Predicting Associations of miRNAs and Candidate Gastric Cancer Genes for Nanomedicine
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. The Characteristics of the miRNA Interactions with 5′UTR mRNAs of Gastric Cancer Candidate Genes
3.2. The Characteristics of the miRNA Interactions with CDS mRNAs of Gastric Cancer Candidate Genes
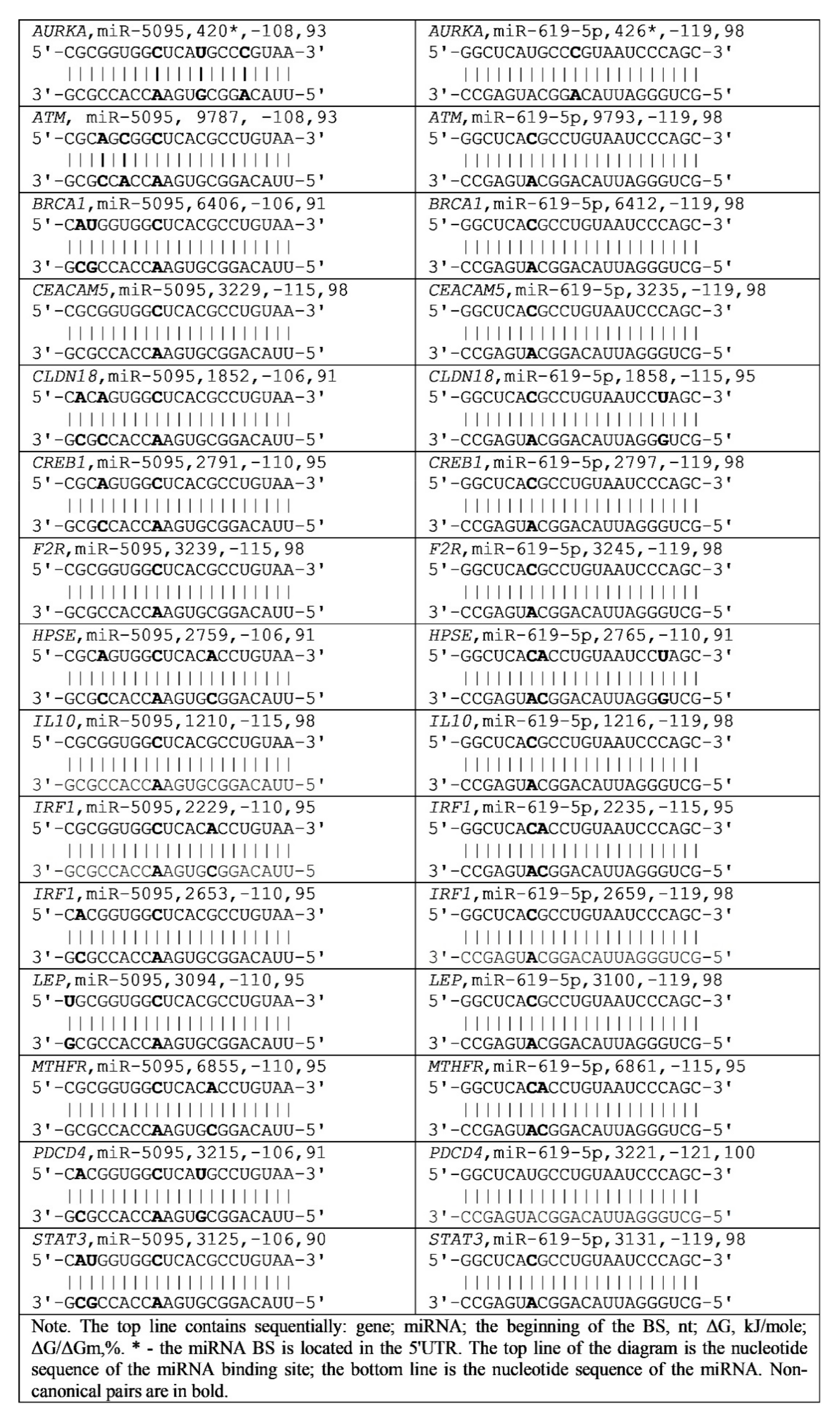
3.3. The Characteristics of miRNA Interactions with 3′UTR mRNAs of Gastric Cancer Candidate Genes
 A) and pyrimidine for pyrimidine (C
A) and pyrimidine for pyrimidine (C  U), which have little effect on the free energy of miRNA interaction with mRNA. The data presented indicate the stability of the interactions of these miRNAs with the mRNA of candidate genes for gastric cancer.
