A.; Rehm, B.H. Recombinant Escherichia coli Produces Tailor-Made Biopolyester Granules for Applications in Fluorescence Activated Cell Sorting: Functional Display of the Mouse Interleukin-2 and Myelin Oligodendrocyte Glycoprotein. BMC Biotechnol. 2007, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Parlane, N.A.; Chen, S.; Jones, G.J.; Vordermeier, H.M.; Wedlock, D.N.; Rehm, B.H.A.; Buddle, B.M. Display of Antigens on Polyester Inclusions Lowers the Antigen Concentration Required for a Bovine Tuberculosis Skin Test. Clin. Vaccine Immunol. 2016, 23, 19–26. [Google Scholar] [CrossRef]
- Dong, Y.; Li, P.; Chen, C.B.; Wang, Z.H.; Ma, P.; Chen, G.Q. The Improvement of Fibroblast Growth on Hydrophobic Biopolyesters by Coating with Polyhydroxyalkanoate Granule Binding Protein PhaP Fused with Cell Adhesion Motif RGD. Biomaterials 2010, 31, 8921–8930. [Google Scholar] [CrossRef]
- You, M.; Peng, G.; Li, J.; Ma, P.; Wang, Z.; Shu, W.; Peng, S.; Chen, G.Q. Chondrogenic Differentiation of Human Bone Marrow Mesenchymal Stem Cells on Polyhydroxyalkanoate (PHA) Scaffolds Coated with PHA Granule Binding Protein PhaP Fused with RGD Peptide. Biomaterials 2011, 32, 2305–2313. [Google Scholar] [CrossRef]
- Xie, H.; Li, J.; Li, L.; Dong, Y.; Chen, G.Q.; Chen, K.C. Enhanced Proliferation and Differentiation of Neural Stem Cells Grown on PHA Films Coated with Recombinant Fusion Proteins. Acta Biomater. 2013, 9, 7845–7854. [Google Scholar] [CrossRef]
- Ke, Y.; Liu, C.; Zhang, X.; Xiao, M.; Wu, G. Surface Modification of Polyhydroxyalkanoates toward Enhancing Cell Compatibility and Antibacterial Activity. Macromol. Mater. Eng. 2017, 302, 1700258. [Google Scholar] [CrossRef]
- Parlane, N.A.; Grage, K.; Lee, J.W.; Buddle, B.M.; Denis, M.; Rehm, B.H.A. Production of a Particulate Hepatitis C Vaccine Candidate by an Engineered Lactococcus Lactis Strain. Appl. Environ. Microbiol. 2011, 77, 8516–8522. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Donato, G.; Piniella, B.; Aguilar, D.; Olivera, S.; Pérez, A.; Castañedo, Y.; Alvarez-Lajonchere, L.; Dueñas-Carrera, S.; Lee, J.W.; Burr, N.; et al. Protective T Cell and Antibody Immune Responses against Hepatitis C Virus Achieved Using a Biopolyester-Bead-Based Vaccine Delivery System. Clin. Vaccine Immunol. CVI 2016, 23, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Xue, Q.; Liu, X.B.; Lao, Y.H.; Wu, L.P.; Wang, D.; Zuo, Z.Q.; Chen, J.Y.; Hou, J.; Bei, Y.Y.; Wu, X.F.; et al. Anti-Infective Biomaterials with Surface-Decorated Tachyplesin I. Biomaterials 2018, 178, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Levy, I.; Shoseyov, O. Cellulose-Binding Domains: Biotechnological Applications. Biotechnol. Adv. 2002, 20, 191–213. [Google Scholar] [CrossRef]
- Sugimoto, N.; Igarashi, K.; Samejima, M. Cellulose Affinity Purification of Fusion Proteins Tagged with Fungal Family 1 Cellulose-Binding Domain. Protein Expr. Purif. 2012, 82, 290–296. [Google Scholar] [CrossRef]
- Kumar, A.; Zhang, S.; Wu, G.; Wu, C.C.; Chen, J.; Baskaran, R.; Liu, Z. Cellulose Binding Domain Assisted Immobilization of Lipase (GSlip–CBD) onto Cellulosic Nanogel: Characterization and Application in Organic Medium. Colloids Surf. B Biointerfaces 2015, 136, 1042–1050. [Google Scholar] [CrossRef]
- Xiao, Z.; Gao, P.; Qu, Y.; Wang, T. Cellulose-Binding Domain of Endoglucanase III from Trichoderma Reesei Disrupting the Structure of Cellulose. Biotechnol. Lett. 2001, 23, 711–715. [Google Scholar] [CrossRef]
- Andrade, F.K.; Moreira, S.M.; Domingues, L.; Gama, F.M. Improving the Affinity of Fibroblasts for Bacterial Cellulose Using Carbohydrate-Binding Modules Fused to RGD. J. Biomed. Mater. Res. A 2010, 92, 9–17. [Google Scholar] [CrossRef]
- Andrade, F.K.; Costa, R.; Domingues, L.; Soares, R.; Gama, M. Improving Bacterial Cellulose for Blood Vessel Replacement: Functionalization with a Chimeric Protein Containing a Cellulose-Binding Module and an Adhesion Peptide. Acta Biomater. 2010, 6, 4034–4041. [Google Scholar] [CrossRef]
- Andrade, F.K.; Silva, J.P.; Carvalho, M.; Castanheira, E.M.; Soares, R.; Gama, M. Studies on the Hemocompatibility of Bacterial Cellulose. J. Biomed. Mater. Res. A 2011, 98, 554–566. [Google Scholar] [CrossRef]
- Pértile, R.; Moreira, S.; Andrade, F.; Domingues, L.; Gama, M. Bacterial Cellulose Modified Using Recombinant Proteins to Improve Neuronal and Mesenchymal Cell Adhesion. Biotechnol. Prog. 2012, 28, 526–532. [Google Scholar] [CrossRef]
