Bioelectronic Nose Based on Single-Stranded DNA and Single-Walled Carbon Nanotube to Identify a Major Plant Volatile Organic Compound (p-Ethylphenol) Released by Phytophthora Cactorum Infected Strawberries
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
- s1-DNA: 5′-CTT CTG TCT TGA TGT TTG TCA AAC-3′
- s2-DNA: 5′-AAA ACC CCC GGG GTT TTT TTT TTT-3′
- s3-DNA: 5′-TTT TTT TTT TTT TTT-3′
- s4-DNA: 5′-GAG TCT GTG GAG GAG GTA GTC-3′
- s5-DNA: 5′-GTT GTT GGT TGT TG-3′
- s6-DNA: 5′-GTG TGT GTG TGT GTG TGT GTG TGT-3′
2.2. Apparatus
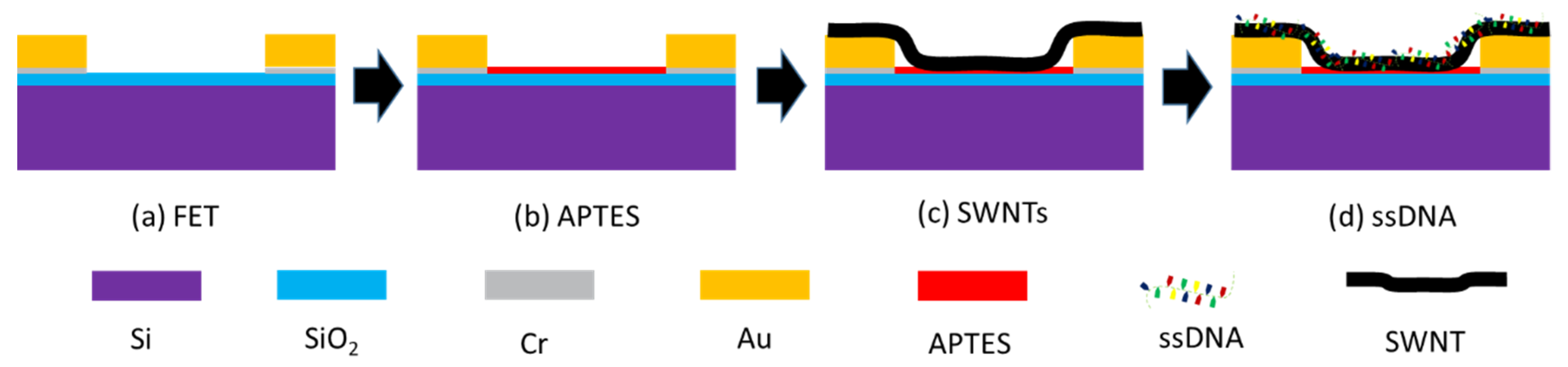
2.3. Fabrication of Biosensor Arrays
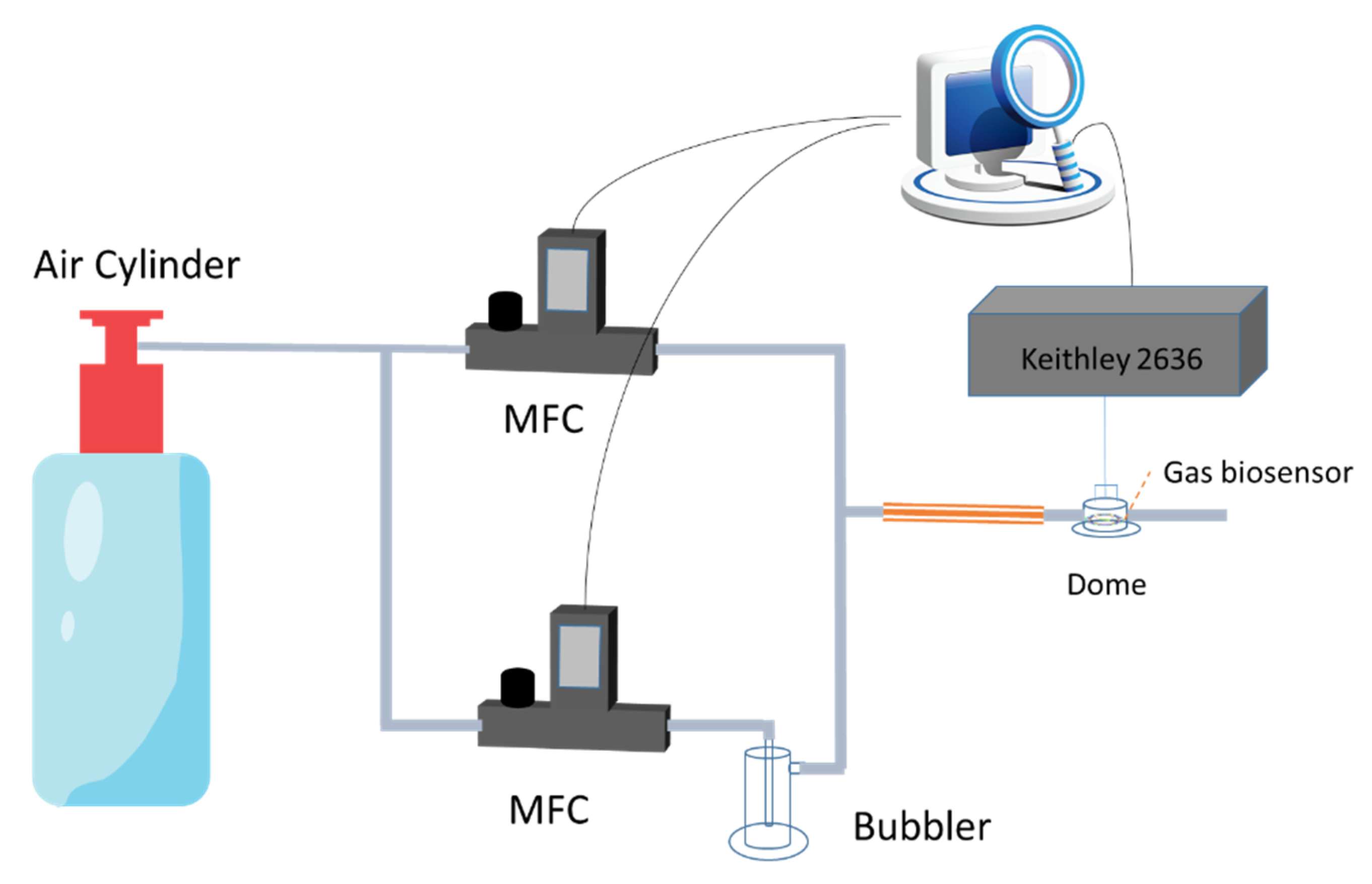
2.4. Gas Generator
2.5. Sensing Protocol
3. Results and Discussion
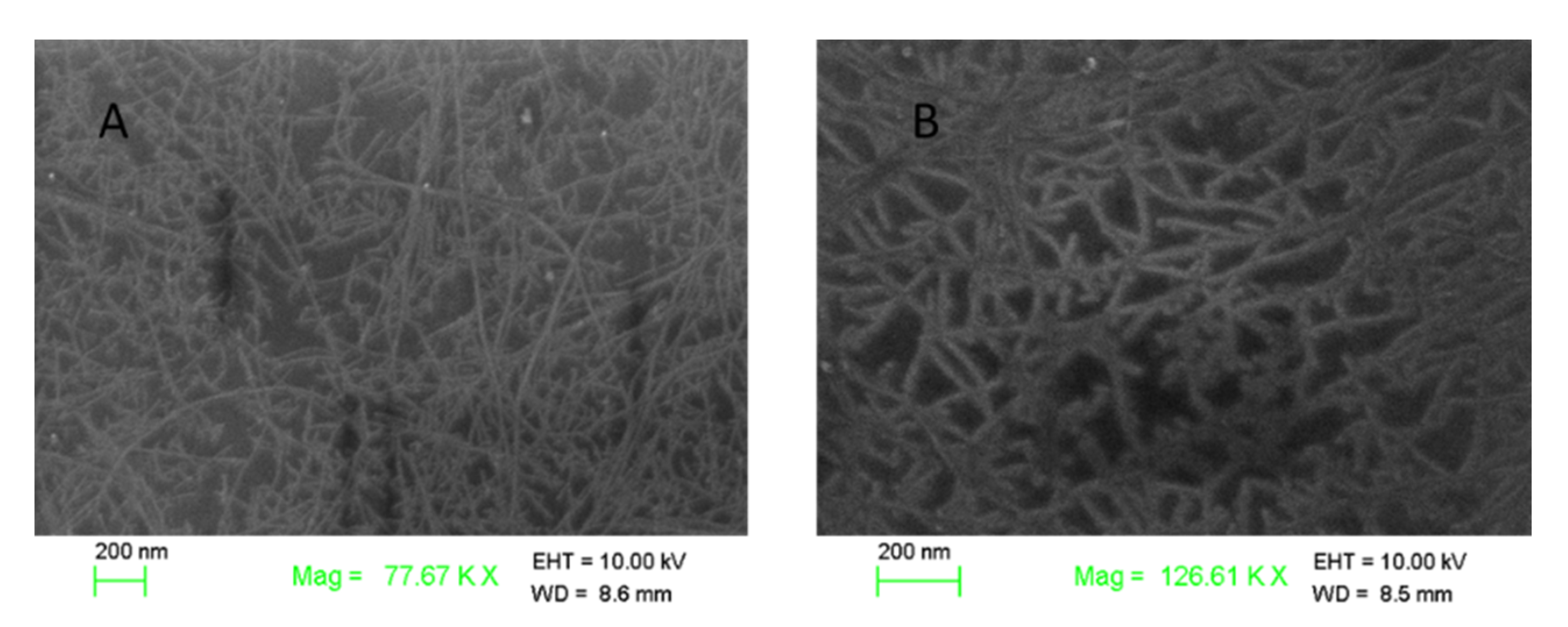
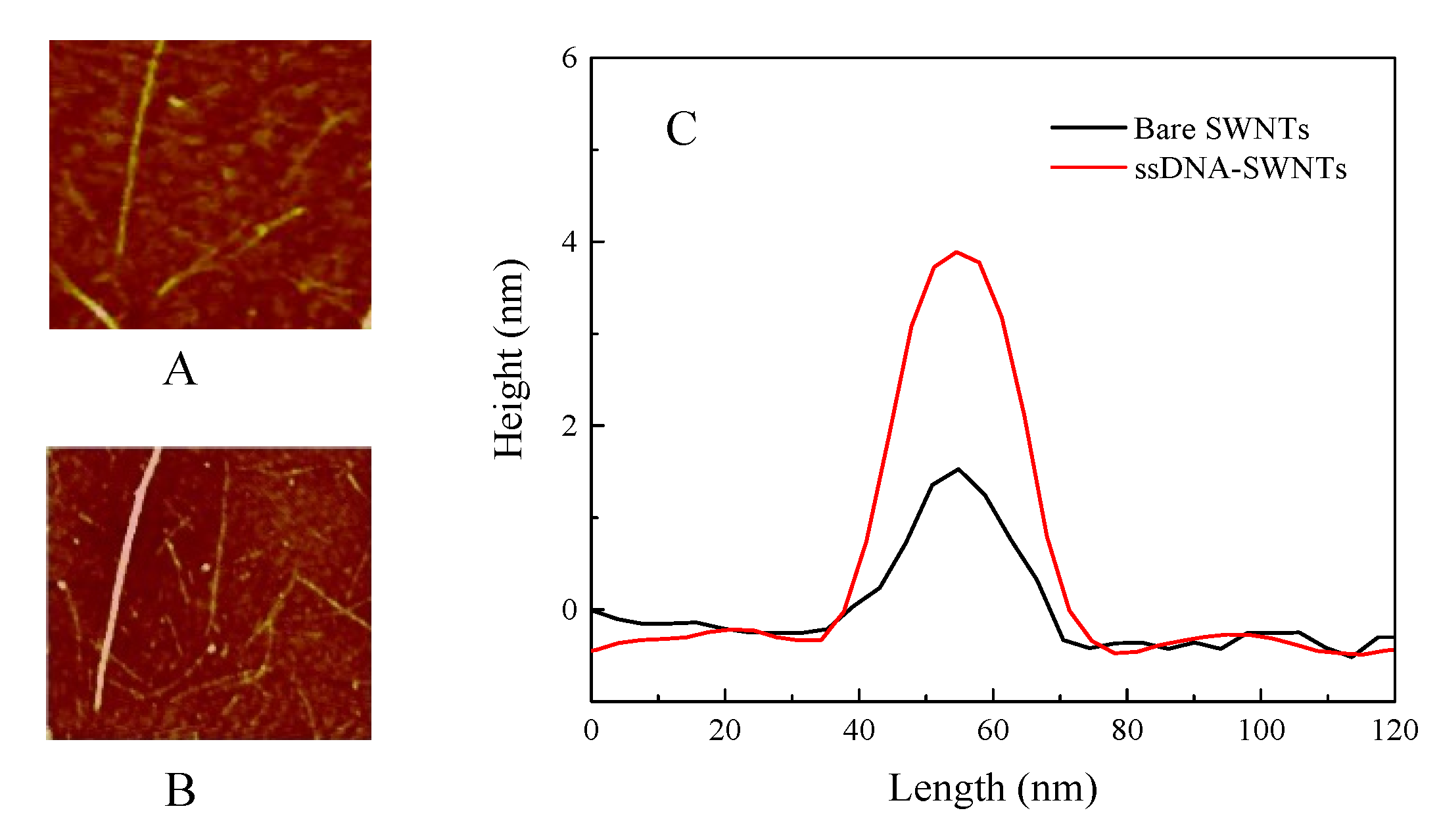
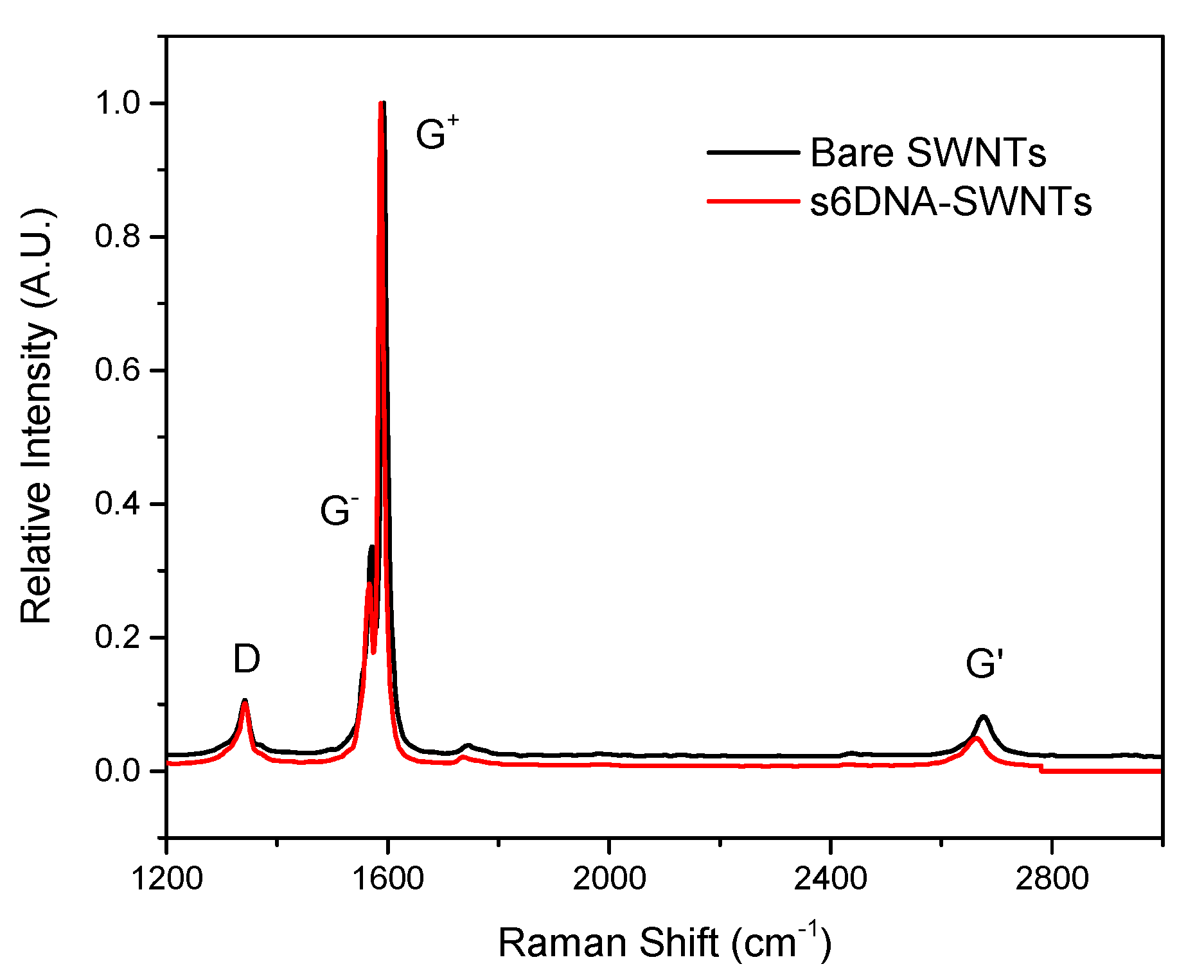
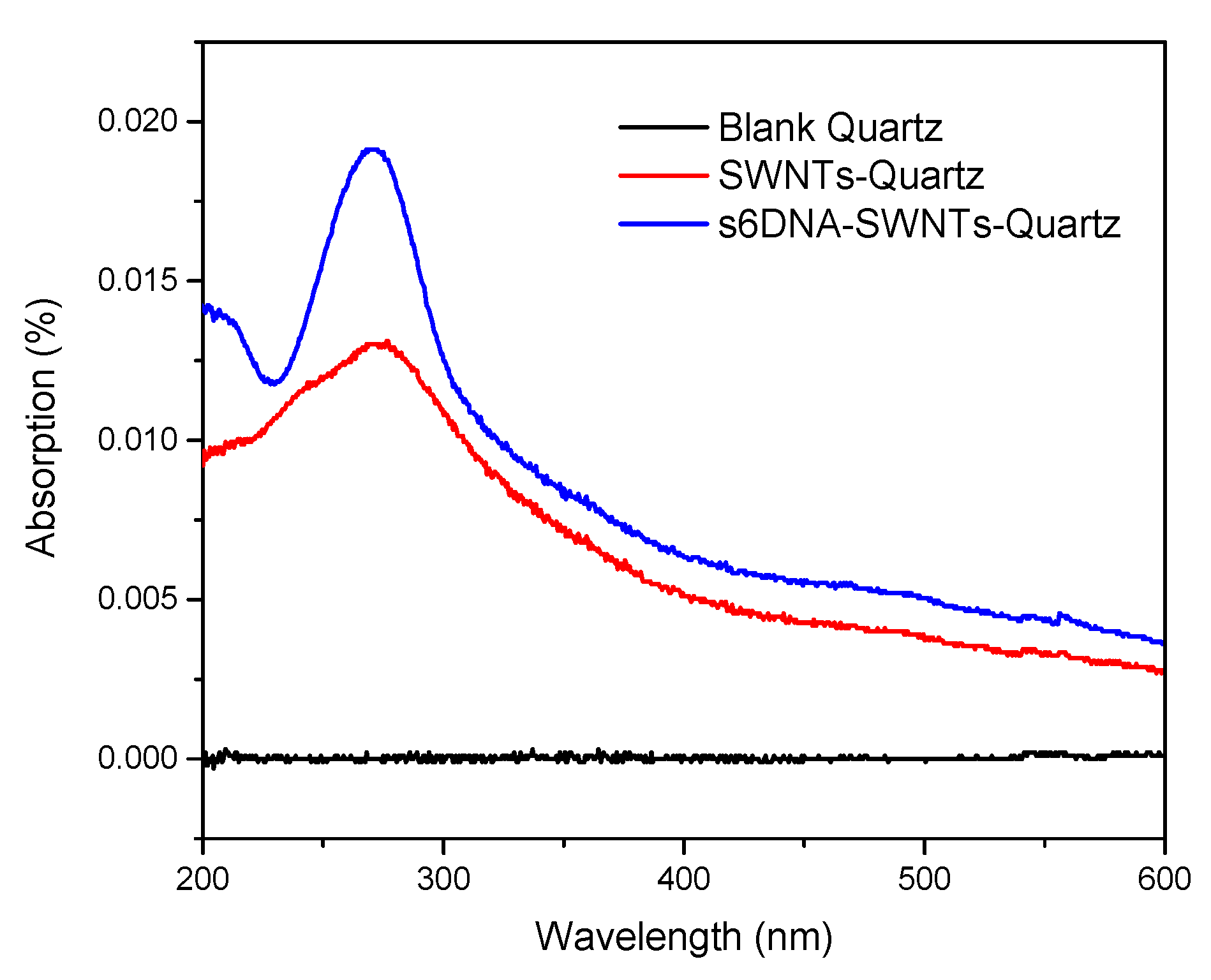
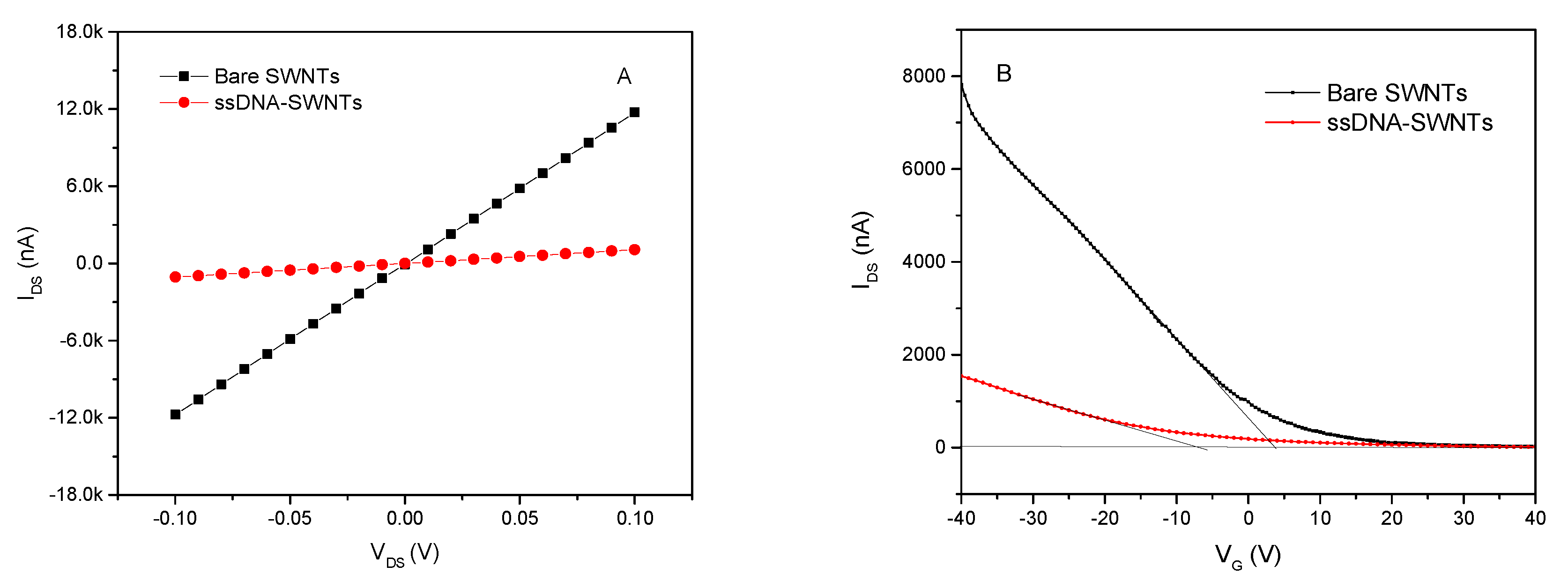
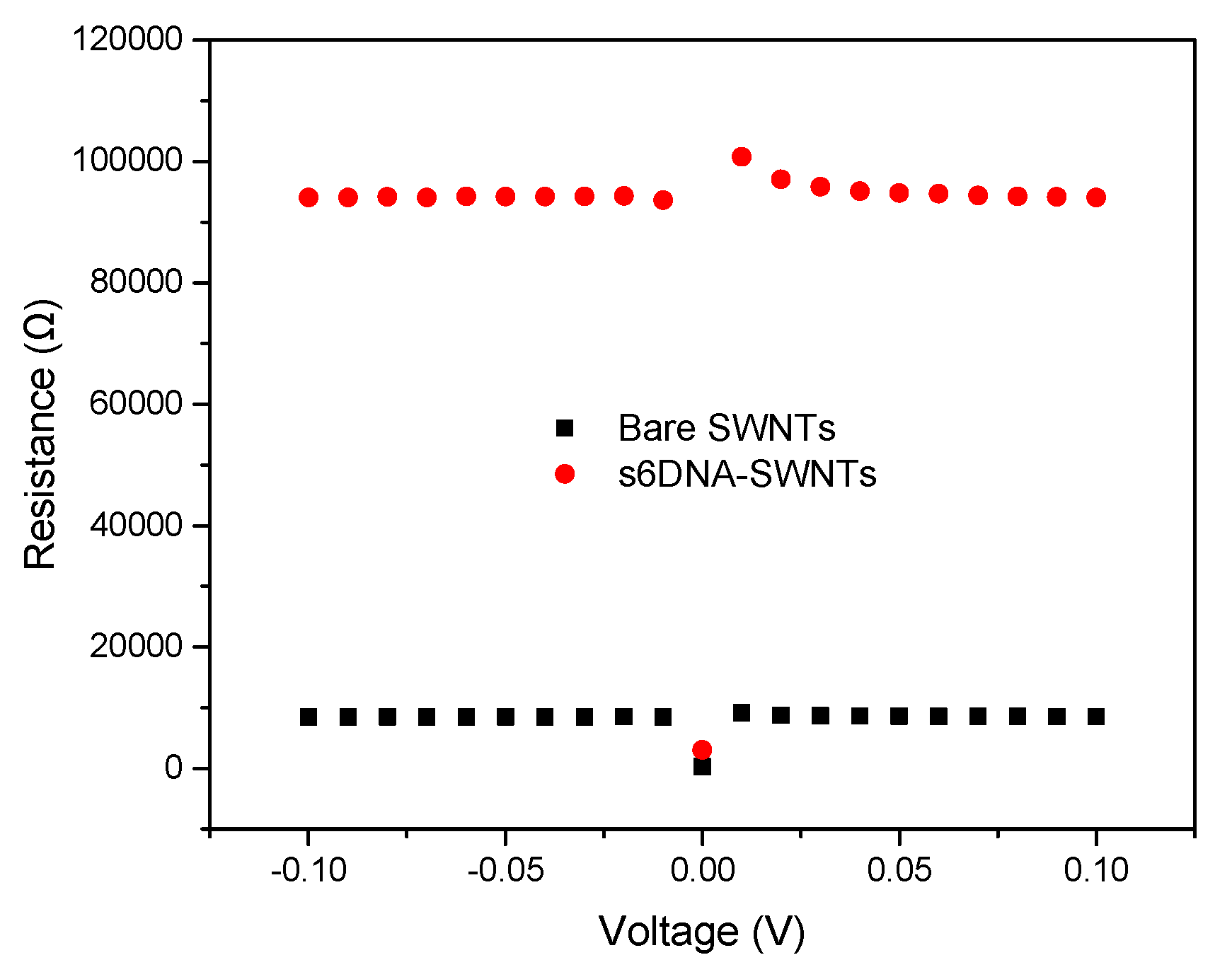
3.1. The Characteristic of ssDNA-SWNTs
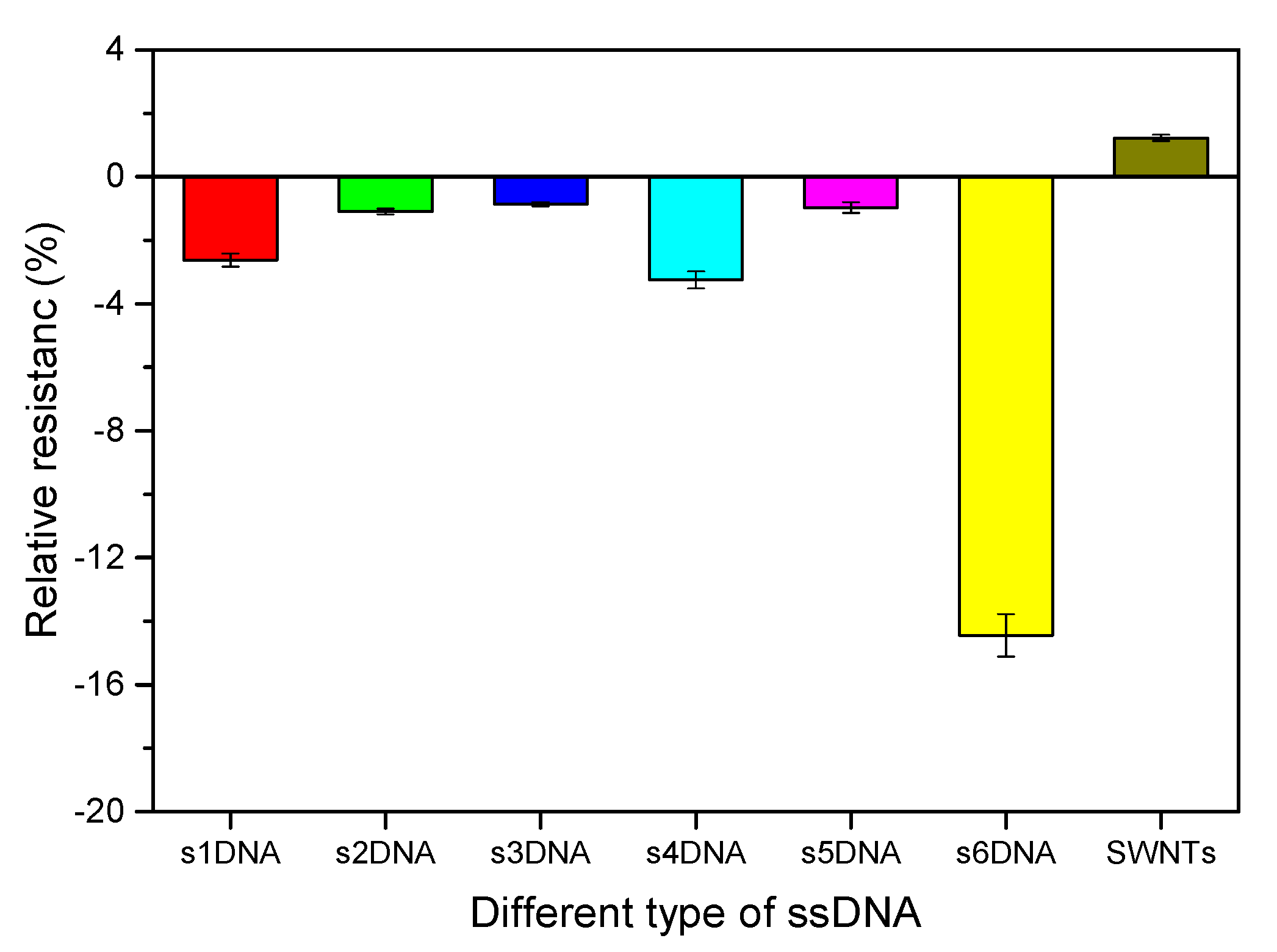
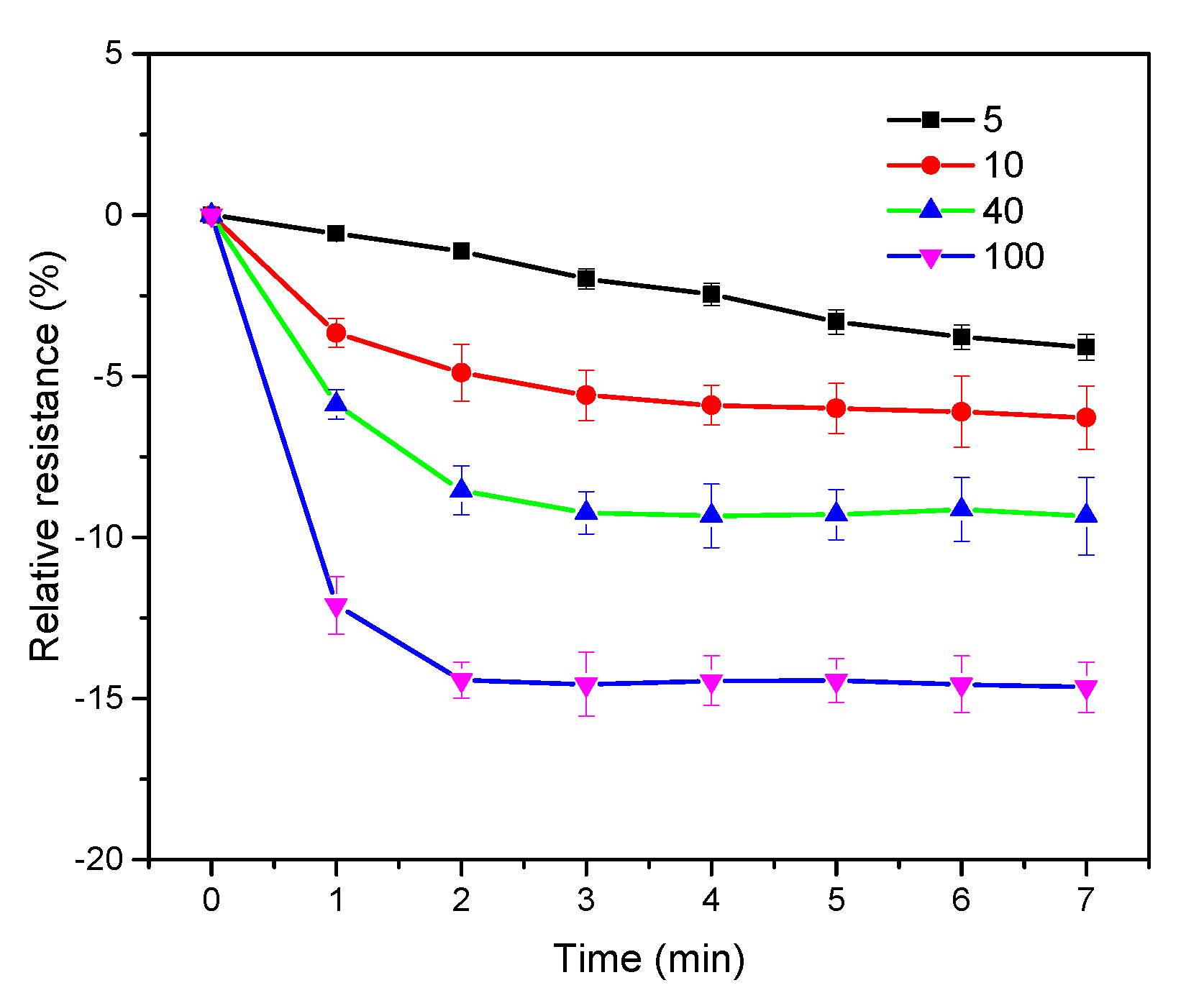
3.2. Optimization
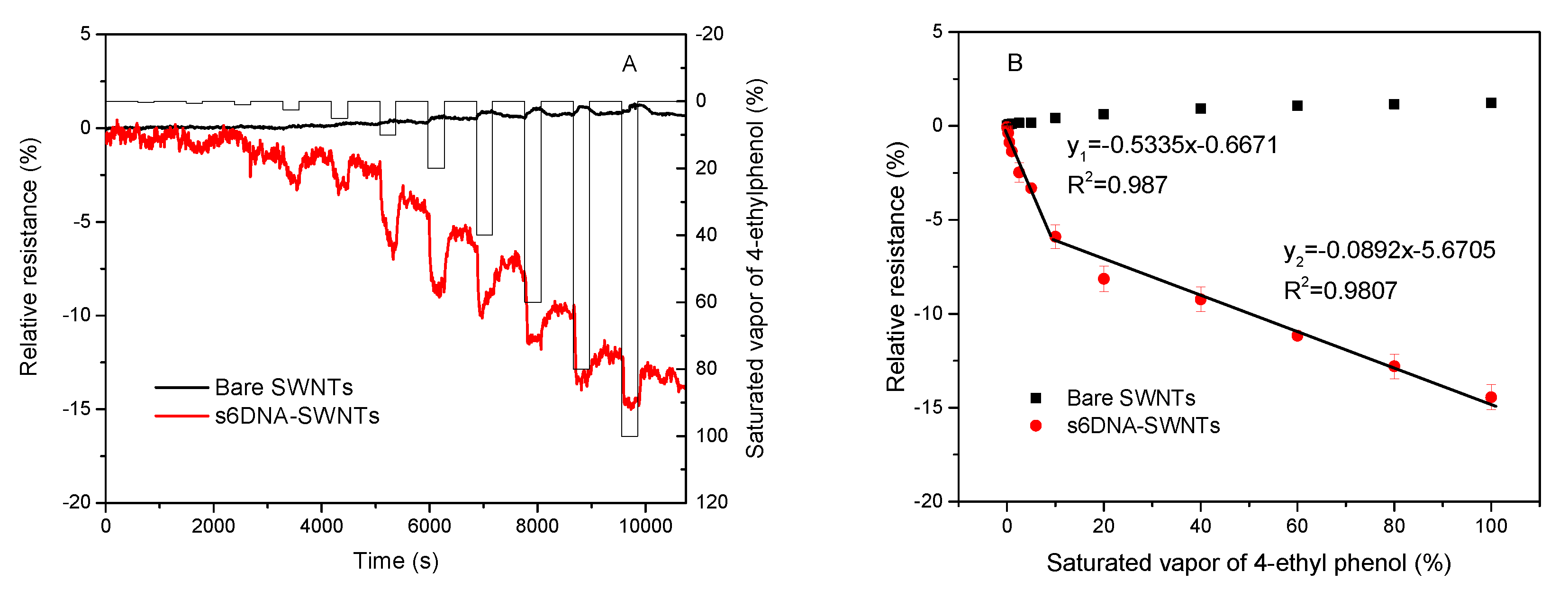
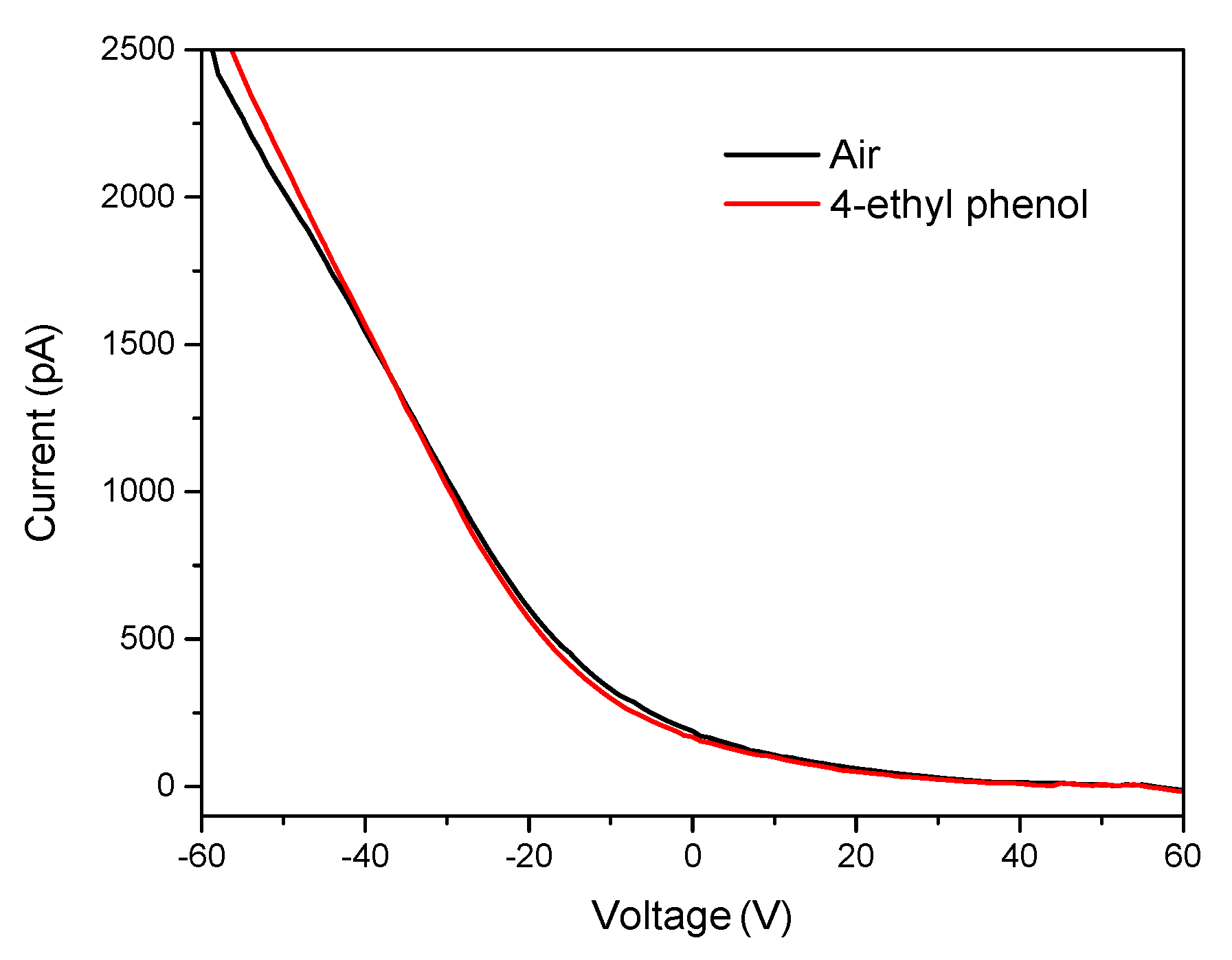
3.3. Sensitivity for 4-ethyl Phenol Detection
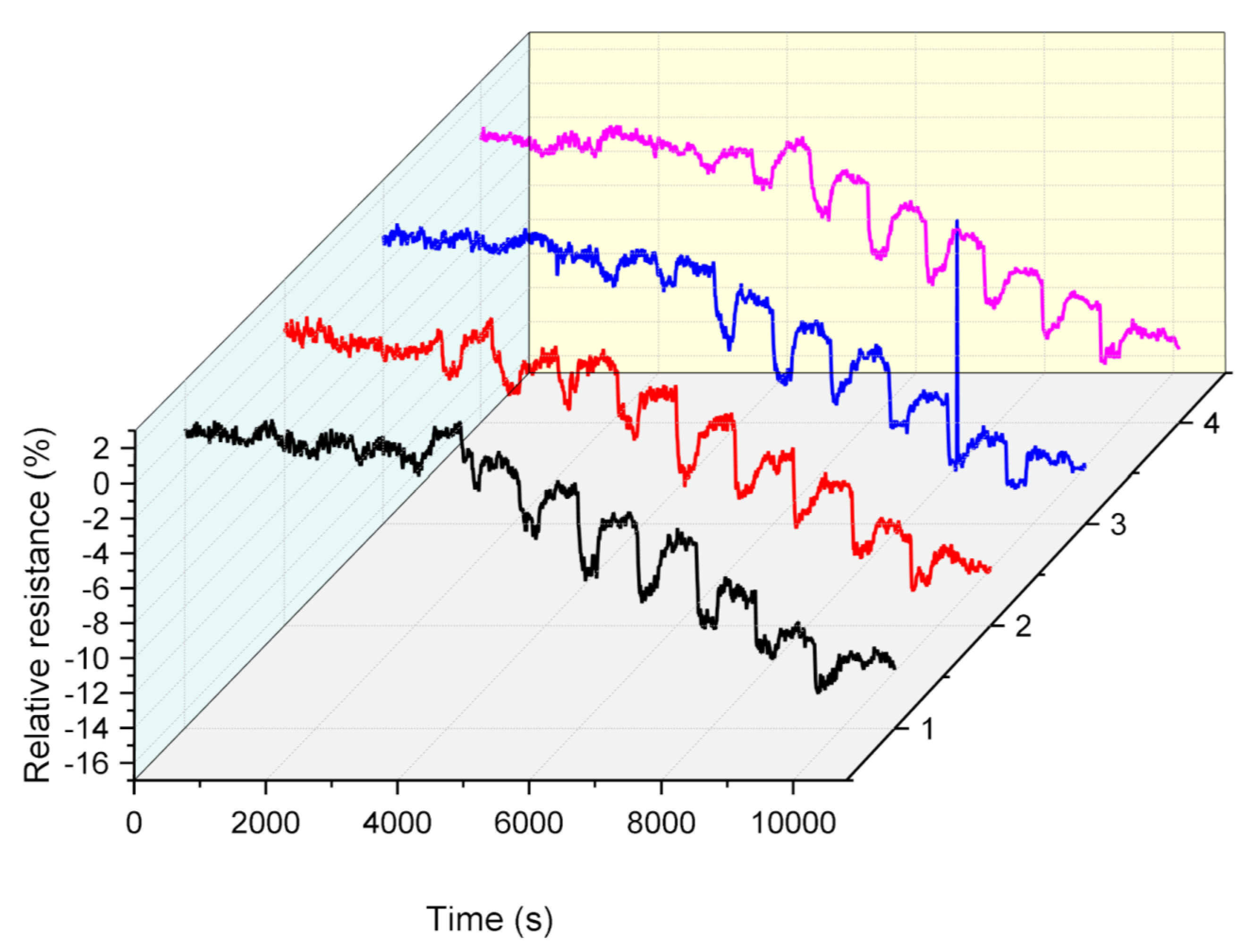
3.4. Repeatability
3.5. Sensing Mechanism
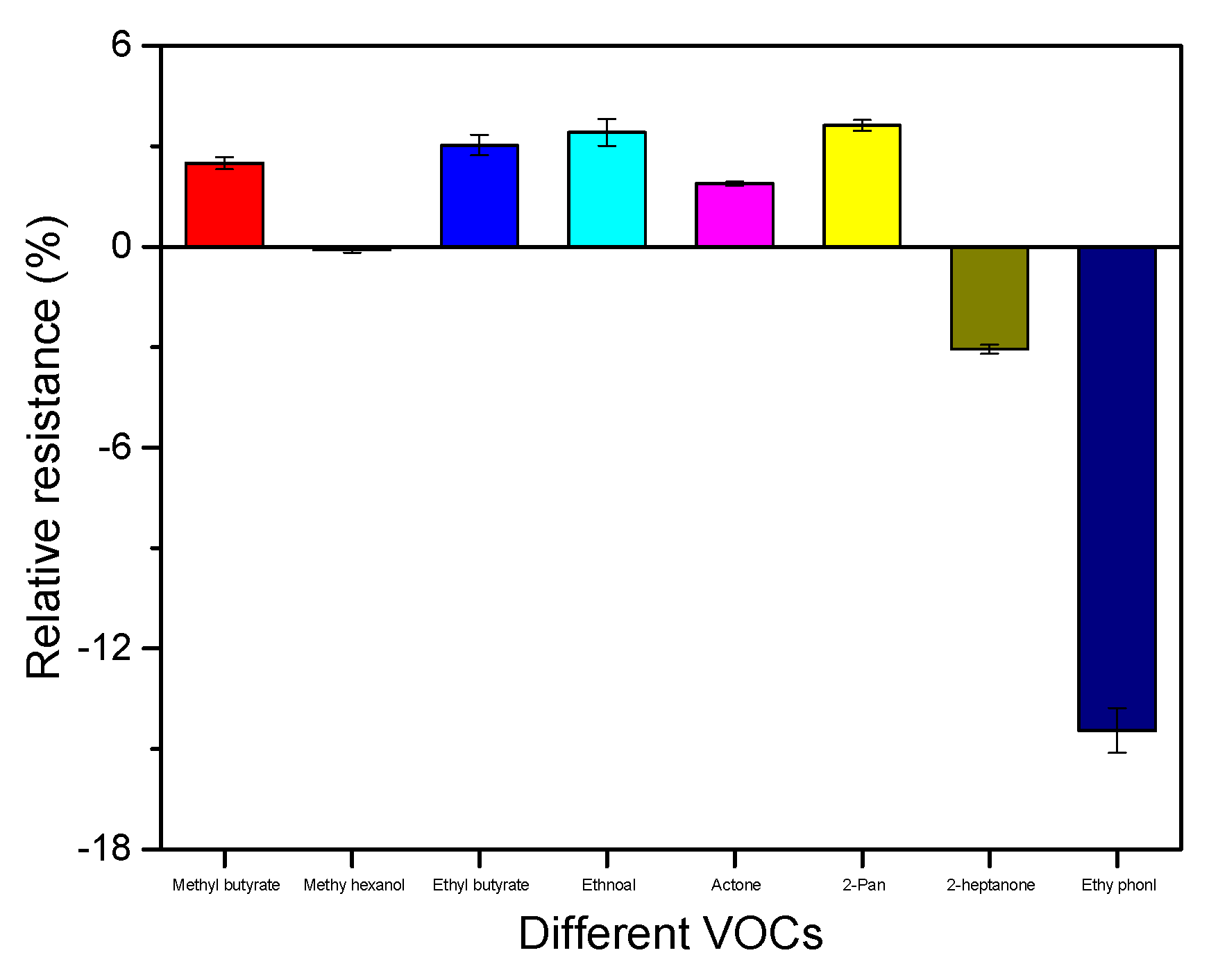
3.6. Interference
