On the Interplay Between Roughness and Elastic Modulus at the Nanoscale: A Methodology Study with Bone as Model Material
Abstract
1. Introduction
- (i)
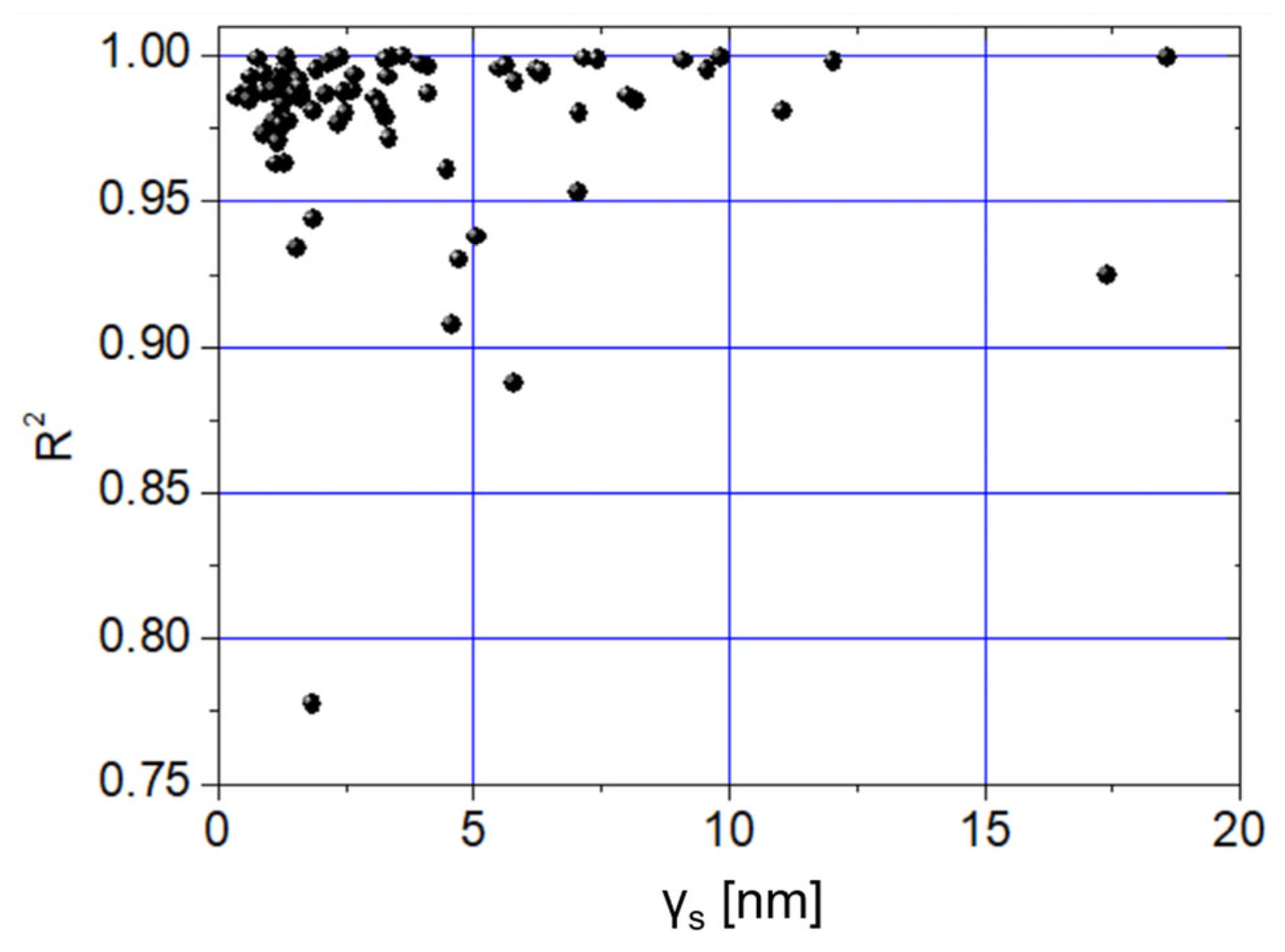
- The impact of local roughness on the reliability of elastic modulus measurements, where reliability is assessed using the coefficient of determination (R2) from the fit of the experimental loading force curve at locations with measured roughness.
- (ii)
- The strength of the correlation between elastic modulus and local roughness, evaluated by non-parametric statistical analysis.