- Abouhmad, A.; Mamo, G.; Dishisha, T.; Amin, M.; Hatti-Kaul, R. T4 Lysozyme Fused with Cellulose Binding Module for Antimicrobial Cellulosic Wound Dressing Materials. J. Appl. Microbiol. 2016, 121. [Google Scholar] [CrossRef]
- Weishaupt, R.; Zünd, J.N.; Heuberger, L.; Zuber, F.; Faccio, G.; Robotti, F.; Ferrari, A.; Fortunato, G.; Ren, Q.; Maniura-Weber, K.; et al. Antibacterial, Cytocompatible, Sustainably Sourced: Cellulose Membranes with Bifunctional Peptides for Advanced Wound Dressings. Adv. Healthc. Mater. 2020, 9, 1901850. [Google Scholar] [CrossRef]
- Raza, Z.A.; Riaz, S.; Banat, I.M. Polyhydroxyalkanoates: Properties and Chemical Modification Approaches for Their Functionalization. Biotechnol. Prog. 2018, 34, 29–41. [Google Scholar] [CrossRef]
- Li, Z.; Yang, J.; Loh, X.J. Polyhydroxyalkanoates: Opening Doors for a Sustainable Future. NPG Asia Mater. 2016, 8, e265. [Google Scholar] [CrossRef]
- López Durán, V.; Larsson, P.A.; Wågberg, L. Chemical Modification of Cellulose-Rich Fibres to Clarify the Influence of the Chemical Structure on the Physical and Mechanical Properties of Cellulose Fibres and Thereof Made Sheets. Carbohydr. Polym. 2018, 182, 1–7. [Google Scholar] [CrossRef]
- Ramachandran, H.; Kannusamy, S. Blends of Polyhydroxyalkanoates (PHAs). In Polyhydroxyalkanoate (PHA) Based Blends, Composites and Nanocomposites; Royal Society of Chemistry: London, UK, 2015; ISBN 9781782622314. [Google Scholar]
- Tavakolian, M.; Jafari, S.M.; van de Ven, T.G.M. A Review on Surface-Functionalized Cellulosic Nanostructures as Biocompatible Antibacterial Materials. Nano-Micro Lett. 2020, 12, 73. [Google Scholar] [CrossRef]
- Reddy, N.; Reddy, R.; Jiang, Q. Crosslinking Biopolymers for Biomedical Applications. Trends Biotechnol. 2015, 33, 362–369. [Google Scholar] [CrossRef]
- Bach, Q.-V.; Manh Vu, C. Bacterial Cellulose Filled Epoxy Resin-Based Green Composites: Fabrication and Characterization. Compos. Interfaces 2019, 1–18. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, Y.; Luo, Z.; Han, W.; Qiu, W.; Zhao, T. Hierarchically Porous Zirconia Monolith Fabricated from Bacterial Cellulose and Preceramic Polymer. ACS Omega 2018, 3, 4688–4694. [Google Scholar] [CrossRef]
- Sudhakar, Y.N.; Selvakumar, M.; Bhat, D.K. Methods of Preparation of Biopolymer Electrolytes. Biopolym. Electrolytes 2018, 35–52. [Google Scholar] [CrossRef]
- Zhao, K.; Yang, X.; Chen, G.Q.; Chen, J.C. Effect of Lipase Treatment on the Biocompatibility of Microbial Polyhydroxyalkanoates. J. Mater. Sci. Mater. Med. 2002, 13, 849–854. [Google Scholar] [CrossRef] [PubMed]
- Kai, Z.; Ying, D.; Guo-Qiang, C. Effects of Surface Morphology on the Biocompatibility of Polyhydroxyalkanoates. Biochem. Eng. J. 2003, 16, 115–123. [Google Scholar] [CrossRef]
- Basnett, P.; Ching, K.Y.; Stolz, M.; Knowles, J.C.; Boccaccini, A.R.; Smith, C.; Locke, I.C.; Keshavarz, T.; Roy, I. Novel Poly(3-Hydroxyoctanoate)/Poly(3-Hydroxybutyrate) Blends for Medical Applications. React. Funct. Polym. 2013, 73, 1340–1348. [Google Scholar] [CrossRef]
- Mousavioun, P.; Halley, P.J.; Doherty, W.O.S. Thermophysical Properties and Rheology of PHB/Lignin Blends. Ind. Crop. Prod. 2013, 50, 270–275. [Google Scholar] [CrossRef]
- Gerard, T.; Budtova, T. Morphology and Molten-State Rheology of Polylactide and Polyhydroxyalkanoate Blends. Eur. Polym. J. 2012, 48, 1110–1117. [Google Scholar] [CrossRef]
- Takagi, Y.; Yasuda, R.; Yamaoka, M.; Yamane, T. Morphologies and Mechanical Properties of Polylactide Blends with Medium Chain Length Poly(3-Hydroxyalkanoate) and Chemically Modified Poly(3-Hydroxyalkanoate). J. Appl. Polym. Sci. 2004, 93, 2363–2369. [Google Scholar] [CrossRef]
- Lim, J.; Chong, M.S.K.; Teo, E.Y.; Chen, G.Q.; Chan, J.K.Y.; Teoh, S.H. Biocompatibility Studies and Characterization of Poly(3-Hydroxybutyrate-Co- 3-Hydroxyhexanoate)/Polycaprolactone Blends. J. Biomed. Mater. Res. Part B Appl. Biomater. 2013, 101 B, 752–761. [Google Scholar] [CrossRef]
- Altun, E.; Aydogdu, M.O.; Koc, F.; Crabbe-Mann, M.; Brako, F.; Kaur-Matharu, R.; Ozen, G.; Kuruca, S.E.; Edirisinghe, U.; Gunduz, O.; et al. Novel Making of Bacterial Cellulose Blended Polymeric Fiber Bandages. Macromol. Mater. Eng. 2018, 303, 1700607. [Google Scholar] [CrossRef]
- Rivero-Buceta, V.; Aguilar, M.R.; Hernández-Arriaga, A.M.; Blanco, F.G.; Rojas, A.; Tortajada, M.; Ramírez-Jiménez, R.A.; Vázquez-Lasa, B.; Prieto, A. Anti-Staphylococcal Hydrogels Based on Bacterial Cellulose and the Antimicrobial Biopolyester Poly(3-Hydroxy-Acetylthioalkanoate-Co-3-Hydroxyalkanoate). Int. J. Biol. Macromol. 2020, 162, 1869–1879. [Google Scholar] [CrossRef]