3.7. Chemometric Analysis
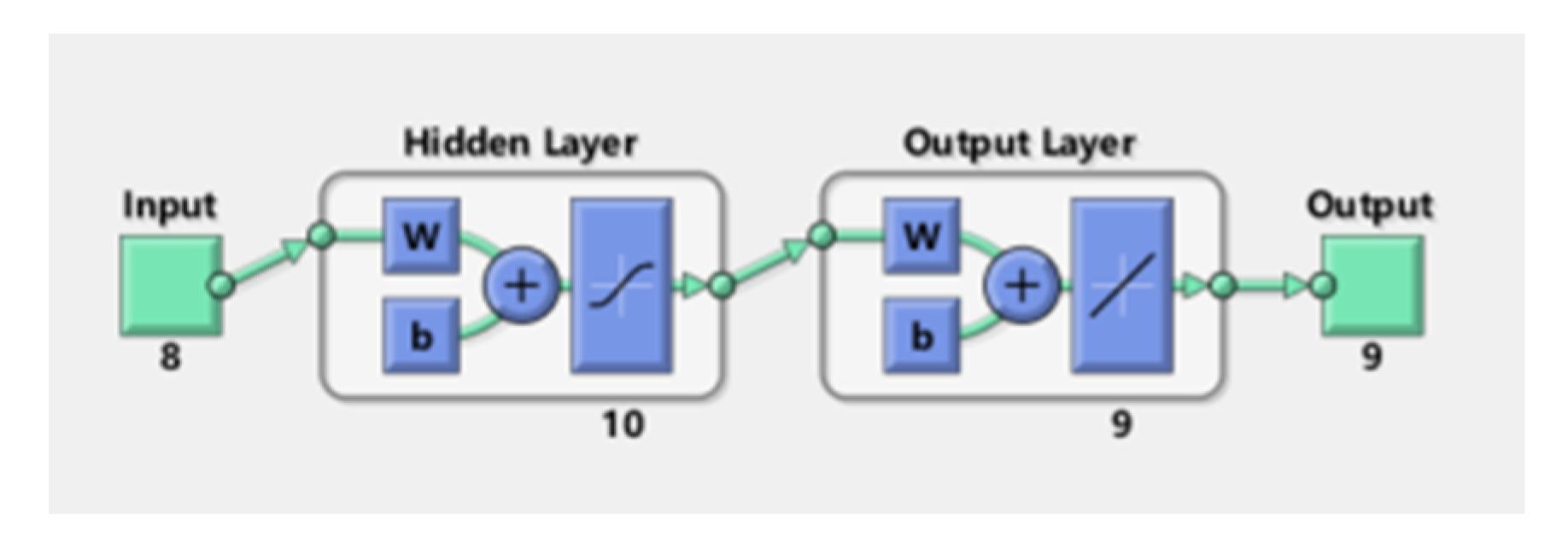
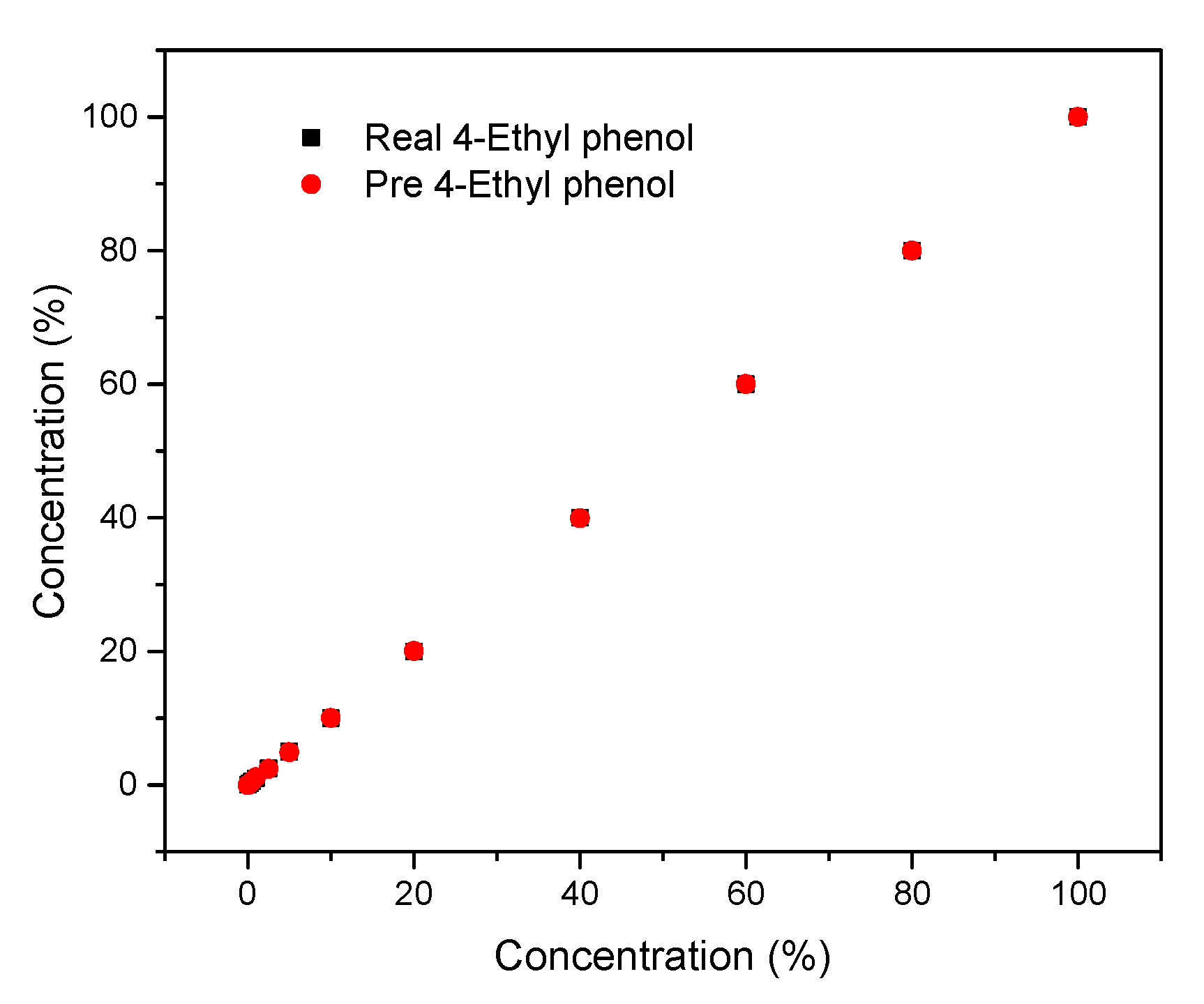
3.7.1. Neural Network Fitting
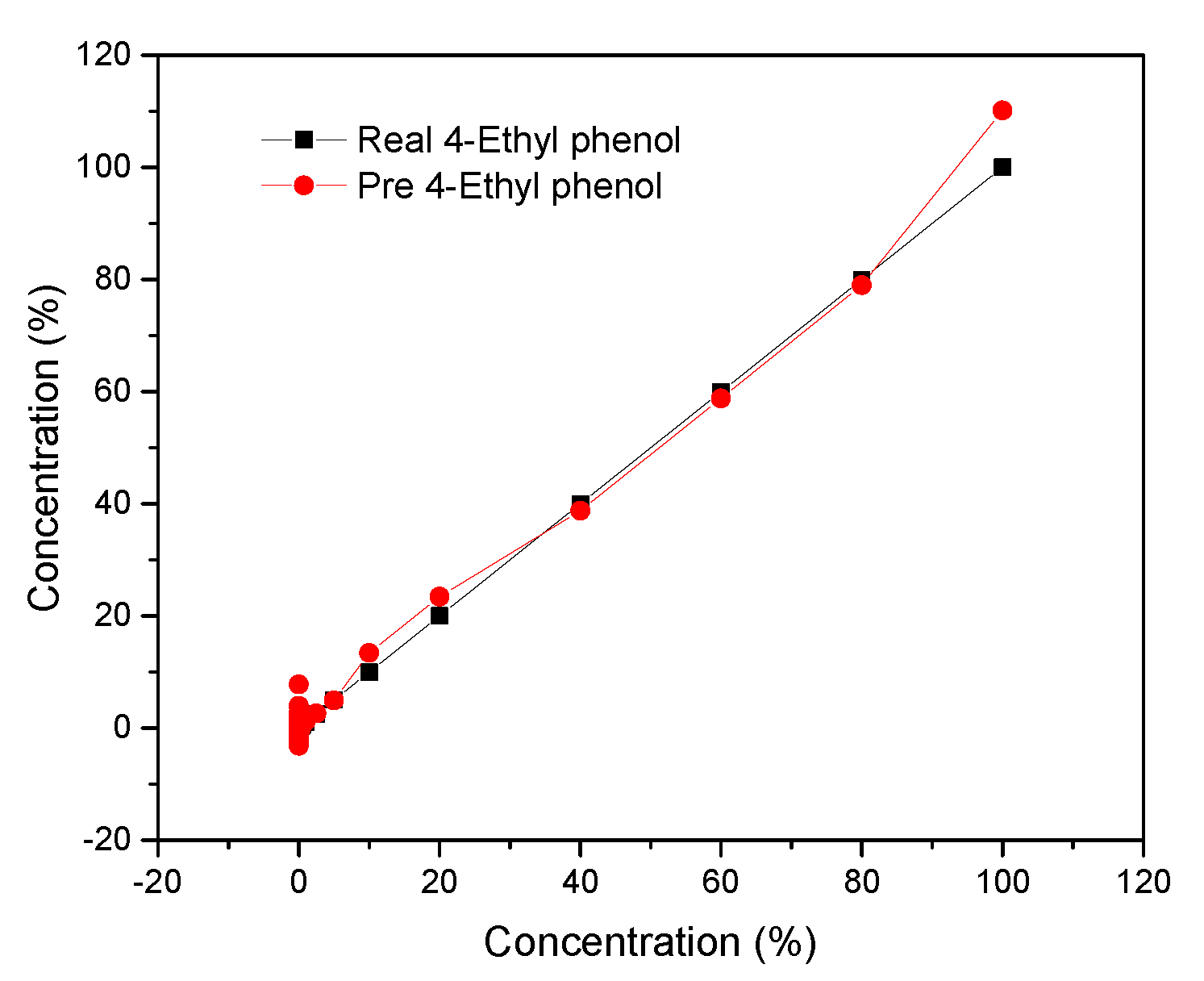
3.7.2. Gaussian Process Regression
4. Conclusion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, Y.; Yeow, J.T. A review of carbon nanotubes-based gas sensors. J. Sens. 2009, 2009, 1–24. [Google Scholar] [CrossRef]
- Moon, H.G.; Jung, Y.; Han, S.D.; Shim, Y.-S.; Shin, B.; Lee, T.; Kim, J.-S.; Lee, S.; Jun, S.C.; Park, H.-H. Chemiresistive electronic nose toward detection of biomarkers in exhaled breath. ACS Appl. Mater. Interfaces 2016, 8, 20969–20976. [Google Scholar] [CrossRef] [PubMed]
- Dinh, T.; Phan, H.-P.; Nguyen, T.-K.; Qamar, A.; Foisal, A.R.M.; Viet, T.N.; Tran, C.-D.; Zhu, Y.; Nguyen, N.-T.; Dao, D.V. Environment-friendly carbon nanotube based flexible electronics for noninvasive and wearable healthcare. J. Mater. Chem. C 2016, 4, 10061–10068. [Google Scholar] [CrossRef]
- Rana, M.M.; Ibrahim, D.S.; Mohd Asyraf, M.; Jarin, S.; Tomal, A. A review on recent advances of CNTs as gas sensors. Sens. Rev. 2017, 37, 127–136. [Google Scholar] [CrossRef]
- Wang, H.; Yin, Y.; Gang, L. Single-gap Microelectrode Functionalized with Single-walled Carbon Nanotubes and Pbzyme for the Determination of Pb2+. Electroanalysis 2019, 31, 1174–1181. [Google Scholar] [CrossRef]