U), which have little effect on the free energy of miRNA interaction with mRNA. The data presented indicate the stability of the interactions of these miRNAs with the mRNA of candidate genes for gastric cancer.4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation. GPB 2009, 7, 147–154. [Google Scholar] [CrossRef] [Green Version]
- Correia de Sousa, M.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Heikkinen, L.; Wang, C.; Yang, Y.; Sun, H.; Wong, G. Trends in the development of miRNA bioinformatics tools. Brief. Bioinform. 2019, 20, 1836–1852. [Google Scholar] [CrossRef] [Green Version]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Stavast, C.J.; Erkeland, S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tafrihi, M.; Hasheminasab, E. MiRNAs: Biology, Biogenesis, their Web-based Tools, and Databases. MicroRNA 2019, 8, 4–27. [Google Scholar] [CrossRef]
- Mishra, S.; Yadav, T.; Rani, V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit. Rev. Oncol. Hematol. 2016, 98, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Vishnoi, A.; Rani, S. MiRNA Biogenesis and Regulation of Diseases: An Overview. Methods Mol. Biol. 2017, 1509, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. J. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Condrat, C.; Thompson, D.; Barbu, M.; Bugnar, O.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.; Voinea, S. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hrovatin, K.; Kunej, T. Classification of miRNA-related sequence variations. Epigenomics 2018, 10, 463–481. [Google Scholar] [CrossRef]
- Bandara, K.V.; Michael, M.Z.; Gleadle, J.M. MicroRNA Biogenesis in Hypoxia. MicroRNA 2017, 6, 80–96. [Google Scholar] [CrossRef]
- Kabekkodu, S.P.; Shukla, V.; Varghese, V.K.; D’Souza, J.; Chakrabarty, S.; Satyamoorthy, K. Clustered miRNAs and their role in biological functions and diseases. Biol. Rev. Camb. Philos. Soc. 2018, 93, 1955–1986. [Google Scholar] [CrossRef]
- Chen, X.; Li, X.; Peng, X.; Zhang, C.; Liu, K.; Huang, G.; Lai, Y. Use of a Four-miRNA Panel as a Biomarker for the Diagnosis of Stomach Adenocarcinoma. Dis. Markers 2020, 2020, 8880937. [Google Scholar] [CrossRef] [PubMed]
- Backes, C.; Meese, E.; Keller, A. Specific miRNA Disease Biomarkers in Blood, Serum and Plasma: Challenges and Prospects. Mol. Diagn. Ther. 2016, 20, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Mellis, D.; Caporali, A. MicroRNA-based therapeutics in cardiovascular disease: Screening and delivery to the target. Biochem. Soc. Trans. 2018, 46, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; She, J.; Chen, X.; Zhang, Q.; Zhang, X.; Wang, Y.; Ye, J.; Shi, J.; Tao, J.; Feng, M.; et al. Exosomal miR-196a-1 promotes gastric cancer cell invasion and metastasis by targeting SFRP1. Nanomedicine 2019, 14, 2579–2593. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Zhang, S.; Yang, L.; Wei, W.; Gao, J.; Guo, N.; Wu, F. Serum miR-101-3p combined with pepsinogen contributes to the early diagnosis of gastric cancer. BMC Med. Genet. 2020, 21, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghafouri-Fard, S.; Vafaee, R.; Shoorei, H.; Taheri, M. MicroRNAs in gastric cancer: Biomarkers and therapeutic targets. Gene 2020, 757, 144937. [Google Scholar] [CrossRef] [PubMed]