- Hsieh, C.Y.; Tsai, S.P.; Wang, D.M.; Chang, Y.N.; Hsieh, H.J. Preparation of γ-PGA/Chitosan Composite Tissue Engineering Matrices. Biomaterials 2005, 26, 5617–5623. [Google Scholar] [CrossRef]
- Hajiali, H.; Shahgasempour, S.; Naimi-Jamal, M.R.; Peirovi, H. Electrospun PGA/Gelatin Nanofibrous Scaffolds and Their Potential Application in Vascular Tissue Engineering. Int. J. Nanomed. 2011, 6, 2133–2141. [Google Scholar] [CrossRef]
- Aghdam, R.M.; Najarian, S.; Shakhesi, S.; Khanlari, S.; Shaabani, K.; Sharifi, S. Investigating the Effect of PGA on Physical and Mechanical Properties of Electrospun PCL/PGA Blend Nanofibers. J. Appl. Polym. Sci. 2012, 124, 123–131. [Google Scholar] [CrossRef]
- Le Fer, G.; Babinot, J.; Versace, D.L.; Langlois, V.; Renard, E. An Efficient Thiol-Ene Chemistry for the Preparation of Amphiphilic PHA-Based Graft Copolymers. Macromol. Rapid Commun. 2012, 33, 2041–2045. [Google Scholar] [CrossRef] [PubMed]
- Ashby, R.D.; Foglia, T.A.; Solaiman, D.K.Y.; Liu, C.-K.; Nuñez, A.; Eggink, G. Viscoelastic Properties of Linseed Oil-Based Medium Chain Length Poly(Hydroxyalkanoate) Films: Effects of Epoxidation and Curing. Int. J. Biol. Macromol. 2000, 27, 355–361. [Google Scholar] [CrossRef]
- Karahalilotlu, Z.; Ercan, B.; Taylor, E.N.; Chung, S.; Denkbąs, E.B.; Webster, T.J. Antibacterial Nanostructured Polyhydroxybutyrate Membranes for Guided Bone Regeneration. J. Biomed. Nanotechnol. 2015, 11, 2253–2263. [Google Scholar] [CrossRef]
- Torres, M.G.; Talavera, J.R.R.; Muñoz, S.V.; Pérez, M.G.; Castro, M.P.C.; Cortes, J.C.; Muñoz, R.A.E. Effects of Solvents on the Radiation Grafting Reaction of Vinyl Compounds on Poly (3-Hydroxybutyrate). Radiat. Phys. Chem. 2015, 108, 87–94. [Google Scholar] [CrossRef]
- Ma, Y.M.; Wei, D.X.; Yao, H.; Wu, L.P.; Chen, G.Q. Synthesis, Characterization and Application of Thermoresponsive Polyhydroxyalkanoate-Graft-Poly(N-Isopropylacrylamide). Biomacromolecules 2016, 17, 2680–2690. [Google Scholar] [CrossRef]
- Tajima, K.; Iwamoto, K.; Satoh, Y.; Sakai, R.; Satoh, T.; Dairi, T. Advanced Functionalization of Polyhydroxyalkanoate via the UV-Initiated Thiol-Ene Click Reaction. Appl. Microbiol. Biotechnol. 2016, 100, 4375–4383. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Lü, L.X.; Shi, J.C.; Wang, H.F.; Xiao, Z.D.; Huang, N.P. Introducing RGD Peptides on PHBV Films through PEG-Containing Cross-Linkers to Improve the Biocompatibility. Biomacromolecules 2011, 12, 551–559. [Google Scholar] [CrossRef]
- Chung, M.G.; Kim, H.W.; Kim, B.R.; Kim, Y.B.; Rhee, Y.H. Biocompatibility and Antimicrobial Activity of Poly(3-Hydroxyoctanoate) Grafted with Vinylimidazole. Int. J. Biol. Macromol. 2012, 50, 310–316. [Google Scholar] [CrossRef]
- Abdelwahab, M.A.; El-Barbary, A.A.; El-Said, K.S.; El Naggar, S.A.; ElKholy, H.M. Evaluation of Antibacterial and Anticancer Properties of Poly(3-Hydroxybutyrate) Functionalized with Different Amino Compounds. Int. J. Biol. Macromol. 2019, 122, 793–805. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.-G.; Jou, C.-H.; Yang, M.-C. Antibacterial and Biodegradable Properties of Polyhydroxyalkanoates Grafted with Chitosan and Chitooligosaccharides via Ozone Treatment. J. Appl. Polym. Sci. 2003, 88, 2797–2803. [Google Scholar] [CrossRef]
- Arslan, H.; Hazer, B.; Yoon, S.C. Grafting of Poly(3-Hydroxyalkanoate) and Linoleic Acid onto Chitosan. J. Appl. Polym. Sci. 2007, 103, 81–89. [Google Scholar] [CrossRef]
- Švorčík, P.S.S.S.N.S.K.S.R.V. Cytocompatibility of Polyhydroxybutyrate Modified by Plasma Discharge. Society 2014, 54, 1231–1238. [Google Scholar] [CrossRef]
- Wang, C.; Sauvageau, D.; Elias, A. Immobilization of Active Bacteriophages on Polyhydroxyalkanoate Surfaces. ACS Appl. Mater. Interfaces 2016, 8, 1128–1138. [Google Scholar] [CrossRef]
- Fernandes, S.C.M.; Sadocco, P.; Alonso-Varona, A.; Palomares, T.; Eceiza, A.; Silvestre, A.J.D.; Mondragon, I.; Freire, C.S.R. Bioinspired Antimicrobial and Biocompatible Bacterial Cellulose Membranes Obtained by Surface Functionalization with Aminoalkyl Groups. ACS Appl. Mater. Interfaces 2013, 5, 3290–3297. [Google Scholar] [CrossRef]
- Sun, B.; Wei, F.; Li, W.; Xu, X.; Zhang, H.; Liu, M.; Lin, J.; Ma, B.; Chen, C.; Sun, D. Macroporous Bacterial Cellulose Grafted by Oligopeptides Induces Biomimetic Mineralization via Interfacial Wettability. Colloids Surf. B Biointerfaces 2019, 183. [Google Scholar] [CrossRef]
- Rouabhia, M.; Asselin, J.; Tazi, N.; Messaddeq, Y.; Levinson, D.; Zhang, Z. Production of Biocompatible and Antimicrobial Bacterial Cellulose Polymers Functionalized by RGDC Grafting Groups and Gentamicin. ACS Appl. Mater. Interfaces 2014, 6, 1439–1446. [Google Scholar] [CrossRef]