- Kavinkumar, T.; Sastikumar, D.; Manivannan, S. Effect of functional groups on dielectric, optical gas sensing properties of graphene oxide and reduced graphene oxide at room temperature. Rsc. Adv. 2015, 5, 10816–10825. [Google Scholar] [CrossRef]
- Wang, H.; Liu, Y.; Liu, G. Label-Free Biosensor Using a Silver Specific RNA-Cleaving DNAzyme Functionalized Single-Walled Carbon Nanotube for Silver Ion Determination. Nanomaterials 2018, 8, 258. [Google Scholar] [CrossRef]
- Wang, H.; Liu, Y.; Liu, G. Electrochemical biosensor using DNA embedded phosphorothioate modified RNA for mercury ion determination. ACS Sens. 2018, 3, 624–631. [Google Scholar] [CrossRef]
- Hartleb, H.; Späth, F.; Hertel, T. Evidence for strong electronic correlations in the spectra of gate-doped single-wall carbon nanotubes. ACS Nano 2015, 9, 10461–10470. [Google Scholar] [CrossRef]
- Abdulrazzak, F.H. Enhance photocatalytic degradation of cobalamin by TiO2 with single walled carbon nanotubes. Int. J. Multi. Curr. Res. 2016, 4, 119–125. [Google Scholar]
- Nag, P.; Sujatha Devi, P. SnO2 based ceramics for hydrogen sensors: current status and perspectives. Trans. Indian Ceram. Soc. 2015, 74, 129–147. [Google Scholar] [CrossRef]