- Pewarchuk, M.E.; Barros-Filho, M.C.; Minatel, B.C.; Cohn, D.E.; Guisier, F.; Sage, A.P.; Marshall, E.A.; Stewart, G.L.; Rock, L.D.; Garnis, C.; et al. Upgrading the Repertoire of miRNAs in Gastric Adenocarcinoma to Provide a New Resource for Biomarker Discovery. Int. J. Mol. Sci. 2019, 20, 5697. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Li, X.; Guo, Z.; Zhou, F. MicroRNA-381 regulates the growth of gastric cancer cell by targeting TWIST1. Mol. Med. Rep. 2019, 20, 4376–4382. [Google Scholar] [CrossRef]
- Ivashchenko, A.; Berillo, O.; Pyrkova, A.; Niyazova, R.; Atambayeva, S. The properties of binding sites of miR-619-5p, miR-5095, miR-5096 and miR-5585-3p in the mRNAs of human genes. Biomed. Res. Int. 2014, e8. [Google Scholar] [CrossRef]
- Ivashchenko, A.; Berillo, O.; Pyrkova, A.; Niyazova, R. Binding sites of miR-1273 family on the mRNA of target genes. Biomed. Res. Int. 2014, e11. [Google Scholar] [CrossRef] [Green Version]
- Atambayeva, S.; Niyazova, R.; Ivashchenko, A.; Pyrkova, A.; Pinsky, I.; Akimniyazova, A.; Labeit, S. The Binding Sites of miR-619-5p in the mRNAs of Human and Orthologous Genes. BMC Genom. 2017, 18, 428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yurikova, O.Y.; Aisina, D.E.; Niyazova, R.E.; Atambayeva, S.A.; Labeit, S.; Ivashchenko, A. The Interaction of miRNA-5p and miRNA-3p with the mRNAs of Orthologous Genes. Mol. Biol. 2019, 53, 692–704. [Google Scholar] [CrossRef]
- Kondybayeva, A.M.; Akimniyazova, A.N.; Kamenova, S.U.; Ivashchenko, A.T. The characteristics of miRNA binding sites in mRNA of ZFHX3 gene and its orthologs. Vavilov. J. Genet. Breed. 2018, 22, 438–444. [Google Scholar] [CrossRef] [Green Version]
- Aisina, D.; Niyazova, R.; Atambayeva, S.; Ivashchenko, A. Prediction of clusters of miRNA binding sites in mRNA candidate genes of breast cancer subtypes. PeerJ 2019, 7, e8049. [Google Scholar] [CrossRef]
- Kondybayeva, A.; Akimniyazova, A.; Kamenova, S.; Duchshanova, G.; Aisina, D.; Goncharova, A.; Ivashchenko, A. Prediction of miRNA interaction with mRNA of stroke candidate genes. Neurol. Sci. 2020, 41, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Mukushkina, D.; Aisina, D.; Pyrkova, A.; Ryskulova, A.; Labeit, S.; Ivashchenko, A. In Silico Prediction of miRNA Interactions with Candidate Atherosclerosis Gene mRNAs. Front. Genet. 2020, 11, 605054. [Google Scholar] [CrossRef]
- Luo, S.S.; Liao, X.W.; Zhu, X.D. Genome-wide analysis to identify a novel microRNA signature that predicts survival in patients with stomach adenocarcinoma. J. Cancer 2019, 10, 6298–6313. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Xie, J.; Liu, Y.; Tang, F.; Long, Z.; Wang, Y.; Luo, J.; Li, J.; Li, G. MicroRNA expression profiling and target gene analysis in gastric cancer. Medicine 2020, 99, e21963. [Google Scholar] [CrossRef]