- Ye, S.; Jiang, L.; Wu, J.; Su, C.; Huang, C.; Liu, X.; Shao, W. Flexible Amoxicillin-Grafted Bacterial Cellulose Sponges for Wound Dressing: In Vitro and in Vivo Evaluation. ACS Appl. Mater. Interfaces 2018, 10, 5862–5870. [Google Scholar] [CrossRef]
- Chuah, C.; Wang, J.; Tavakoli, J.; Tang, Y. Novel Bacterial Cellulose-Poly (Acrylic Acid) Hybrid Hydrogels with Controllable Antimicrobial Ability as Dressings for Chronic Wounds. Polymers 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Y.; Cheng, Z.; Sheng, J.; Yang, R. Nano-Sized Fibrils Dispersed from Bacterial Cellulose Grafted with Chitosan. Carbohydr. Polym. 2019, 214, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Pertile, R.A.N.; Andrade, F.K.; Alves, C.; Gama, M. Surface Modification of Bacterial Cellulose by Nitrogen-Containing Plasma for Improved Interaction with Cells. Carbohydr. Polym. 2010, 82, 692–698. [Google Scholar] [CrossRef]
- Kurniawan, H.; Lai, J.T.; Wang, M.J. Biofunctionalized Bacterial Cellulose Membranes by Cold Plasmas. Cellulose 2012, 19, 1975–1988. [Google Scholar] [CrossRef]
- Zhang, K.; Wu, J.; Zhang, W.; Yan, S.; Ding, J.; Chen, X.; Cui, L.; Yin, J. In Situ Formation of Hydrophobic Clusters to Enhance Mechanical Performance of Biodegradable Poly(l-Glutamic Acid)/Poly(ε-Caprolactone) Hydrogel towards Meniscus Tissue Engineering. J. Mater. Chem. B 2018, 6, 7822–7833. [Google Scholar] [CrossRef]
- Chung, S.; Gentilini, C.; Callanan, A.; Hedegaard, M.; Hassing, S.; Stevens, M.M. Responsive Poly (γ-Glutamic Acid) Fibres for Biomedical Applications. J. Mater. Chem. B 2013, 1, 1397–1401. [Google Scholar] [CrossRef]
- Michiya, M.; Ken-ichiro, H.; Mariko, H.; Tatsuo, K.; Mitsuru, A. Stably-Dispersed and Surface-Functional Bionanoparticles Prepared by Self-Assembling Amphipathic Polymers of Hydrophilic Poly(γ-Glutamic Acid) Bearing Hydrophobic Amino Acids. Chem. Lett. 2004, 33, 398–399. [Google Scholar] [CrossRef]
- Clarke, D.E.; Pashuck, E.T.; Bertazzo, S.; Weaver, J.V.M.; Stevens, M.M. Self-Healing, Self-Assembled β-Sheet Peptide-Poly(γ-Glutamic Acid) Hybrid Hydrogels. J. Am. Chem. Soc. 2017, 139, 7250–7255. [Google Scholar] [CrossRef]
- Cacicedo, M.L.; Pacheco, G.; Islan, G.A.; Alvarez, V.A.; Barud, H.S.; Castro, G.R. International Journal of Biological Macromolecules Chitosan-Bacterial Cellulose Patch of Ciprofloxacin for Wound Dressing: Preparation and Characterization Studies. Int. J. Biol. Macromol. 2020, 147, 1136–1145. [Google Scholar] [CrossRef]
- Wahid, F.; Hu, X.H.; Chu, L.Q.; Jia, S.R.; Xie, Y.Y.; Zhong, C. Development of Bacterial Cellulose/Chitosan Based Semi-Interpenetrating Hydrogels with Improved Mechanical and Antibacterial Properties. Int. J. Biol. Macromol. 2019, 122, 380–387. [Google Scholar] [CrossRef]
- Fürsatz, M.; Skog, M.; Sivlér, P.; Palm, E.; Aronsson, C.; Skallberg, A.; Greczynski, G.; Khalaf, H.; Bengtsson, T.; Aili, D. Functionalization of Bacterial Cellulose Wound Dressings with the Antimicrobial Peptide ϵ-Poly-L-Lysine. Biomed. Mater. Bristol 2018, 13. [Google Scholar] [CrossRef]
- Levine, A.C.; Sparano, A.; Twigg, F.F.; Numata, K.; Nomura, C.T. Influence of Cross-Linking on the Physical Properties and Cytotoxicity of Polyhydroxyalkanoate (PHA) Scaffolds for Tissue Engineering. ACS Biomater. Sci. Eng. 2015, 1, 567–576. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Z.; Che, X.; Yu, L.; Jia, W.; Shen, R.; Chen, J.; Ma, Y.; Chen, G.Q. Synthesis and Characterization of Polyhydroxyalkanoate Organo/Hydrogels. Biomacromolecules 2019, 20, 3303–3312. [Google Scholar] [CrossRef]
- Park, S.J.; Uyama, H.; Kwak, M.S.; Sung, M.H. Comparison of the Stability of Poly-γ-Glutamate Hydrogels Prepared by UV and γ-Ray Irradiation. J. Microbiol. Biotechnol. 2019, 29, 1078–1082. [Google Scholar] [CrossRef]
- Meftahi, A.; Khajavi, R.; Rashidi, A.; Rahimi, M.K.; Bahador, A. Preventing the Collapse of 3D Bacterial Cellulose Network via Citric Acid. J. Nanostruct. Chem. 2018, 8, 311–320. [Google Scholar] [CrossRef]
- Wang, C.; Wang, H.; Zou, F.; Chen, S.; Wang, Y. Development of Polyhydroxyalkanoate-Based Polyurethane Withwater-Thermal Response Shape-Memory Behavior as New 3D Elastomers Scaffolds. Polymers 2019, 11, 1927. [Google Scholar] [CrossRef]
- Bakó, J.; Kerényi, F.; Hrubi, E.; Varga, I.; Daróczi, L.; Dienes, B.; Csernoch, L.; Gáll, J.; Hegedus, C. Poly- γ -Glutamic Acid Nanoparticles Based Visible Light-Curable Hydrogel for Biomedical Application. J. Nanomater. 2016, 2016. [Google Scholar] [CrossRef]