2. Materials and Methods
2.1. Histological Processing for Polymethyl Methacrylate Embedding
2.2. FTIR Setup
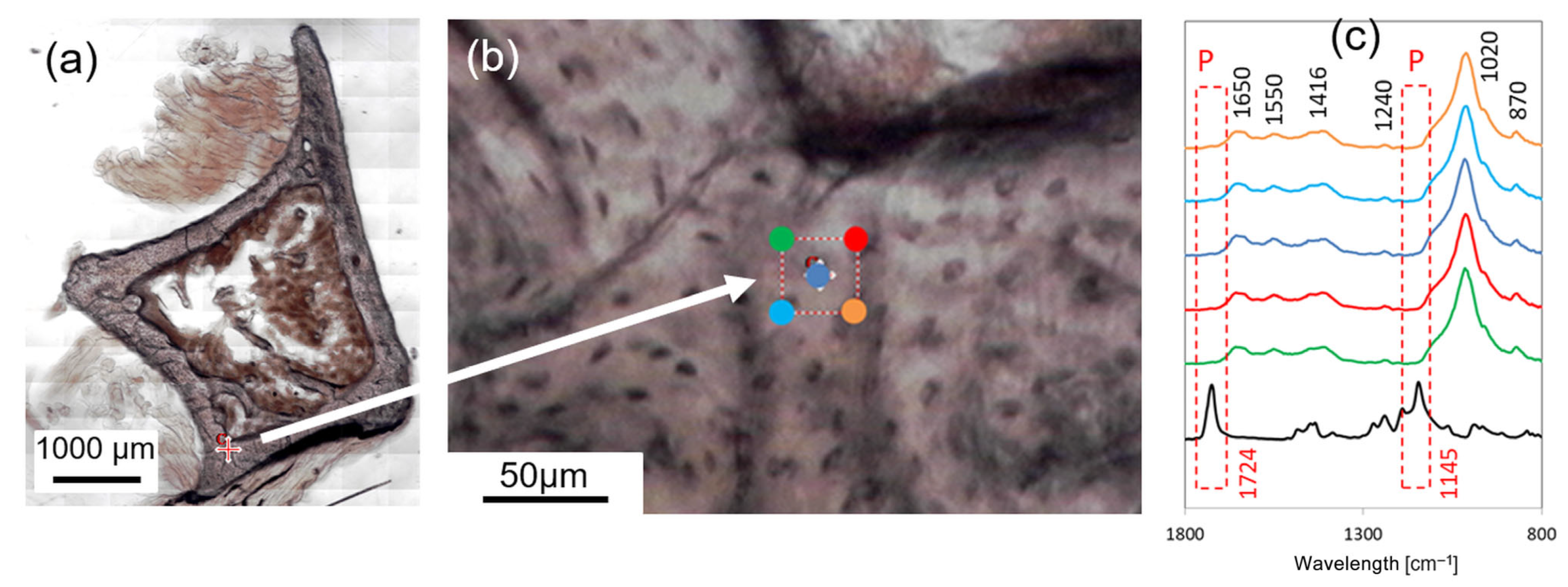
2.3. Targeting the ROI: FTIR Preliminary Analysis
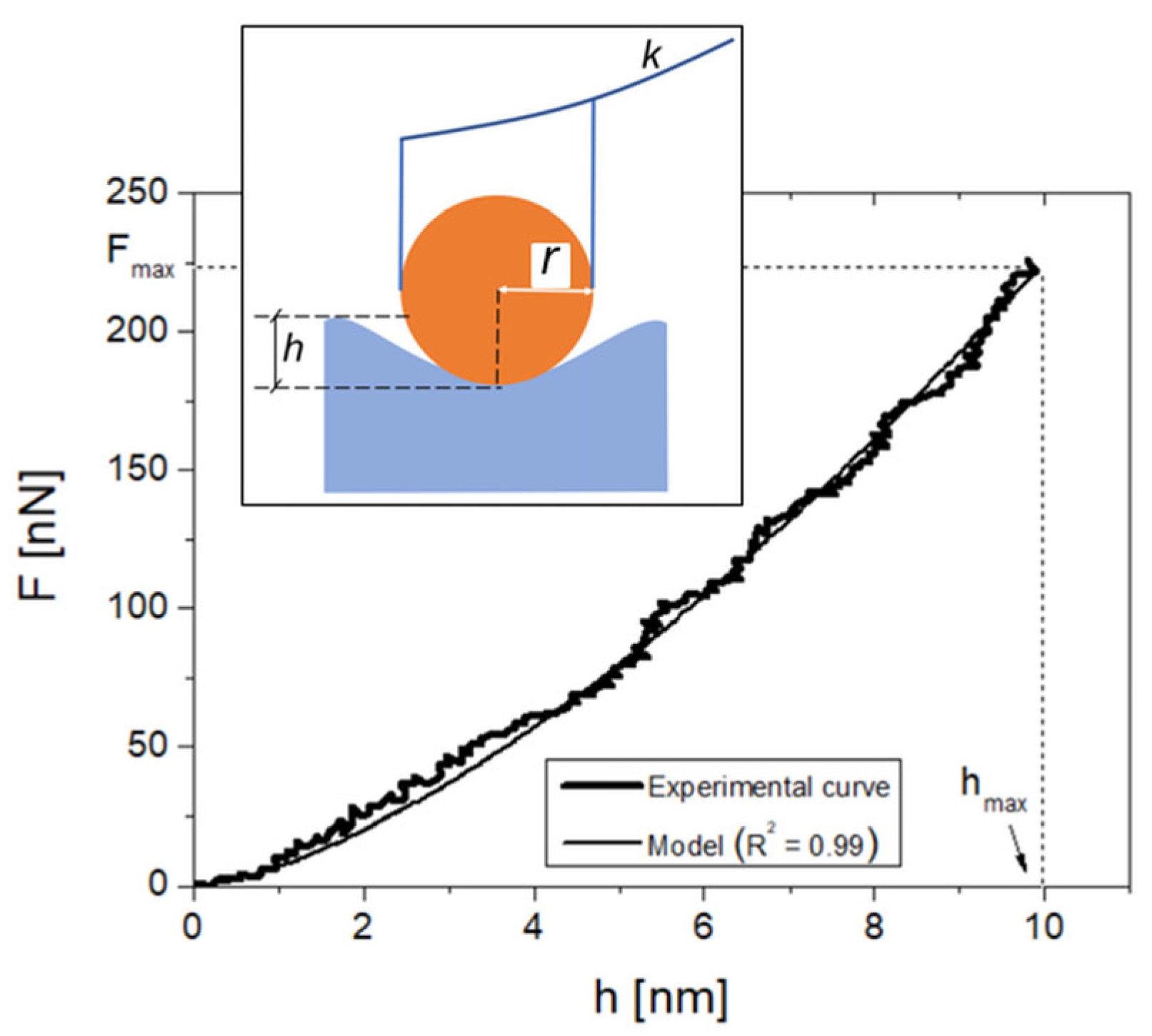
2.4. AFM Setup Calibration and Operation
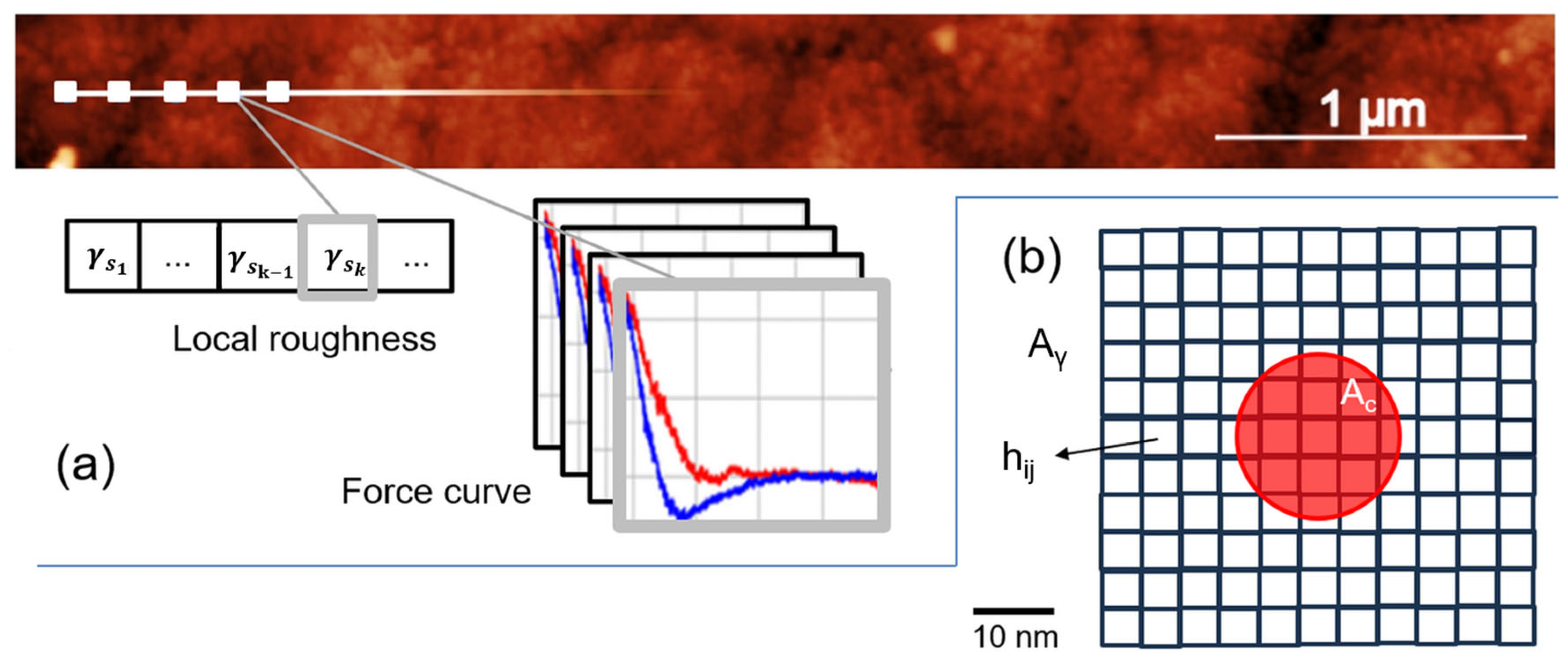
2.5. AFM Data Processing
2.6. Software and Statistics
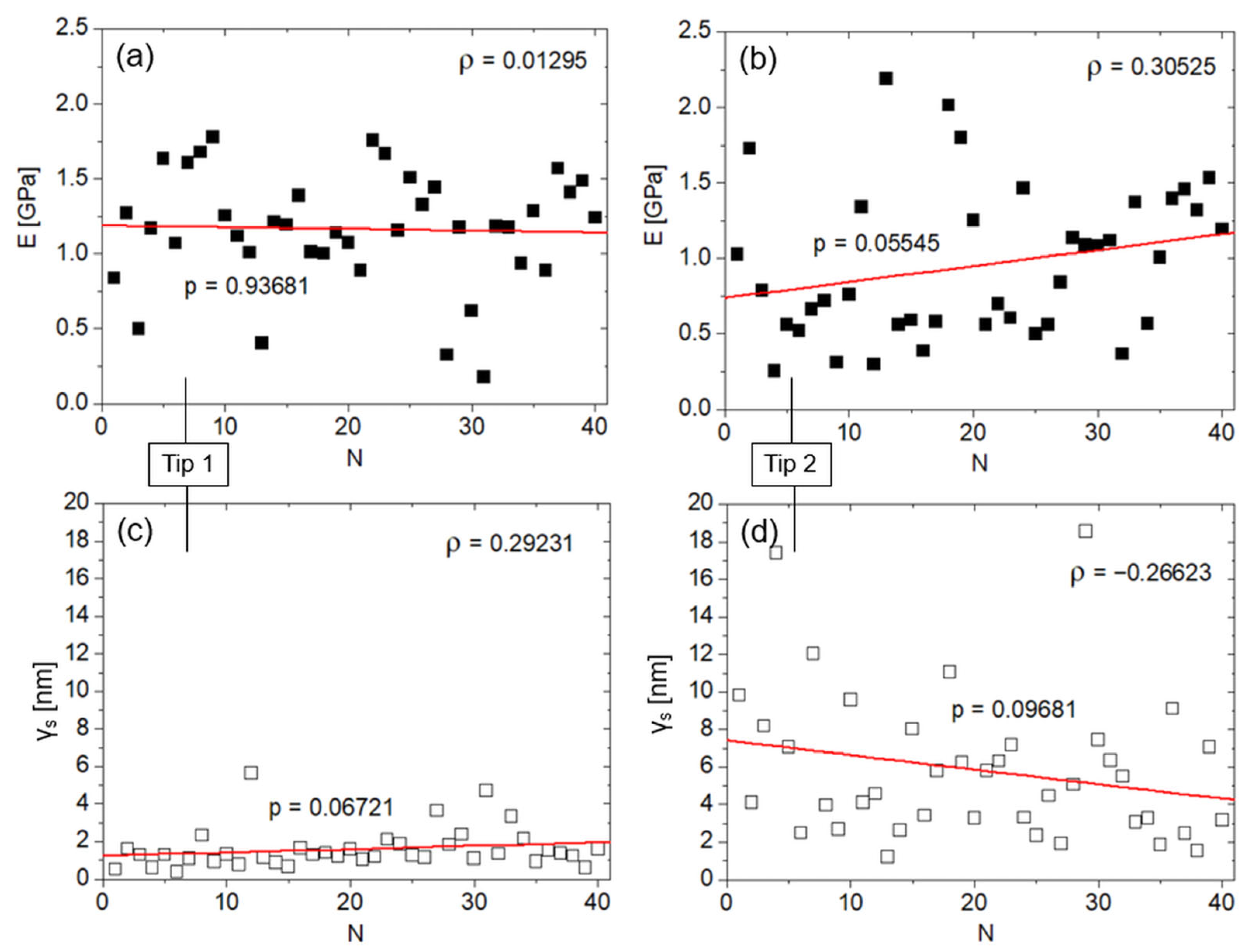
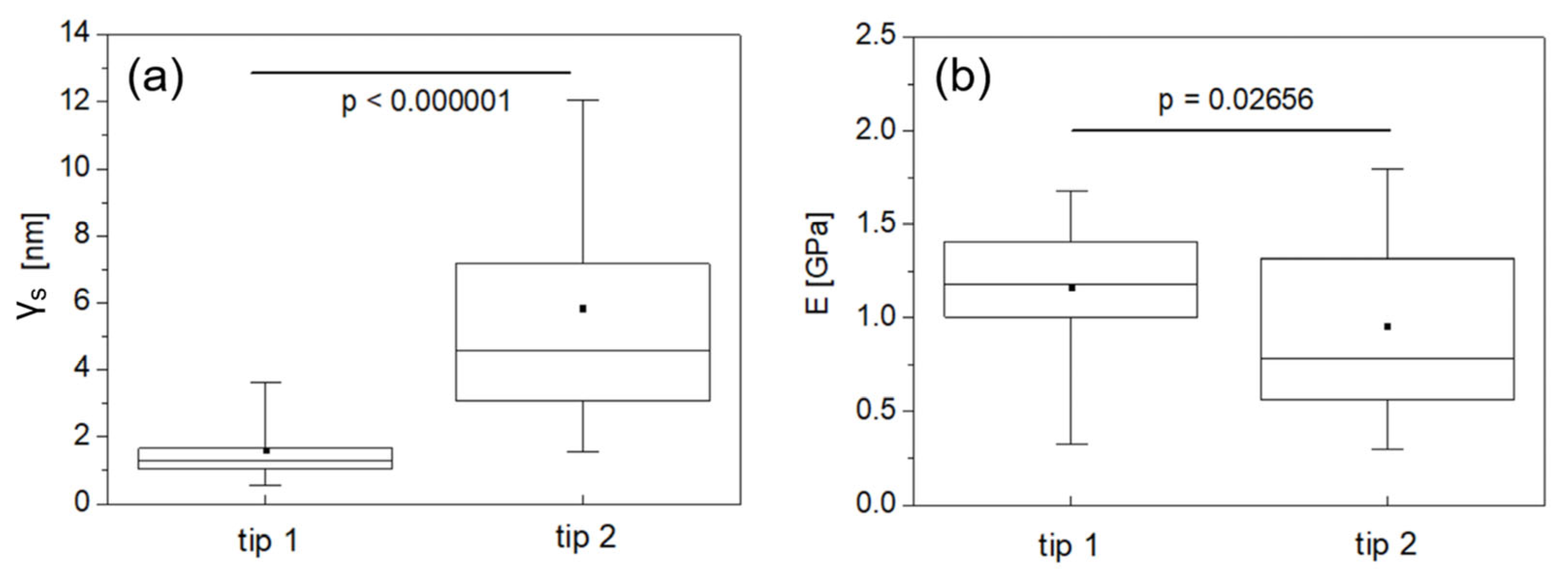
3. Results
4. Discussion
4.1. Comparison with Literature on Shallow Nanoindentation of Bone
4.2. General Considerations, Limitations, and Benefits of the Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AFM | Atomic Force Microscopy |
| ROI | Region of Interest |
| FTIR | Fourier Transform InfraRed |
| PMMA | Poly Methyl Methacrylate |
| F | Force |
| h | Penetration Depth |
| r | Tip Curvature Radius |
| k | Cantilever Stiffness |
| E | Elastic Modulus |
| Ac | Maximum Contact Area |
| Aγ | Sampled Area |
| γs | Local Roughness |
| N | Indentation Number |
| PTH | parathyroid Hormone |
References
- Rosenhek-Goldian, I.; Ram-On, M.; Weiner, S. Nanomechanics of biomaterials—From cells to shells. Isr. J. Chem. 2020, 60, 1171. [Google Scholar] [CrossRef]
- Holuigue, H.; Nacci, L.; Di Chiaro, P.; Chighizola, M.; Locatelli, I.; Schulte, C.; Alfano, M.; Diaferia, G.R.; Podestà, A. Native extracellular matrix probes to target patient- and tissue-specific cell–microenvironment interactions by force spectroscopy. Nanoscale 2023, 15, 15382–15395. [Google Scholar] [CrossRef]
- Sun, A.R.; Hengst, R.M.; Young, J.L. All the small things: Nanoscale matrix alterations in aging tissues. Curr. Opin. Cell Biol. 2024, 102, 102322. [Google Scholar] [CrossRef] [PubMed]