- Mao, Q.Q.; Chen, J.J.; Xu, W.J.; Zhao, X.Z.; Sun, X.; Zhong, L. miR-92a-3p promotes the proliferation and invasion of gastric cancer cells by targeting KLF2. J. Biol. Regul. Homeost. Agents 2020, 34, 1333–1341. [Google Scholar] [CrossRef]
- Zhang, X.; Zheng, P.; Li, Z.; Gao, S.; Liu, G. The Somatic Mutation Landscape and RNA Prognostic Markers in Stomach Adenocarcinoma. Onco Targets Ther. 2020, 13, 7735–7746. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhang, F.; Dong, L.; Wu, H.; Xu, J.; Li, H.; Wang, J.; Zhou, Z.; Liu, C.; Wang, Y.; et al. SIDT1-dependent absorption in the stomach mediates host uptake of dietary and orally administered microRNAs. Cell Res. 2020, 1–12. [Google Scholar] [CrossRef]
- Gu, E.; Song, W.; Liu, A.; Wang, H. SCDb: An integrated database of stomach cancer. BMC Cancer 2020, 20, 490. [Google Scholar] [CrossRef]
- Qu, X.; Zhao, L.; Zhang, R.; Wei, Q.; Wang, M. Differential microRNA expression profiles associated with microsatellite status reveal possible epigenetic regulation of microsatellite instability in gastric adenocarcinoma. Ann. Transl. Med. 2020, 8, 484. [Google Scholar] [CrossRef]
- Feng, X.; Zhu, M.; Liao, B.; Tian, T.; Li, M.; Wang, Z.; Chen, G. Upregulation of miR-552 Predicts Unfavorable Prognosis of Gastric Cancer and Promotes the Proliferation, Migration, and Invasion of Gastric Cancer Cells. Oncol. Res. Treat. 2020, 43, 103–111. [Google Scholar] [CrossRef]
- Zhou, J.; Su, M.; Zhang, H.; Wang, J.; Chen, Y. miR-539-3P inhibits proliferation and invasion of gastric cancer cells by targeting CTBP1. Int. J. Clin. Exp. Pathol. 2019, 12, 1618–1625. [Google Scholar]
- Ishikawa, D.; Yoshikawa, K.; Takasu, C.; Kashihara, H.; Nishi, M.; Tokunaga, T.; Higashijima, J.; Shimada, M. Expression Level of MicroRNA-449a Predicts the Prognosis of Patients with Gastric Cancer. Anticancer Res. 2020, 40, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Guo, H.; Wu, X.; Li, J.; Duan, X.; Ba, Q.; Wang, H. Epigenetic silencing of microRNA-204 by Helicobacter pylori augments the NF-κB signaling pathway in gastric cancer development and progression. Carcinogenesis 2020, 41, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, G.; Sun, S.; Qin, J. miR-1294 alleviates epithelial-mesenchymal transition by repressing FOXK1 in gastric cancer. Genes Genom. 2020, 42, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Chen, Z.; Jiang, Z.; Huang, T.; Hu, J.; Luo, P.; Zhang, H.; Huang, M.; Huang, L.; Chen, Y.; et al. MiR-122-5p suppresses the proliferation, migration, and invasion of gastric cancer cells by targeting LYN. Acta Biochim. Biophys. Sin. 2020, 52, 49–57. [Google Scholar] [CrossRef]
- Lv, H.; Hou, H.; Lei, H.; Nie, C.; Chen, B.; Bie, L.; Han, L.; Chen, X. MicroRNA-6884-5p Regulates the Proliferation, Invasion, and EMT of Gastric Cancer Cells by Directly Targeting S100A16. Oncol. Res. 2020, 28, 225–236. [Google Scholar] [CrossRef]