- Schmutz, M.; Borges, O.; Jesus, S.; Borchard, G.; Perale, G.; Zinn, M.; Sips, Ä.A.J.A.M.; Soeteman-Hernandez, L.G.; Wick, P.; Som, C. A Methodological Safe-by-Design Approach for the Development of Nanomedicines. Front. Bioeng. Biotechnol. 2020, 8, 258. [Google Scholar] [CrossRef]
- Som, C.; Schmutz, M.; Borges, O.; Jesus, S.; Borchard, G.; Nguyen, V.; Perale, G.; Casalini, T.; Zinn, M.; Amstutz, V.; et al. Guidelines for Implementing a Safe-by-Design Approach for Medicinal Polymeric Nanocarriers; Empa: St. Gallen, Switzerland, 2019. [Google Scholar]
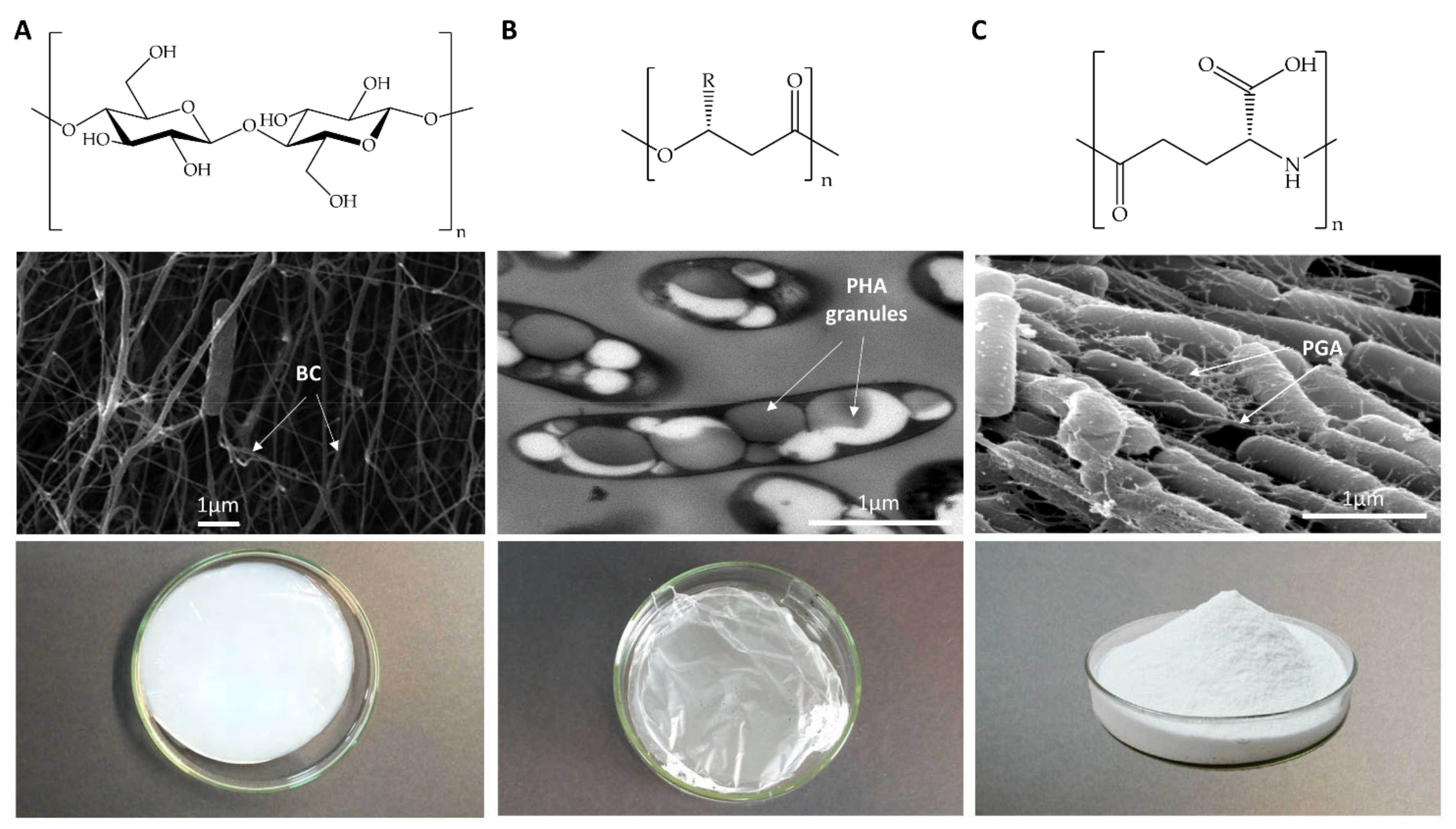


| BC | PHA | PGA | |
|---|---|---|---|
| Chemical structure | Polysaccharide (Figure 1A) Glucose (glc) homopolymer. Properties of the polymer depend on culture conditions Hydrophilic | Polyester (Figure 1B) High diversity. Polymer properties rely on its monomer combination Hydrophobic | Polyamide (Figure 1C) Anionic. D- or L-glutamic acid (glu) homopolymers, or D-/L-glu copolymers Hydrophilic |
| Industrial production prototype bacteria | Species belonging to Komagataeibacter genus, K. xylinus | High diversity Scl-PHA Cupriavidus necator, Halomonas spp. Mcl-PHA Pseudomonas spp. | Bacillus spp., B. subtilis |
| Precursors at industrial production level | Direct: sugars, preference depends on the species | Direct: fatty acids | Direct: glutamic acid |
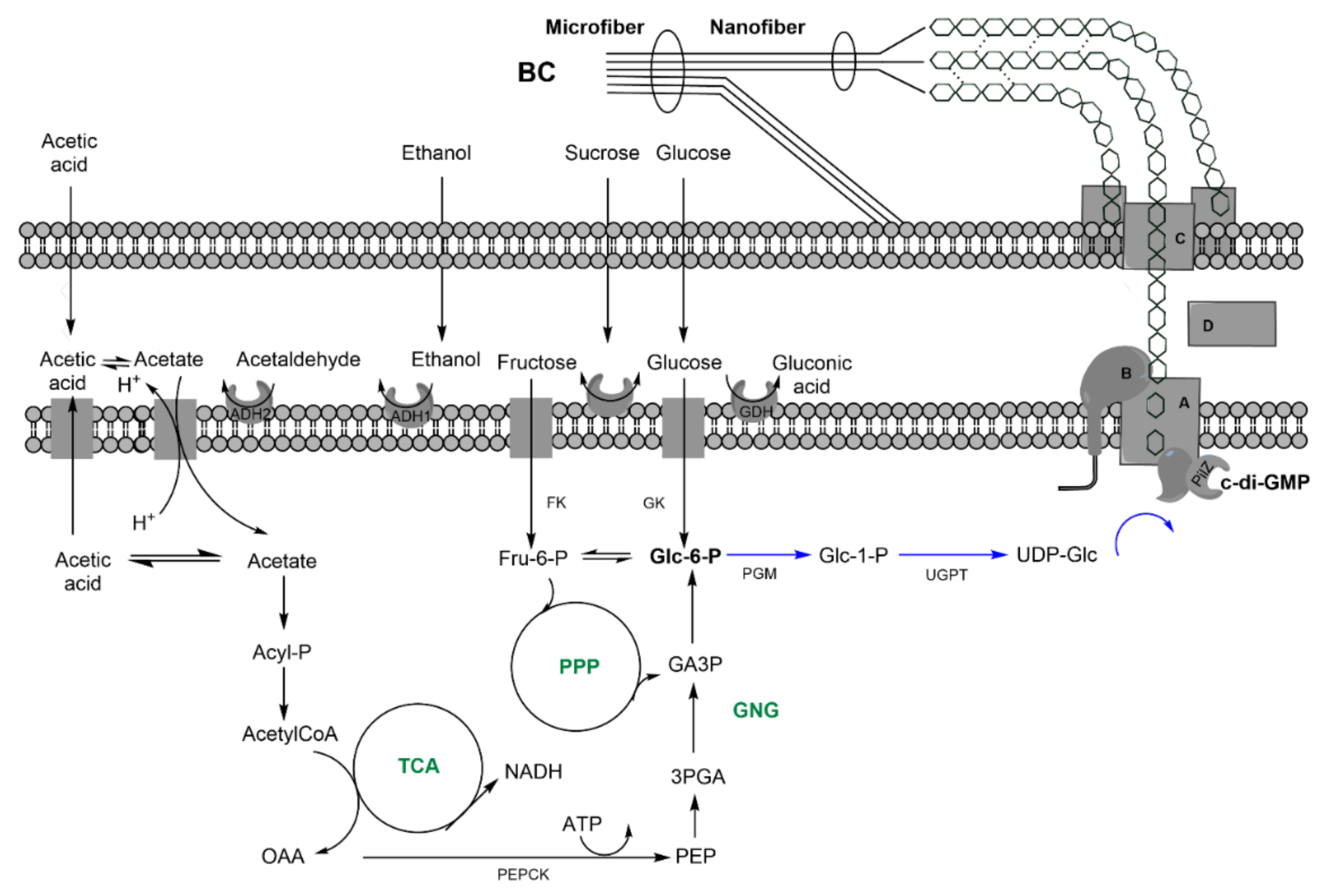
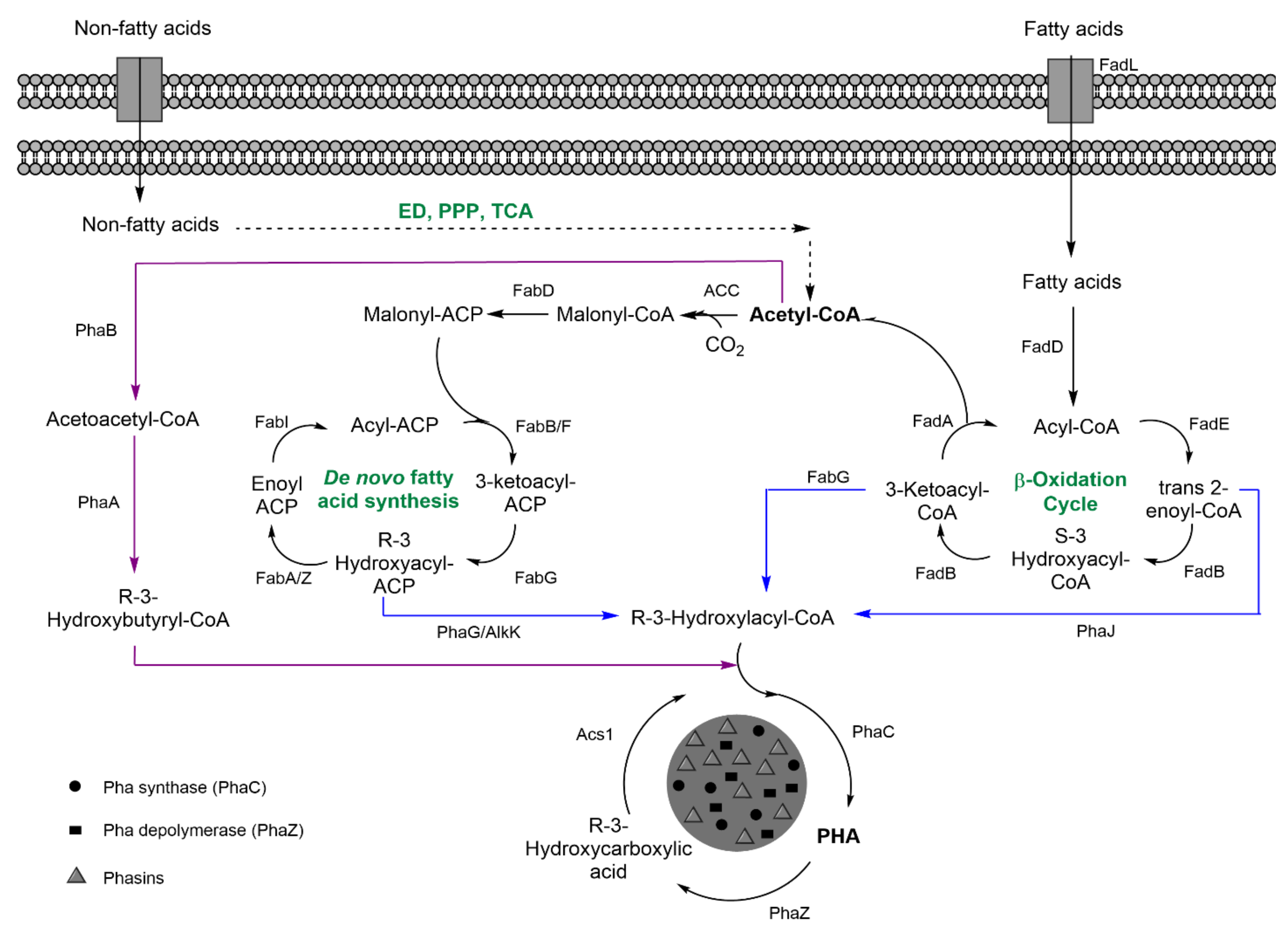
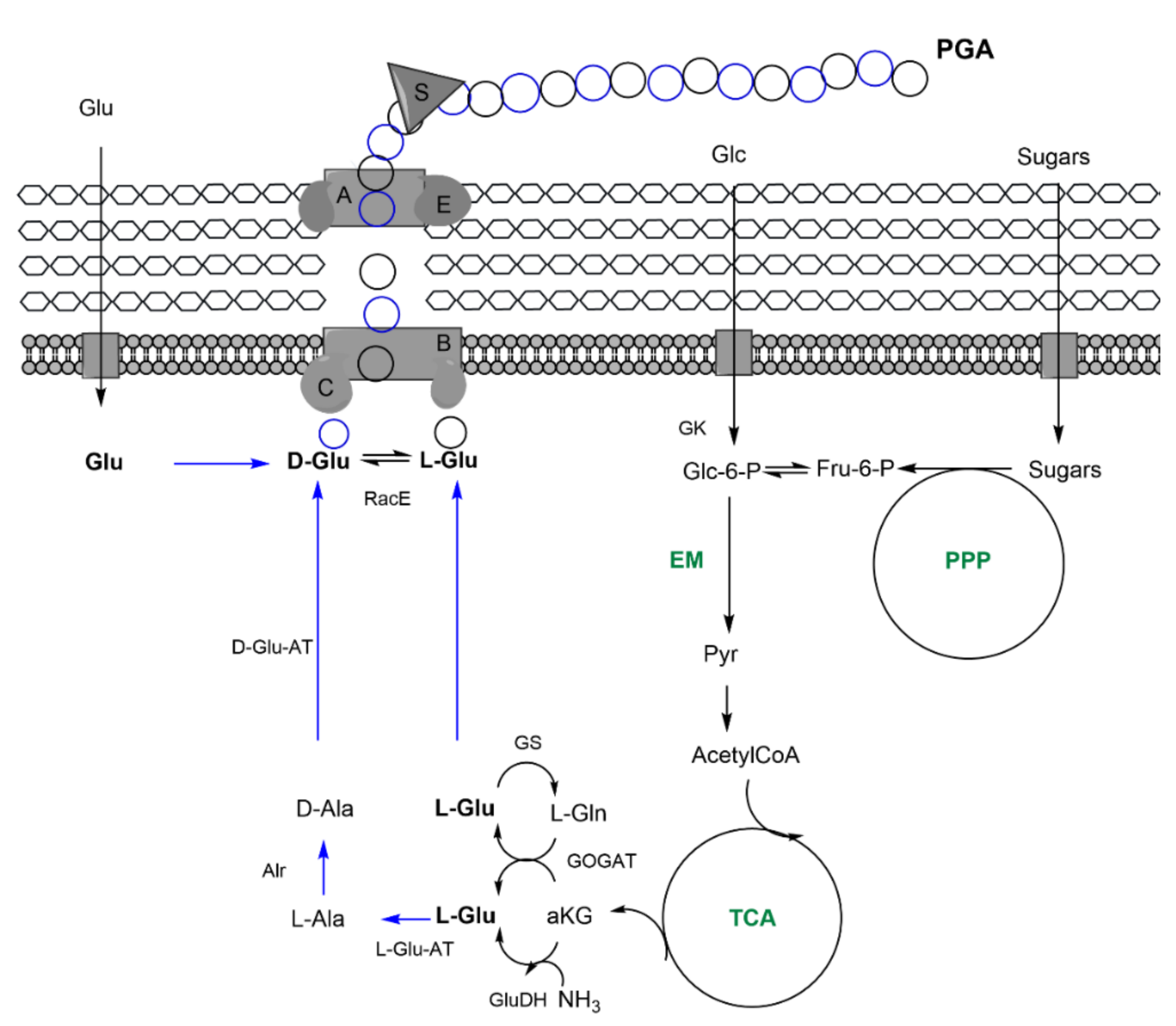
| Indirect: ethanol, converted into acetate, and, finally, glc through tricarbolxylic acid cycle (TCA) and gluconeogenesis (GNG) (Figure 2) | Indirect: sugars through TCA and de novo synthesis of fatty acids (Figure 3) | Indirect: sugars, through TCA and alpha-ketoglutarate (α–KG) conversion into glutamic acid (Figure 4) | |
| Culture conditions for pure cultures industrial production | Submerged fermentation Mainly in static conditions for biomedical applications | Submerged fermentation Batch and Fed-Batch strategies | Submerged and solid-state fermentation |
| Downstream processing | Extracellular polymer. Easy, cheap purification, isolation, and alkali treatment | Intracellular polymers. Costly purification, cell lysis, release, and polymer isolation | Extracellular polymer. Precipitation by chelation, solubility reduction or filtration |
| Producing Species | WHC 1 (%) | E2 (MPa) | σt 3 (MPa) | εb 4 (%) | CIXRD 5 | Ref. |
|---|---|---|---|---|---|---|
| K. xylinus | ||||||
| B12068 | NR | 3.73 | NR | 12.5 | 0.65 | [58] |
| ATCC 10245 | 5400 | 2.87 | 0.36 | 18.6 | NR | [59] |
| NBRC 13693 | 16,500 | 3.1 | 0.62 | 18.7 | NR | [59] |
| ATCC 53524 | NR | 9.09 | 1.68 | 26.9 | NR | [60] |
| ATCC 23760 | 18,000 | 1.5 | 46.9 | 2.5 | 0.85 | [61] |
| K. medellinensis | ||||||
| ID13488 | 7218 | 6.75 | 50 | NR | 0.89 | [62] |
| K. sucrofermentans | ||||||
| ATCC 700178 | 52,600 | 1.1 | 0.15 | 20.72 | NR | [59] |
| DSM 15973 | 260 | NR | NR | NR | 0.87 | [63] |
| K. hansenii | ||||||
| ATCC 23769 | 45,000 | 1.26 | 0.12 | 17.9 | NR | [59] |
| GA2016 | 700 | NR | NR | NR | 0.87 | [64] |
| K. rhaeticus | ||||||
| AF-1 | 14,000 | 3.2 | 46.9 | 1.5 | 0.89 | [61] |
| Polymer | Copolymer Content (%) | E1 (MPa) | σt 2 (MPa) | εb 3 (%) | Tm4 (° C) | Tg5 (° C) | Ref. |
|---|---|---|---|---|---|---|---|
| PHA (R)-alkanoic | |||||||
| P(3HB) | 100 | 3500–4000 | 40 | 3–8 | 173–180 | 5–9 | [68] |
| P(4HB) | 100 | 70 | 50 | 1000 | 60 | −51 | [69] |
| P(3HB-co-3HV) | (97:3) (91:9) (75:25) | 2900 1900 700 | 38 37 30 | - - - | 170 162 137 | - - - | [70] |
| P(3HB-co-4HB) | (97:3) | NR 6 | 28 | 45 | 166 | [71] | |
| (90:10) | NR | 24 | 252 | 159 | |||
| P(3HB-co-3HHx) | (88:12) | 1286 | 18.3 | 3.6 | 170 | [72] | |
| (45:55) | 1207 | 21.6 | 4.1 | 167 | |||
| P(3HD-co-3HDD) | (15.7:84.3) | 103.13 | 5.24 | 88.30 | 77.62 | 32.49 | [67] |
| (R)-Aromatic | |||||||
| P(3HPhHHx) | 100 | NR | NR | NR | NR | −1.3 | [73] |
| P(3HB-co-3H3PhP) | (89.5:8.9) | NR | NR | NR | 135, 149 | 14.6 | [74] |
| P(3HDD-co-3H5PhV) | (97.1:2.9) | 93.9 | 2 | 37.38 | 81 | −33.4 | [75] |
| (68.1:31.9) | 48.7 | 3.15 | 32.2 | 75.84 | −35.2 | [75] | |
| (R)-Nitrogen | |||||||
| P(3H-p-nitroPV-co-3HN) | (4–7% N) | NR | NR | NR | 56.4 | −35.9, 28.7 | [76] |
| (R)-Sulfur | |||||||
| PHACOS | (16.5 to 77% thiolated side chains) | NR | NR | NR | -- | −5 | [77] |
| (R)-Halogenated | |||||||
| P(3HB)-Cl | (22% Cl) | NR | NR | NR | 134 | 2 | [78] |
| P(FHB-co-HB)-F | (7% F) | NR | NR | NR | 160.5 173.6 | −0.8 | [79] |