- Yi, B.; Xu, Q.; Liu, W. An overview of substrate stiffness guided cellular response and its applications in tissue regeneration. Bioact. Mater. 2022, 15, 82–102. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, A.K.; Sankar, D.; Amirthalingam, S.; Kim, H.D.; Rangasamy, J.; Hwang, N.S. Trends in mechanobiology guided tissue engineering and tools to study cell-substrate interactions: A brief review. Biomater. Res. 2023, 27, 55. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tian, L.; Zhang, W.; Wang, Z.; Liu, X. Effect of AFM Nanoindentation Loading Rate on the Characterization of Mechanical Properties of Vascular Endothelial Cell. Micromachines 2020, 11, 562. [Google Scholar] [CrossRef]
- Ku, C.-H.; Chamberlain, J.; Kao, A.P.; Han, I.-A.; Wang, H.; Tsimbouri, P.M.; Tsiridis, E.; Dalby, M.J. Nanoindentation Characteristics of Stem Cells Towards Osteogenic Differentiation in a Nanoscale Vibratory Bioreactor. Cell Biochem. Biophys. 2024, 14, 2435–2445. [Google Scholar] [CrossRef]
- Olubowale, O.H.; Biswas, S.; Azom, G.; Prather, B.L.; Owoso, S.D.; Rinee, K.C.; Marroquin, K.; Gates, K.A.; Chambers, M.B.; Xu, A.; et al. “May the Force Be with You!” Force–Volume Mapping with Atomic Force Microscopy. ACS Omega 2021, 6, 25860–25875. [Google Scholar] [CrossRef]
- Charvátová Campbell, A.; Buršíková, V.; Martinek, J.; Klapetek, P. Modeling the influence of roughness on nanoindentation data using finite element analysis. Int. J. Mech. Sci. 2019, 161–162, 105015. [Google Scholar] [CrossRef]
- Del Alamo, J.C.; Meili, R.; Alvarez-Gonzalez, B.; Alonso-Latorre, B.; Bastounis, E.; Firtel, R.; Lasheras, J.C. Three-Dimensional Quantification of Cellular Traction Forces and Mechanosensing of Thin Substrata by Fourier Traction Force Microscopy. PLoS ONE 2013, 8, e69850. [Google Scholar] [CrossRef]