- Liao, L.; Zhang, Z.; Yan, B.; Zheng, Z.; Bao, Q.; Wu, T.; Li, C.M.; Shen, Z.; Zhang, J.; Gong, H. Multifunctional CuO nanowire devices: p-type field effect transistors and CO gas sensors. Nanotechnology 2009, 20, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Jiang, Y.; Pan, W.; Li, D.; Wu, Z.; Li, Y. Fabrication and characterization of polyaniline-based gas sensor by ultra-thin film technology. Sens. Actuators B 2002, 81, 158–164. [Google Scholar] [CrossRef]
- Korposh, S.; Kodaira, S.; Selyanchyn, R.; Ledezma, F.H.; James, S.W.; Lee, S.-W. Porphyrin-nanoassembled fiber-optic gas sensor fabrication: Optimization of parameters for sensitive ammonia gas detection. Opt. Laser Technol. 2018, 101, 1–10. [Google Scholar] [CrossRef]
- Yang, Y.; Qin, P.; Zhang, J.; Li, W.; Zhu, J.; Lu, M.; Cai, Z. Fabrication of nanoscale graphitic carbon nitride/copper oxide hybrid composites coated solid-phase microextraction fibers coupled with gas chromatography for determination of polycyclic aromatic hydrocarbons. J. Chromatogr. A 2018, 1570, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Pérez, E.M.; Martín, N. π–π interactions in carbon nanostructures. Chem. Soc. Rev. 2015, 44, 6425–6433. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, M.L. DNA-functionalized single-walled carbon nanotube-based sensor array for breath analysis. Int. J. Electron. Electron. Eng. 2016, 4, 177–180. [Google Scholar] [CrossRef][Green Version]
- Wu, C.; Du, L.; Zou, L. DNA-Decorated Devices as Smell Sensors. In Bioinspired Smell and Taste Sensors; Springer: Dordrecht, The Netherlands, 2015; pp. 145–165. [Google Scholar]