- Wang, Z.; Yao, L.; Li, Y.; Hao, B.; Wang, M.; Wang, J.; Gu, W.; Zhan, H.; Liu, G.; Wu, Q. miR-337-3p inhibits gastric tumor metastasis by targeting ARHGAP10. Mol. Med. Rep. 2020, 21, 705–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Londin, E.; Loher, E.; Telonis, A.G.; Quann, K.; Clark, P.; Jing, Y.; Hatzimichael, E.; Kirino, Y.; Honda, S.; Lally, M.; et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc. Natl. Acad. Sci. USA 2015, 112, 1106–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivashchenko, A.; Berillo, O.; Pyrkova, A.; Niyazova, R.; Atambayeva, S. MiR-3960 binding sites with mRNA of human genes. Bioinformation 2014, 10, 423–427. [Google Scholar] [CrossRef] [Green Version]
- Ivashchenko, A.T.; Pyrkova, A.Y.; Niyazova, R.Y.; Alybayeva, A.; Baskakov, K. Prediction of miRNA binding sites in mRNA. Bioinformation 2016, 12, 237–240. [Google Scholar] [CrossRef]
- Leontis, N.B.; Stombaugh, J.; Westhof, E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002, 30, 3497–3531. [Google Scholar] [CrossRef]
- Kool, E.T. Hydrogen bonding, base stacking, and steric effects in DNA replication. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Lemieux, S.; Major, F. RNA canonical and non-canonical base pairing types: A recognition method and complete repertoire. Nucleic Acids Res. 2002, 30, 4250–4263. [Google Scholar] [CrossRef] [Green Version]
- Garg, A.; Heinemann, U. A novel form of RNA double helix based on G·U and C·A+ wobble base pairing. RNA 2018, 24, 209–218. [Google Scholar] [CrossRef] [Green Version]

| Gene; NX | miRNA | Start of Site, nt | ΔG, kJ/mole | ΔG/ΔGm, % | Length, nt |
|---|---|---|---|---|---|
| ARID1A; 19.5 | ID02106.3p-miR | 136 | −123 | 89 | 23 |
| ID01778.3p-miR | 140 | −134 | 90 | 24 | |
| ID00296.3p-miR | 141, 166 | −138 | 88 | 25 | |
| miR-6081 | 143 | −125 | 89 | 24 | |
| ID01702.3p-miR | 147 | −134 | 89 | 24 | |
| ID00465.5p-miR | 148 | −113 | 93 | 20 | |
| ID01377.3p-miR | 152 | −117 | 92 | 20 | |
| ID02592.5p-miR | 164, 167 | −123 | 89 | 23 | |
| miR-3960 | 167 | −115 | 92 | 20 | |
| ID03065.3p-miR | 172 | −115 | 92 | 21 | |
| E2F1; 3.4 | ID02574.3p-miR | 84 | −115 | 93 | 20 |
| ID02052.5p-miR | 85 | −149 | 100 | 24 | |
| ID01873.3p-miR | 87 | −123 | 94 | 21 | |
| miR-3960 | 90 | −115 | 92 | 20 | |
| ID00071.3p-miR | 90 | −117 | 92 | 20 | |
| ID00722.5p-miR | 90 | −113 | 93 | 20 | |
| ID02064.5p-miR | 95 | −132 | 91 | 23 | |
| ODC1; 19.2 | ID00756.3p-miR | 9 | −129 | 94 | 23 |
| ID01804.3p-miR | 13 | −132 | 90 | 23 | |
| ID02187.5p-miR | 14 | −123 | 89 | 23 | |
| ID00457.3p-miR | 15, 21 | −123, −125 | 91, 92 | 22 | |
| ID02084.3p-miR | 17 | −140 | 93 | 24 | |
| ID02064.5p-miR | 17 ÷ 23(3) | −129 ÷ −140 | 90 ÷ 97 | 23 | |
| miR-3960 | 18 ÷ 21(3) | −115 | 92 | 20 | |
| ID01652.3p-miR | 19 | −125 | 89 | 23 | |
| ID02538.3p-miR | 19 | −123 | 92 | 22 | |
| ID01702.3p-miR | 19, 22 | −134, −142 | 89, 94 | 24 | |
| ID02229.3p-miR | 21 | −121 | 92 | 21 | |
| ID02499.3p-miR | 21 | −119 | 92 | 21 | |
| ID01157.5p-miR | 22 | −117 | 93 | 20 | |
| ID01377.3p-miR | 23 | −121 | 95 | 20 | |
| ID00061.3p-miR | 24 | −125 | 91 | 22 | |