- Kamml, J.; Acevedo, C.; Kammer, D. Mineral and Cross-Linking in Collagen Fibrils: The Mechanical Behavior of Bone Tissue at the Nano-Scale. Biomolecules 2024, 159, 106697. [Google Scholar] [CrossRef]
- Zhang, G.; Zhen, C.; Yang, J.; Wang, J.; Wang, S.; Fang, Y.; Shang, P. Recent Advances of Nanoparticles on Bone Tissue Engineering and Bone Cells. Nanoscale Adv. 2024, 6, 1957–1973. [Google Scholar] [CrossRef]
- Wan, T.; Zhang, M.; Jiang, H.-R.; Zhang, Y.-C.; Zhang, X.-M.; Wang, Y.-L.; Zhang, P.-X. Tissue-Engineered Nanomaterials Play Diverse Roles in Bone Injury Repair. Nanomaterials 2023, 13, 1449. [Google Scholar] [CrossRef] [PubMed]
- Bauso, L.V.; La Fauci, V.; Longo, C.; Calabrese, G. Bone Tissue Engineering and Nanotechnology: A Promising Combination for Bone Regeneration. Biology 2024, 13, 237. [Google Scholar] [CrossRef] [PubMed]
- Tertuliano, O.A.; Edwards, B.W.; Meza, L.R.; Deshpande, V.S.; Greer, J.R. Nanofibril-Mediated Fracture Resistance of Bone. Bioinspir. Biomim. 2021, 16, 035001. [Google Scholar] [CrossRef] [PubMed]
- Dee, P.; You, H.Y.; Teoh, S.-H.; Le Ferrand, H. Bioinspired Approaches to Toughen Calcium Phosphate-Based Ceramics for Bone Repair. J. Mech. Behav. Biomed. Mater. 2020, 112, 104078. [Google Scholar] [CrossRef]
- Blázquez-Carmona, P.; Mora-Macías, J.; Pajares, A.; Mármol, Á.; Reina-Romo, E. On the Influence of Structural and Chemical Properties on the Elastic Modulus of Woven Bone Under Healing. Front. Bioeng. Biotechnol. 2024, 12, 1476473. [Google Scholar] [CrossRef]
- Semenov, A.S.; Grishchenko, A.I.; Melnikov, B.E. Computational Study of Bone Tissue Mechanical Properties Dependence on Nanoscale Morphological Characteristics. Appl. Mech. Mater. 2015, 752–753, 103–108. [Google Scholar] [CrossRef]
- Wang, X.; Yang, X.; Zhang, S.; Jiang, T.; Zhou, Y.; Wang, Y. Bone Tissue Response to the Bone-Like Tissue Coating on Titanium. J. Wuhan Univ. Technol. Mater. Sci. Ed. 2015, 30, 203–209. [Google Scholar] [CrossRef]
