In Vitro Biological Properties Assessment of 3D-Printed Hydroxyapatite–Polylactic Acid Scaffolds Intended for Bone Regeneration
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
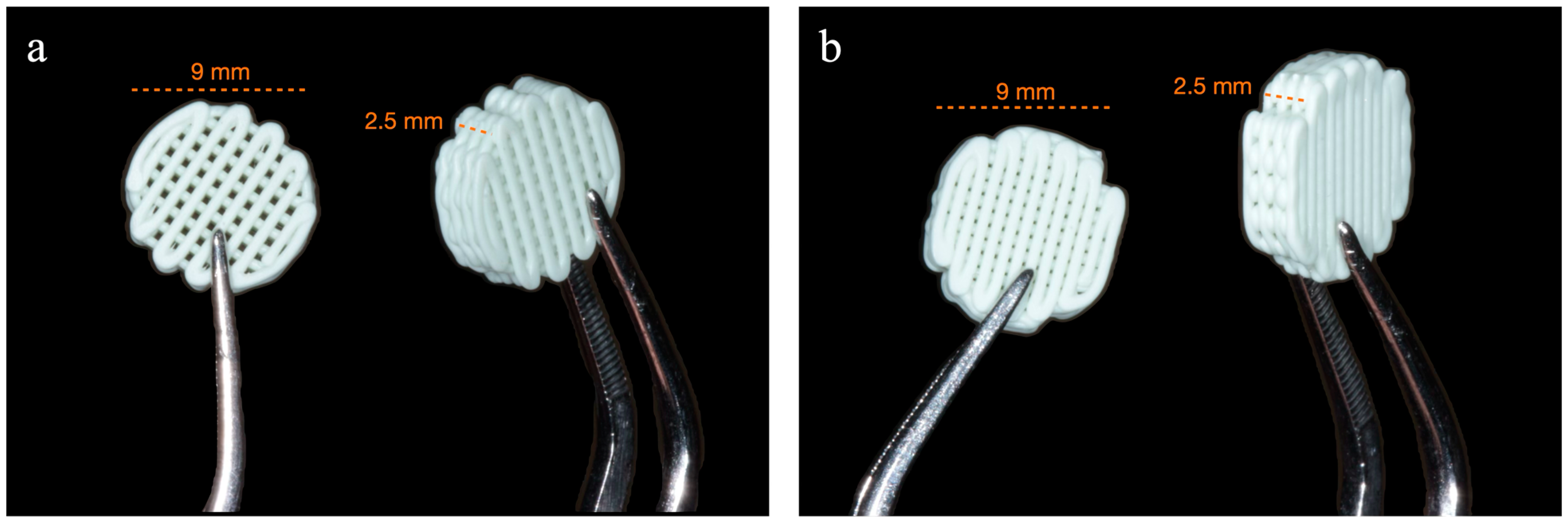
2.2. Fabrication and Design of Scaffolds
2.3. Cell Culture