| PIK3CA; 10.6 | ID00296.3p-miR | 1 | −134 | 85 | 25 |
| ID01190.5p-miR | 4 | −140 | 92 | 24 | |
| ID01702.3p-miR | 4 | −140 | 93 | 24 | |
| ID01895.5p-miR | 4 | −132 | 89 | 24 | |
| ID01641.3p-miR | 5 | −127 | 86 | 24 | |
| ID00966.5p-miR | 6 | −136 | 90 | 24 | |
| ID00030.3p-miR | 7 | −121 | 90 | 22 | |
| ID02294.5p-miR | 7 | −125 | 86 | 24 | |
| ID01804.3p-miR | 8 | −134 | 91 | 23 | |
| ID01873.3p-miR | 8 | −123 | 94 | 21 | |
| ID02064.5p-miR | 9 | −127 | 88 | 23 | |
| ID02084.3p-miR | 9 | −129 | 86 | 24 | |
| TBC1D9; 10.0 | ID01895.5p-miR | 125 | −134 | 90 | 24 |
| ID02187.5p-miR | 130 | −127 | 92 | 23 | |
| ID03229.5p-miR | 130, 133 | −121 | 90 | 22 | |
| ID01041.5p-miR | 132 | −132 | 90 | 24 | |
| ID01702.3p-miR | 136 | −140 | 93 | 24 | |
| ID02084.3p-miR | 137 | −136 | 90 | 24 | |
| ID00457.3p-miR | 138 | −125 | 92 | 22 |
| Gene; NX | miRNA | Start of Site, nt | ΔG, kJ/mole | ΔG/ΔGm, % | Length, nt |
|---|---|---|---|---|---|
| ARID1A; 19.5 | ID02052.5p-miR | 410 | −132 | 89 | 24 |
| ID00522.5p-miR | 410 | −127 | 91 | 23 | |
| ID02187.5p-miR | 411 | −127 | 92 | 23 | |
| ID02692.5p-miR | 413 | −127 | 90 | 23 | |
| ID00457.3p-miR | 415 | −125 | 92 | 22 | |
| ID02064.5p-miR | 417 | −134 | 93 | 23 | |
| ID02084.3p-miR | 418 | −138 | 92 | 24 | |
| ID02538.3p-miR | 420 | −125 | 94 | 22 | |
| ID01704.5p-miR | 471 | −123 | 89 | 23 | |
| ID02761.3p-miR | 487, 493 | −134, −140 | 90, 94 | 24 | |
| ID00756.3p-miR | 843 | −125 | 91 | 23 | |
| ID02294.5p-miR | 851 | −129 | 88 | 24 | |
| ID00061.3p-miR | 852 | −125 | 91 | 22 | |
| E2F1; 3.4 | ID02051.3p-miR | 291 | −153 | 100 | 24 |
| ID03448.3p-miR | 292 | −123 | 91 | 22 | |
| ID01157.5p-miR | 295 | −117 | 93 | 20 | |
| MAPK1; 17.2 | ID03332.3p-miR | 243 | −134 | 90 | 24 |
| ID01310.3p-miR | 244 | −121 | 92 | 22 | |
| ID00798.3p-miR | 246 | −136 | 91 | 24 | |
| ID01546.5p-miR | 246 | −132 | 90 | 24 | |
| TERT; 0.4 | ID01098.3p-miR | 3224 | −125 | 89 | 24 |
| ID01338.5p-miR | 3236 | −132 | 91 | 24 | |
| ID01816.3p-miR | 3237 | −138 | 92 | 24 | |
| VEGFC; 0.4 | ID02052.5p-miR | 498 | −132 | 89 | 24 |
| ID02187.5p-miR | 500 | −123 | 89 | 23 | |
| ID01041.5p-miR | 501 | −132 | 90 | 24 | |
| ID01873.3p-miR | 501 | −123 | 94 | 21 | |
| ID00457.3p-miR | 503 | −127 | 94 | 22 | |
| miR-3960 | 504 | −115 | 92 | 20 | |
| ID02064.5p-miR | 505 | −129 | 90 | 23 |
| Gene; NX | miRNA | Start of Site, nt | ΔG, kJ/mole | ΔG/ΔGm, % | Length, nt |
|---|---|---|---|---|---|
| ATM; 8.9 | ID03006.5p-miR | 9778 | −121 | 89 | 24 |
| miR-5095 | 9787 | −108 | 93 | 21 | |
| miR-619-5p | 9793 | −119 | 98 | 22 | |
| miR-5096 | 9882 | −104 | 92 | 21 | |
| miR-5585-3p | 9950 | −110 | 95 | 22 | |
| miR-1273a | 11,054 | −119 | 90 | 25 | |
| miR-1273g-3p | 11,076 | −113 | 96 | 21 | |
| miR-1273e | 11,119 | −108 | 93 | 22 | |
| miR-5585-5p | 11,156 | −106 | 91 | 22 | |
| FLT1; 4.6 | ID01030.3p-miR | 6909 ÷ 6923 (7) | −108 ÷ −110 | 89 ÷ 91 | 23 |
| miR-466 | 6911 ÷ 6937 (9) | −106 ÷ −108 | 89 ÷ 93 | 23 | |
| ID00436.3p-miR | 6913 ÷ 6925 (7) | −104 | 89 | 23 | |