- Maglio, M.; Salamanna, F.; Brogini, S.; Borsari, V.; Pagani, S.; Nicoli Aldini, N.; Giavaresi, G.; Fini, M. Histological, Histomorphometrical, and Biomechanical Studies of Bone-Implanted Medical Devices: Hard Resin Embedding. Biomed. Res. Int. 2020, 2020, 1804630. [Google Scholar] [CrossRef]
- Kourkoumelis, N.; Zhang, X.; Lin, Z.; Wang, J. Fourier Transform Infrared Spectroscopy of Bone Tissue: Bone Quality Assessment in Preclinical and Clinical Applications of Osteoporosis and Fragility Fracture. Clin. Rev. Bone Miner. Metab. 2019, 17, 24–39. [Google Scholar] [CrossRef]
- Lebon, M.; Müller, K.; Bahain, J.J.; Fröhlich, F.; Falguères, C.; Bertrand, L.; Sandt, C.; Reiche, I. Imaging Fossil Bone Alterations at the Microscale by SR-FTIR Microspectroscopy. J. Anal. At. Spectrom. 2011, 26, 922–929. [Google Scholar] [CrossRef]
- Bontempi, M.; Salamanna, F.; Capozza, R.; Visani, A.; Fini, M.; Gambardella, A. Nanomechanical Mapping of Hard Tissues by Atomic Force Microscopy: An Application to Cortical Bone. Materials 2022, 15, 7512. [Google Scholar] [CrossRef] [PubMed]
- Butt, H.-J.; Cappella, B.; Kappl, M. Force Measurements with the Atomic Force Microscope: Technique, Interpretation and Applications. Surf. Sci. Rep. 2005, 59, 1–152. [Google Scholar] [CrossRef]
- Kontomaris, S.V.; Malamou, A. Hertz model or Oliver & Pharr analysis? Tutorial regarding AFM nanoindentation experiments on biological samples. Mater. Res. Express 2020, 7, 033001. [Google Scholar] [CrossRef]
- Asgari, M.; Abi-Rafeh, J.; Hendy, G.N.; Pasini, D. Material anisotropy and elasticity of cortical and trabecular bone in the adult mouse femur via AFM indentation. J. Mech. Behav. Biomed. Mater. 2019, 93, 81–92. [Google Scholar] [CrossRef]