- Chen, X.-R.; Huang, S.-X.; Zhang, Y.; Sheng, G.-L.; Zhang, B.-Y.; Li, Q.-Y.; Zhu, F.; Xu, J.-Y. Transcription profiling and identification of infection-related genes in Phytophthora cactorum. Mol. Genet. Genomics 2018, 293, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Jeleń, H.; Krawczyk, J.; Larsen, T.; Jarosz, A.; Gołębniak, B. Main compounds responsible for off-odour of strawberries infected by Phytophthora cactorum. Lett. Appl. Microbiol. 2005, 40, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Munawar, M.; Toljamo, A.; Martin, F.; Kokko, H. Recombinase Polymerase Amplification Assay for fast, sensitive and on-site detection of Phytophthora cactorum without DNA extraction. Eur. J. Horticultural Sci. 2019, 84, 14–19. [Google Scholar] [CrossRef]
- Fang, Y.; Ramasamy, R. Current and prospective methods for plant disease detection. Biosensors 2015, 5, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Rivero, V.; Giayetto, A.; Rossini, M.; Vera, D. Detection of Phytophthora cactorum in the irrigation water in commercial orchards of’Bartlett’pear in Villa Regina, Rio Negro, Argentina. In Proceedings of the XI International Pear Symposium, Rio Negro, Argentina, 23–26 November 2010; Volume 909, pp. 521–526. [Google Scholar]