| Waste Origin | Strain | Productivity (g L−1 day−1) | Type of Polymer | Ref. |
|---|---|---|---|---|
| Cheese whey | ||||
| Alcaligenes latus | 2.64 | P(3HB) | [112] | |
| Caulobacter segnis | 4.56 | P(3HB) | [97] | |
| Haloferax mediterranei | 4.04 | P(3HB) | [113] | |
| C. necator DSM 545 (recombinant strain) | 0.82 | P(3HB) | [114] | |
| K. sucrofermentans DSM No 15973 | 2.7 | BC | [115] | |
| Acetobacter strain ITDI 2.1 (recombinant strain) | 0.1 | BC | [116] | |
| Cane molasses | ||||
| Pseudomonas | 2.17 | (P3HO-co-3-HHx) | [117] | |
| B. megaterium BA-019 | 30.48 | P(3HB) | [102] | |
| Mixed culture | 10.93 | mcl-PHA | [118] | |
| A. xylinum BPR 2001 | 1.77 | BC | [103] | |
| A. pasteurianus RSV-4 yielded | 0.51 | BC | [119] | |
| B. subtilis NX-2 | 12.96 | PGA | [104] | |
| B. subtilis NX-2 | 25.92 | PGA | [105] | |
| Crops | ||||
| Vinasse | H. mediterranei | 5.04 | P(3HB-co-3HV) | [120] |
| H. marismortui | 0.48 | P(3HB) | [120] | |
| Sugarcane bagasse | C. necator | 3.16 | P(3HB) | [121] |
| B. cepacia IPT 048 | 11.28 | P(3HB) | [122] | |
| Grape pomace | P. putida KT2440 | 1.2 | P(3HO-co-3-HHx) | [123] |
| Waste beer yeast | G. hanseii CGMCC 3917 | 0.514 | BC | [124] |
| Apple pomace | K. medenillensis ID13488 | 0.177 | BC | [62] |
| Potato peel | G. xylinum ATCC 10245 | 0.65 | BC | [100] |
| Citrus peel | K. xilynus CICC No 10529 | 0.712 | BC | [125] |
| Orange juice | A. xylinum NBRC 13693 | 0.421 | BC | [126] |
| Litchi extract | G. xylinus CH001 | 0.18 | BC | [127] |
| Citrus waste | K. sucrofermentans DSM 15973 | 0.515 | BC | [128] |
| Coffee cherry husk | G. hanseii UAC 09 | 0.547 | BC | [129] |
| Olive oil mills | G. sacchari | 0.212 | BC | [130] |
| Tomato juice | A. pasteurianus RSV-4 | 0.68 | BC | [119] |
| Rice straw | B. cepcecea USM (JMC 15050) | 1.95 | P(3HB) | [99] |
| B. subtilis NX-2 | 0.87 | PGA | [92] | |
| Soybean meal and Corn straw | B. amiloliquefaciens JX-6 | 116 (g kg−1) | PGA | [91] |
| Soybean straw | B. amiloliquefaciens NX-2S | 65.79 (g kg−1) | PGA | [131] |
| Household and industrial oils | ||||
| Sesame | C. necator H16 | 31.32 | P(3HB) | [107,109] |
| Sunflower | C. necator H16 | 35.04 | P(3HB) | [107,109] |
| Canola | Wautersia eutropha ATCC 17699 | 10.96 | P(3HB) | [132] |
| Cooking | P.aeruginosa 42A2 | 0.76 | P(3HB) | [110] |
| Palm | C. necator H16 | 4.2 | P(3HB) | [108] |
| Rapeseed | K. xylinus DSM 46602 | 6 | BC | [111] |
| Waste water | ||||
| Fruit processing | Halomonas i4786 | 1.8 | P(3HB) | [133] |
| Alcohol distillery | K. saccharivorans BC1 | 0.155 | BC | [134] |
| Rice wine distillery | G. xilynus BCRC12334 | 1 | BC | [135] |
| Lipid fermentation | G. xylinus CH001 | 0.1 | BC | [136] |
| Hot water wood sugar extraction | A. xylinus 23769 | 0.019 | BC | [137] |
| Butanol fermentation | G. xylinus CH001 | 0.17 | BC | [138] |
| Jujube | G. xylinys CGMC2955 | 0.375 | BC | [139] |
| WW anaerobically fermented to VFAs | ||||
| Municipal | Activated sludge | 1.37 | P(3HB-co-3HV) | [140] |
| Paperboard mill | Activated sludge | 3 | P(3HB-co-3HV) | [141] |
| Candy factory f | Plasticicumulans acidivorans | 0.05 (gPHA/gVSS) | P(3HB-co-3HV) | [142] |
| Urban waste | Activated sludge | 0.65 (gPHA/gVSS) | P(3HB-co-3HV) | [143] |
| Sewage sludge and municipal | Activated sludge | 8.64 | scl-PHA | [144] |
| Blend Composition | Key Features | Ref. | |
|---|---|---|---|
| PHA | P(3HB)/P(3HB-co-3HHx) | Better cell biocompatibility on blend polymer scaffolds of PHBHHx/PHB. PHB crystallization degree decreased with increasing PHBHHx content. | [227,228] |
| P(3HB)/P(3HO-co-3HHx) | Higher Young’s modulus, tensile strength, thermal stability, tailorable biodegradability, and improved biocompatibility with HMEC-1 cells when compared with P(3HO-co-3HHx) films. | [229] | |
| P(3HB)/lignin | Lignin contents ≤30 wt % reduce the crystallinity of PHB. At higher lignin contents, the blends have higher dynamic storage and loss modulus than pure PHB. | [230] | |
| P(3HB-co-3HV)/PLA | Blends were immiscible for all compositions. Improved thermal stability and significant ductile plastic deformation. | [231] | |
| mcl-PHA/PLA | Improved elongation at break, lower crystallization, and higher biocompatibility. | [232] | |
| P(3HB-co-3HHx)/polycaprolactone (PCL) | Improved degradation and mechanical and biocompatibility properties. | [233] | |
| BC | BC/poly(methylmethacrylate) (PMMA) | Improved mechanical properties and biocompatibility. | [234] |
| BC/antimicrobial PHA (PHACOS) | Antibacterial activity against S. aureus. | [235] | |