- Sadeghzadeh, S.; Korayem, M.H. Role of Mechanical and Thermal Nonlinearities in Imaging by Atomic Force Microscope. Int. J. Mech. Sci. 2017, 122, 255–266. [Google Scholar] [CrossRef]
- Mordue, C.W.; Weaver, J.M.R.; Dobson, P.S. Thermal Induced Deflection in Atomic Force Microscopy Cantilevers: Analysis & Solution. Meas. Sci. Technol. 2023, 34, 125013. [Google Scholar] [CrossRef]
- Wu, Y.; Fan, Z.; Fang, Y.; Liu, C. An Effective Correction Method for AFM Image Distortion due to Hysteresis and Thermal Drift. IEEE Trans. Instrum. Meas. 2021, 70, 1–12. [Google Scholar] [CrossRef]
- Wang, W.; Ma, N.; Kratz, K.; Xu, X.; Li, Z.; Roch, T.; Bieback, K.; Jung, F.; Lendlein, A. The influence of polymer scaffolds on cellular behaviour of bone marrow derived human mesenchymal stem cells. Clin. Hemorheol. Microcirc. 2012, 52, 357–373. [Google Scholar] [CrossRef]
- Xu, X.; Kratz, K.; Wang, W.; Li, Z.; Roch, T.; Jung, F.; Lendlein, A.; Ma, N. Cultivation and spontaneous differentiation of rat bone marrow-derived mesenchymal stem cells on polymeric surfaces. Clin Hemorheol Microcirc. 2013, 55, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Katsamenis, O.L.; Jenkins, T.; Thurner, P.J. Toughness and damage susceptibility in human cortical bone is proportional to mechanical inhomogeneity at the osteonal-level. Bone 2015, 76, 158–168. [Google Scholar] [CrossRef]
- Grue, B.H.; Vincent, L.C.; Kreplak, L.; Veres, S.P. Alternate soaking enables easy control of mineralized collagen scaffold mechanics from nano- to macro-scale. J. Mech. Behav. Biomed. Mater. 2020, 110, 103863. [Google Scholar] [CrossRef] [PubMed]