- Fang, Y.; Umasankar, Y.; Ramasamy, R.P. Electrochemical detection of p-ethylguaiacol, a fungi infected fruit volatile using metal oxide nanoparticles. Analyst 2014, 139, 3804–3810. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, G.C.; Fang, X.D.; Lu, H.L.; Zhou, X.; Meng, X.; Yu, S.K.; Guo, M.F.; Xia, Y.L. Repellency of Five Mineral Oils against Diaphorina Citri (Hemiptera: Liviidae). Florida Entomologist 2013, 96, 974–982. [Google Scholar] [CrossRef]
- Peirano, D.J.; Pasamontes, A.; Davis, C.E. Supervised semi-automated data analysis software for gas chromatography/differential mobility spectrometry (GC/DMS) metabolomics applications. Int. J. Ion. Mobil. Spec. 2016, 19, 155–166. [Google Scholar] [CrossRef]
- Pasamontes, A.; Cheung, W.H.K.; Simmons, J.; Aksenov, A.A.; Peirano, D.J.; Grafton-Cardwell, E.E.; Kapaun, T.; Dandekar, A.M.; Fiehn, O.; Davis, C.E. Citrus tristeza virus infection in sweet orange trees and a mandarin x tangor cross alters low molecular weight metabolites assessed using gas chromatography mass spectrometry (GC/MS). Metabolomics 2016, 12, 1–10. [Google Scholar] [CrossRef]
- Lu, Y.; Goldsmith, B.R.; Kybert, N.J.; Johnson, A.C. DNA-decorated graphene chemical sensors. Appl. Phys. Lett. 2010, 97, 1–3. [Google Scholar] [CrossRef]
- Rhodes, P.A.; Khamis, S.M. Hybrid Sensor Array. US Patents US20120028820, 2 February 2012. [Google Scholar]