| IGF1; 5.9 | ID00470.5p-miR | 4042 ÷ 4058 (9) | −108 | 89 | 23 |
| miR-574-5p | 4042 ÷ 4062 (11) | −108 ÷ −113 | 89 ÷ 93 | 23 | |
| miR-1273g-3p | 6009 | −113 | 96 | 21 | |
| miR-1273f | 6042 | −102 | 98 | 19 | |
| miR-1273d | 6043 | −119 | 87 | 25 | |
| miR-1273e | 6052 | −108 | 93 | 22 | |
| IGF2; 3.2 | ID00470.5p-miR | 2286 ÷ 2351 (6) | −108 ÷ −113 | 89 ÷ 93 | 23 |
| miR-574-5p | 2288, 2290 | −108 | 89 | 23 | |
| miR-574-5p | 2397 ÷ 2408 (4) | −108 ÷ −113 | 89 ÷ 93 | 23 | |
| ID00470.5p-miR | 2404, 2412 | −110 | 91 | 23 | |
| ID00470.5p-miR | 2442 ÷ 2463 (3) | −108 ÷ −113 | 89 ÷ 93 | 23 | |
| miR-574-5p | 2465, 2484 | −108 | 89 | 23 | |
| ID00470.5p-miR | 2520 ÷ 2539 (3) | −108 | 89 | 23 | |
| miR-574-5p | 2522 | −108 | 89 | 23 | |
| ID00470.5p-miR | 2655 ÷ 2672 (3) | −108 ÷ −110 | 89 ÷ 91 | 23 | |
| ID00470.5p-miR | 2704, 2725 | −108 | 89 | 23 | |
| miR-574-5p | 2727, 2731 | −108, −110 | 89, 91 | 23 | |
| JAK2; 14.1 | miR-466 | 5182 ÷ 5200 (10) | −104 ÷ −106 | 89 ÷ 91 | 23 |
| ID01030.3p-miR | 5184 ÷ 5200 (9) | −108 | 89 | 23 | |
| ID00436.3p-miR | 5184 ÷ 5202 (10) | −104 ÷ −106 | 89 ÷ 91 | 23 | |
| SP1; 18.8 | miR-466 | 4145 ÷ 4161 (9) | −104 ÷ −106 | 89 ÷ 91 | 23 |
| ID01030.3p-miR | 4147 ÷ 4159 (7) | −108 | 89 | 23 | |
| ID00436.3p-miR | 4147 ÷ 4161 (8) | −104 ÷ −106 | 89 ÷ 91 | 23 | |
| XRCC1; 17.2 | miR-574-5p | 2033 ÷ 2055 (11) | −108 ÷ −113 | 89 ÷ 93 | 23 |
| ID00470.5p-miR | 2035 ÷ 2055 (9) | −108 ÷ −113 | 89 ÷ 93 | 23 | |
| ZEB1; 14.8 | miR-574-5p | 3587 ÷ 3605 (10) | −113 | 93 | 23 |
| ID00470.5p-miR | 3587 ÷ 3605 (10) | −108 | 89 | 23 |
| miRNA | Candidate Genes |
|---|---|
| ID02064.5p-miR | ARID1A, E2F1, NFKB1, ODC1, TDFB1, VEGFC |
| miR-3960 | ARID1A, CDX2, E2F1, ODC1, VEGFC |
| ID02052.5p-miR | CDX2, DNMT1, E2F1, VEGFC |
| ID01041.5p-miR | CDX2, TBC1D9, VEGFC |
| ID02761.3p-miR | ARID1A, EZH2, PTEN |
| ID01702.3p-miR | ARID1A, PIC3CA, TBC1D9 |
| ID01895.5p-miR | PIC3CA, CDX2, TBC1D9 |
| ID03332.3p-miR | KRAS, MAPK1, SIRT1 |
| ID00296.3p-miR | ARID1A, TGFB1 |
| ID02084.5p-miR | ARID1A, ODC1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akimniyazova, A.; Pyrkova, A.; Uversky, V.; Ivashchenko, A. Predicting Associations of miRNAs and Candidate Gastric Cancer Genes for Nanomedicine. Nanomaterials 2021, 11, 691. https://doi.org/10.3390/nano11030691
Akimniyazova A, Pyrkova A, Uversky V, Ivashchenko A. Predicting Associations of miRNAs and Candidate Gastric Cancer Genes for Nanomedicine. Nanomaterials. 2021; 11(3):691. https://doi.org/10.3390/nano11030691
Chicago/Turabian StyleAkimniyazova, Aigul, Anna Pyrkova, Vladimir Uversky, and Anatoliy Ivashchenko. 2021. "Predicting Associations of miRNAs and Candidate Gastric Cancer Genes for Nanomedicine" Nanomaterials 11, no. 3: 691. https://doi.org/10.3390/nano11030691
APA StyleAkimniyazova, A., Pyrkova, A., Uversky, V., & Ivashchenko, A. (2021). Predicting Associations of miRNAs and Candidate Gastric Cancer Genes for Nanomedicine. Nanomaterials, 11(3), 691. https://doi.org/10.3390/nano11030691