- Hongo, H.; Hasegawa, T.; Saito, M.; Tsuboi, K.; Yamamoto, T.; Sasaki, M.; Abe, M.; de Freitas, P.H.L.; Yurimoto, H.; Udagawa, N.; et al. Osteocytic Osteolysis in PTH-Treated Wild-Type and Rankl−/− Mice Examined by Transmission Electron Microscopy, Atomic Force Microscopy, and Isotope Microscopy. J. Histochem. Cytochem. 2020, 68, 651–668. [Google Scholar] [CrossRef] [PubMed]
- Cisneros, T.; Sevostianov, I.; Drach, B. Elasticity and Material Anisotropy of Lamellar Cortical Bone in Adult Bovine Tibia Characterized via AFM Nanoindentation. J. Mech. Behav. Biomed. Mater. 2023, 144, 105992. [Google Scholar] [CrossRef] [PubMed]
- Walter, C.; Antretter, T.; Daniel, R.; Mitterer, C. Finite element simulation of the effect of surface roughness on nanoindentation of thin films with spherical indenters. Surf. Coat. Technol. 2007, 202, 1103–1107. [Google Scholar] [CrossRef]
- Chen, L.; Ahadi, A.; Zhou, J.; Jan-Eric Ståhl, J.E. Modeling Effect of Surface Roughness on Nanoindentation Tests. Procedia CIRP 2013, 8, 334–339. [Google Scholar] [CrossRef]
- Tai, K.; Dao, M.; Suresh, S.; Palazoglu, A.; Ortiz, C. Nanoscale Heterogeneity Promotes Energy Dissipation in Bone. Nat. Mater. 2007, 6, 454–462. [Google Scholar] [CrossRef]
- Berni, M.; Marchiori, G.; Cassiolas, G.; Grassi, A.; Zaffagnini, S.; Fini, M.; Lopomo, N.F.; Maglio, M. Anisotropy and Inhomogeneity of Permeability and Fibrous Network Response in the Pars Intermedia of the Human Lateral Meniscus. Acta Biomater. 2021, 135, 393–402. [Google Scholar] [CrossRef]
- Casanova, M.; Balmelli, A.; Carnelli, D.; Courty, D.; Schneider, P.; Müller, R. Nanoindentation analysis of the micromechanical anisotropy in mouse cortical bone. R. Soc. Open Sci. 2017, 4, 160971. [Google Scholar] [CrossRef]