- Glamazda, A.Y.; Plokhotnichenko, A.M.; Leontiev, V.S.; Karachevtsev, V.A. DNA-wrapped carbon nanotubes aligned in stretched gelatin films: polarized resonance Raman and absorption spectroscopy study. Physica E 2017, 93, 92–96. [Google Scholar] [CrossRef]
- Nepal, D.; Sohn, J.I.; Aicher, W.K.; Lee, S.; Geckeler, K.E. Supramolecular conjugates of carbon nanotubes and DNA by a solid-state reaction. Biomacromolecules 2005, 6, 2919–2922. [Google Scholar] [CrossRef]
- Nakashima, N.; Okuzono, S.; Murakami, H.; Nakai, T.; Yoshikawa, K. DNA dissolves single-walled carbon nanotubes in water. Chem. Lett. 2003, 32, 456–457. [Google Scholar] [CrossRef]
- Wang, H.; Liu, Y.; Liu, G. Reusable resistive aptasensor for Pb (II) based on the Pb (II)-induced despiralization of a DNA duplex and formation of a G-quadruplex. Microchim. Acta 2018, 185, 1–12. [Google Scholar] [CrossRef]
- Kybert, N.J.; Lerner, M.B.; Yodh, J.S.; Preti, G.; Johnson, A.C. Differentiation of complex vapor mixtures using versatile DNA–carbon nanotube chemical sensor arrays. ACS Nano 2013, 7, 2800–2807. [Google Scholar] [CrossRef] [PubMed]
- Heller, I.; Janssens, A.M.; Männik, J.; Minot, E.D.; Lemay, S.G.; Dekker, C. Identifying the mechanism of biosensing with carbon nanotube transistors. Nano Lett. 2008, 8, 591–595. [Google Scholar] [CrossRef] [PubMed]

| R0 | RSMT | |
|---|---|---|
| NNF | 0.98321 | 4.82493 |