| γ-PGA | γ-PGA/chitosan (CS) | Improved hydrophilic, cytocompatibility, and mechanical properties. | [236] |
| PGA/gelatin | Gelatin stabilizes PGA molecules. Improved mechanical properties and biocompatibility with vascular cells. | [237] | |
| PCL/PGA | Hydrophilicity and water uptake of the nanofibrous scaffolds increased with PGA content. | [238] | |
| Polymer | Grafting Molecule | Key Features | Ref. |
| P(3HO-co-3HU) | Thiolation with Jeffamine® | Water-soluble amphiphilic copolymers with thermoresponsive behavior. | [239] |
| unsaturated PHA | Epoxidation | Epoxidation sped up the crosslinking reaction and resulted in a strong, tear-resistant film with increased tensile strength and Young’s modulus. | [240] |
| P(3HO), P(3HB) | Chlorination | Increased Tg and Tm, while the same procedure on P(3HB) translates to a decrease in Tm and increase in Tg. Modulation of the hydrophobicity of the polymers. | [78] |
| P(3HB) | Alkali treatment with NaOH | Treatment of P(3HB) surfaces with NaOH enhanced proliferation of human osteoblasts and inhibited S. aureus growth | [241] |
| PHB | PVA | Decreased crystallinity and enhanced biodegradability of the final polymer. | [242] |
| unsaturated PHA | PNIPAm oligomers | Improved surface hydrophilicity and thermoresponsive properties. Good biocompatibility for cell growth and thermoresponsive cell detachment ability. | [243] |
| PHA | Fibronectin active fragment (GRGDS peptide) | Exhibited cell adhesiveness and improved biocompatibility. | [244] |
| P(3HB-co-3HV) | RGD-containing peptides | Increased hydrophilicity of the surface of the film and improved cellular compatibility. | [245] |
| P(3HO) | Vinyl imidazole | Increased hydrophilicity and biocompatibility and showed antibacterial activity against E. coli and S. aureus. | [246] |
| P(3HB) | Different amino compounds | Amino-PHB polymers showed antibacterial, antioxidant, and anticancer activities. PHB-ethylendiamine displayed better growth-inhibitory antibacterial activity against S. aureus, K. pneumoniae, P. aeruginosa, and E. coli. PHB-piperazine showed a potent anticancer effect against in vivo Ehrlich ascitic carcinoma-bearing mice. | [247] |
| P(3HB), P(3HB-co-3HV) | CS and CS oligosaccharides | Decreased thermal stability of the chitosan backbone. | [248] |
| P(3HO), P(3HB-co-3HV) | CS | Solubilization of chitosan-g-PHA graft depends on grafting percentage. | [249] |
| P(3HB) | Ar plasma | Increased surface polarity; improved cell adhesion, proliferation, and spreading homogeneity on the PHB surface | [250] |
| P(3HB) | O2 plasma | Enhanced hydrophilicity. Ability to directly immobilize T4 bacteriophages, resulting in an antimicrobial material against E. coli. | [251] |
| BC | Aminoalkyl groups | Improved mechanical and thermal properties. Antimicrobial properties against S. aureus and E. coli and was nontoxic to human adipose-derived mesenchymal stem cells. | [252] |
| Oligo peptides, glycyl-L-glutamine or glycyl-glycyl-glycine | Enhanced its interfacial wettability, boosted mineralization induction, and improved affinity between polymeric and mineral phases. | [253] | |
| RGDC peptides and gentamicin | Growth inhibition of S. mutans and promoted fibroblast adhesion and proliferation. | [254] | |
| Amoxicillin (AM) | Good porosity and swelling behaviors. Antibacterial activities against E. coli, C. albicans, and S. aureus and nontoxic to HEK293 cells | [255] | |
| AM | Good stability with a slight reduction in swelling capabilities. pH responsiveness with an increase in drug swelling and release at higher pH. | [256] | |
| CS | Better uniformity of nanosized fibrils, with better acid and temperature stability. Enhanced BC dispersion. | [257] | |
| N2 plasma | Increased porosity and the number of functional groups on the surface of BC, which improved the cell adhesion. | [258] | |
| O2 and N2 plasmas | Increased surface hydrophilicity due to the incorporation of carbon–oxygen and amide and amino groups. | [259] | |
| CF4 plasma | Surface presented hydrophobic properties and potential to promote cell adhesion and proliferation due to greater adsorption of proteins on the BC surface. | [259] | |
| PGA | PCL | Improved shear elasticity and compressive strength. Provided effective protection for femoral condyle and tibial plateau cartilage when applied in a rabbit model and regenerated a meniscus-like tissue. | [260] |
| Benzyl groups | Novel electrospun biopolymer exhibited rapid shrinkage upon induction by heat or a series of solvents. | [261] | |