- Gregorio, M.; Alessandro, G.; Melania, M.; Marco, B.; Marco, B.; Matteo, M.; Jessika, B.; Stefano, B.; Nicola, B.; Gianluca, G. Targeting the Micrometers Scale on The Relation of Bone Crystallinity, Elastic Modulus and Mineral Density. In Proceedings of the 30th Congress of the European Society of Biomechanics, Zürich, Switzerland, 6–9 July 2025; Available online: https://www.conftool.com/esb2025/index.php/ESB2025_D2_10_Marchiori.pdf?page=downloadPaper&filename=ESB2025_D2_10_Marchiori.pdf&form_id=840 (accessed on 16 July 2025).


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gambardella, A.; Marchiori, G.; Maglio, M.; Boi, M.; Montesissa, M.; Bertacchini, J.; Biressi, S.; Baldini, N.; Giavaresi, G.; Bontempi, M. On the Interplay Between Roughness and Elastic Modulus at the Nanoscale: A Methodology Study with Bone as Model Material. J. Funct. Biomater. 2025, 16, 276. https://doi.org/10.3390/jfb16080276
Gambardella A, Marchiori G, Maglio M, Boi M, Montesissa M, Bertacchini J, Biressi S, Baldini N, Giavaresi G, Bontempi M. On the Interplay Between Roughness and Elastic Modulus at the Nanoscale: A Methodology Study with Bone as Model Material. Journal of Functional Biomaterials. 2025; 16(8):276. https://doi.org/10.3390/jfb16080276
Chicago/Turabian StyleGambardella, Alessandro, Gregorio Marchiori, Melania Maglio, Marco Boi, Matteo Montesissa, Jessika Bertacchini, Stefano Biressi, Nicola Baldini, Gianluca Giavaresi, and Marco Bontempi. 2025. "On the Interplay Between Roughness and Elastic Modulus at the Nanoscale: A Methodology Study with Bone as Model Material" Journal of Functional Biomaterials 16, no. 8: 276. https://doi.org/10.3390/jfb16080276
APA StyleGambardella, A., Marchiori, G., Maglio, M., Boi, M., Montesissa, M., Bertacchini, J., Biressi, S., Baldini, N., Giavaresi, G., & Bontempi, M. (2025). On the Interplay Between Roughness and Elastic Modulus at the Nanoscale: A Methodology Study with Bone as Model Material. Journal of Functional Biomaterials, 16(8), 276. https://doi.org/10.3390/jfb16080276