| GPR | 0.99883 | 1.33108 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Wang, Y.; Hou, X.; Xiong, B. Bioelectronic Nose Based on Single-Stranded DNA and Single-Walled Carbon Nanotube to Identify a Major Plant Volatile Organic Compound (p-Ethylphenol) Released by Phytophthora Cactorum Infected Strawberries. Nanomaterials 2020, 10, 479. https://doi.org/10.3390/nano10030479
Wang H, Wang Y, Hou X, Xiong B. Bioelectronic Nose Based on Single-Stranded DNA and Single-Walled Carbon Nanotube to Identify a Major Plant Volatile Organic Compound (p-Ethylphenol) Released by Phytophthora Cactorum Infected Strawberries. Nanomaterials. 2020; 10(3):479. https://doi.org/10.3390/nano10030479
Chicago/Turabian StyleWang, Hui, Yue Wang, Xiaopeng Hou, and Benhai Xiong. 2020. "Bioelectronic Nose Based on Single-Stranded DNA and Single-Walled Carbon Nanotube to Identify a Major Plant Volatile Organic Compound (p-Ethylphenol) Released by Phytophthora Cactorum Infected Strawberries" Nanomaterials 10, no. 3: 479. https://doi.org/10.3390/nano10030479
APA StyleWang, H., Wang, Y., Hou, X., & Xiong, B. (2020). Bioelectronic Nose Based on Single-Stranded DNA and Single-Walled Carbon Nanotube to Identify a Major Plant Volatile Organic Compound (p-Ethylphenol) Released by Phytophthora Cactorum Infected Strawberries. Nanomaterials, 10(3), 479. https://doi.org/10.3390/nano10030479