| Phenylalanine (Phe) or leucine (Leu) | Production of bionanoparticles derived from γ-PGA and phenylalanine ethyl ester, with excellent water dispersibility, 200 nm diameters, and surface chemical functionality of carboxyl groups, | [262] | |
| β-sheet peptides | Hydrogel stiffness can be controlled by changing the β-sheet peptide graft density, the bulk hydrogel concentration, and the ratio of covalently coupled and free peptides. Additional functionality can be incorporated into this self-healing hybrid hydrogel. | [263] | |
| Material | Cross-Linking Agent | Key Features | Ref. |
| BC BC-CS membranes | Tripolyphosphate (TPP) | Optimum water vapor permeability. BC-CS exhibited local and peripheral inhibition of bacterial growth. The presence of ciprofloxacin effectively produced a stronger inhibition effect on both tested bacteria, P. aeruginosa and S. aureus | [264] |
| BC/CS blends | Glutaraldehyde | Showed flexibility, high thermal stability, and high mechanical properties. Showed antibacterial properties against tested Gram-positive and Gram-negative bacteria. | [265] |
| Functionalization of BC with ε-poly-L-Lysine (ε-PLL) | Carbodiimide chemistry | Preserved the good structural and mechanical properties of BC. Inhibited growth of S. epidermidis on the membranes and was cytocompatible with human fibroblasts. | [266] |
| Unsaturated copolyester (PHBU) | Thiol-ene click chemistry | Enhanced tensile strength without affecting cytotoxicity and biocompatibility towards human mesenchymal stem cells. | [267] |
| Unsaturated PHA copolymer poly[(R)-3-hydroxyundecanoate-co-(R)-3-hydroxy-10-undecenoate] P(HU10U) | Polyethylene glycol dithiol (PDT) | Swelling behavior in different solvents; mechanical and morphological properties could be tuned by varying the ratio of P(HU10U) to PDT. Good biocompatibility. | [268] |
| PGA-conjugated cysteamine (PGA-SH) and methacrylate-PGA (PGA-GMA) | Michael-addition reactions | Mechanical properties, porous structure, swelling, and degradation process of the hydrogels could be controlled by adjusting modified PGA polymer component. Exhibited good biocompatibility and high stability. Promoted chondrogenesis of loaded BMSCs and facilitated cartilage reconstruction in the defected area in a rabbit auricular cartilage defect model. | [157] |
| Hydrogels of PGA | N,N,N-trimethyl-3-[(2-methylacryloyl)amino]propan-1-aminium (METH) | Maintained a stable form during the nine weeks of the study. Useful for preparing particles. | [269] |
| Cured Material | Key Features | Ref. | |
| BC crosslinked with citric acid | Improved rehydration capacity; showed higher porosity, wettability, and water swelling. | [270] | |
| PHA combined with segments of polyurethane (PHP), telechelic-hydroxylated polyhydroxyalkanoate (PHA-diols), and polyethylene glycol (PEG) | Good shape–memory effect (SME) and rapid recovery. Possess thermo- and water-responsive properties, properties of triggering shape-morphing, enabling self-folding and self-expansion of shapes into three-dimensional (3D) scaffolds. | [271] | |
| Light cured methacrylated PGA nanoparticle-created hydrogel system (PGA nanogel) | Good swelling and mechanical properties. Good antibiotic release behavior. Good biocompatibility. | [272] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blanco, F.G.; Hernández, N.; Rivero-Buceta, V.; Maestro, B.; Sanz, J.M.; Mato, A.; Hernández-Arriaga, A.M.; Prieto, M.A. From Residues to Added-Value Bacterial Biopolymers as Nanomaterials for Biomedical Applications. Nanomaterials 2021, 11, 1492. https://doi.org/10.3390/nano11061492
Blanco FG, Hernández N, Rivero-Buceta V, Maestro B, Sanz JM, Mato A, Hernández-Arriaga AM, Prieto MA. From Residues to Added-Value Bacterial Biopolymers as Nanomaterials for Biomedical Applications. Nanomaterials. 2021; 11(6):1492. https://doi.org/10.3390/nano11061492
Chicago/Turabian StyleBlanco, Francisco G., Natalia Hernández, Virginia Rivero-Buceta, Beatriz Maestro, Jesús M. Sanz, Aránzazu Mato, Ana M. Hernández-Arriaga, and M. Auxiliadora Prieto. 2021. "From Residues to Added-Value Bacterial Biopolymers as Nanomaterials for Biomedical Applications" Nanomaterials 11, no. 6: 1492. https://doi.org/10.3390/nano11061492
APA StyleBlanco, F. G., Hernández, N., Rivero-Buceta, V., Maestro, B., Sanz, J. M., Mato, A., Hernández-Arriaga, A. M., & Prieto, M. A. (2021). From Residues to Added-Value Bacterial Biopolymers as Nanomaterials for Biomedical Applications. Nanomaterials, 11(6), 1492. https://doi.org/10.3390/nano11061492