2.4. Experimental Assays
2.4.1. Sterilization Method
2.4.2. Cell Proliferation
2.4.3. Scanning Electron Microscopy (SEM)
2.4.4. Cell Viability—Confocal Laser Scanning Microscopy (CLSM)
2.4.5. Cytotoxicity
2.4.6. Gene Expression
2.4.7. Protein Synthesis
2.5. Data Analysis
2.5.1. Outcome Variables
2.5.2. Statistical Analysis
3. Results
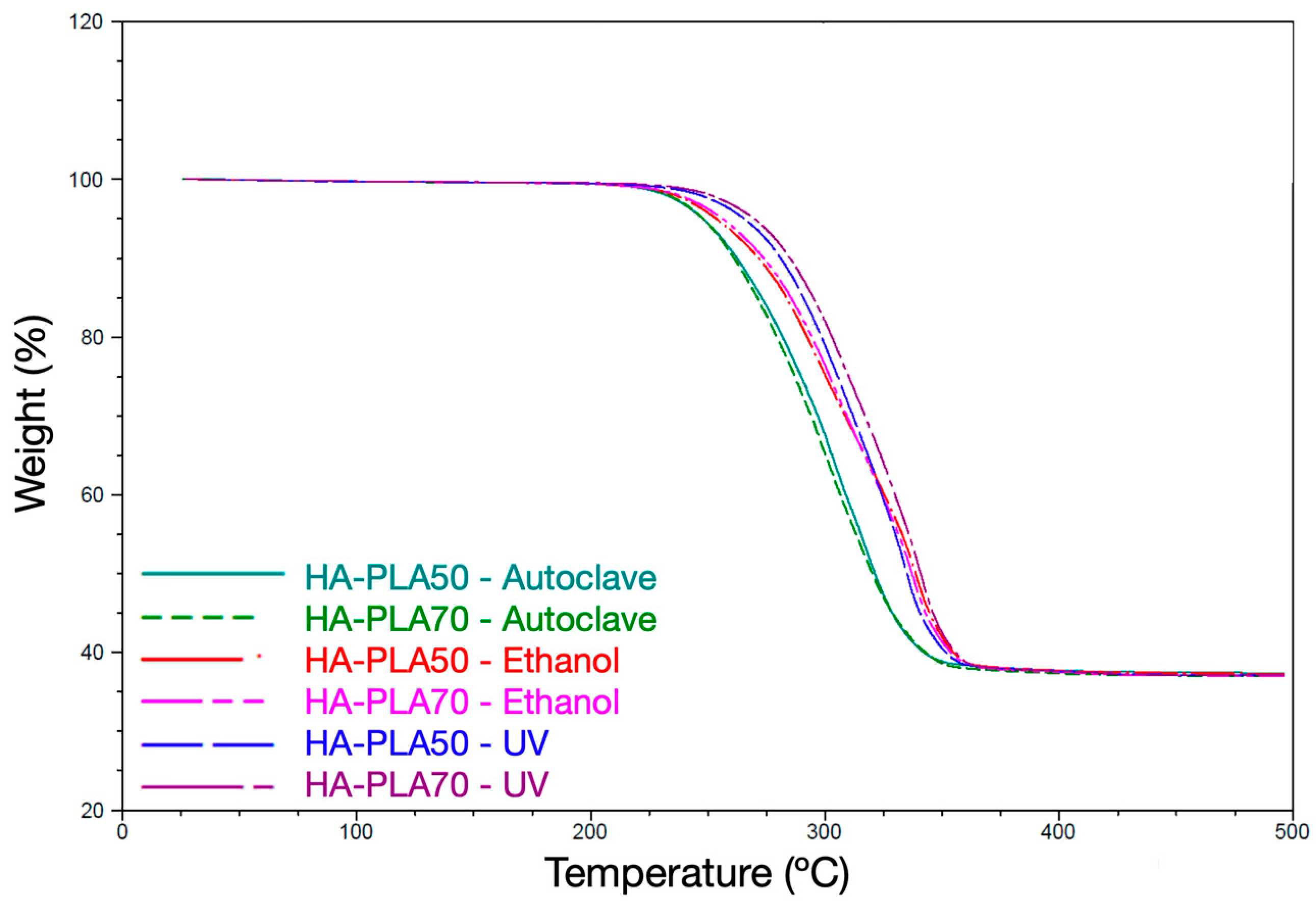
3.1. Sterilization Method
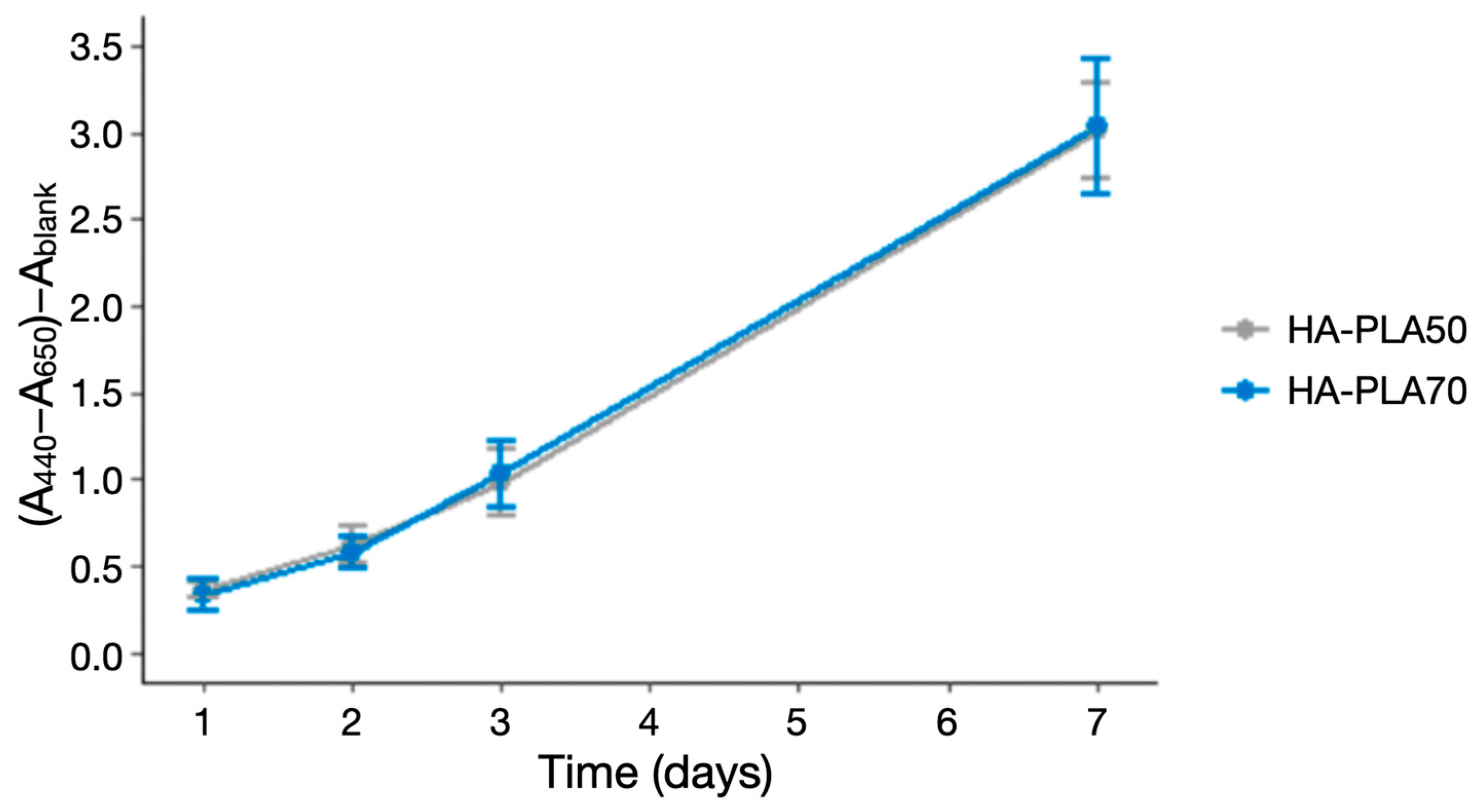
3.2. Cell Proliferation
3.3. Scanning Electron Microscopy
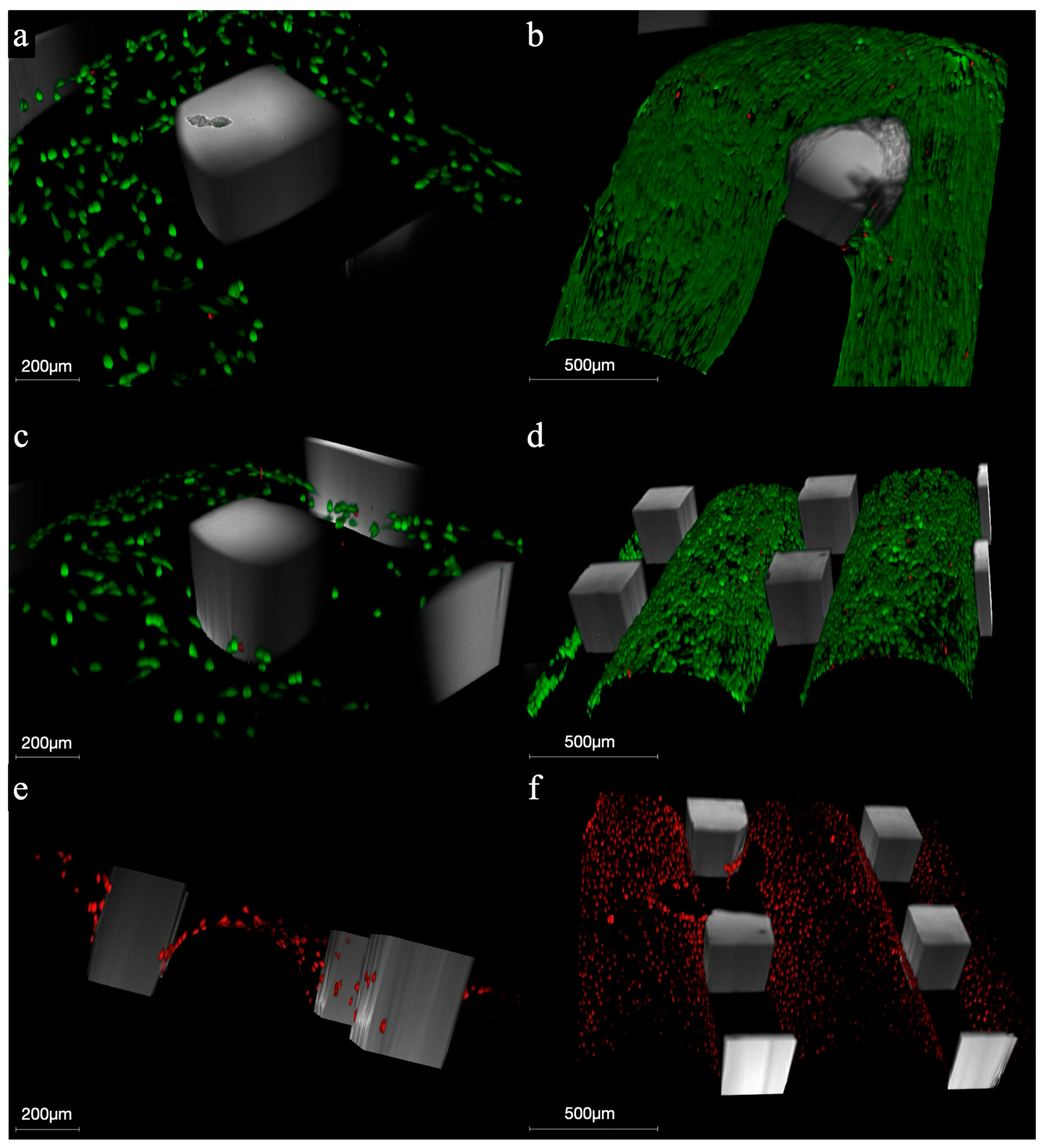
3.4. Cell Viability–CLSM
3.5. Cytotoxicity
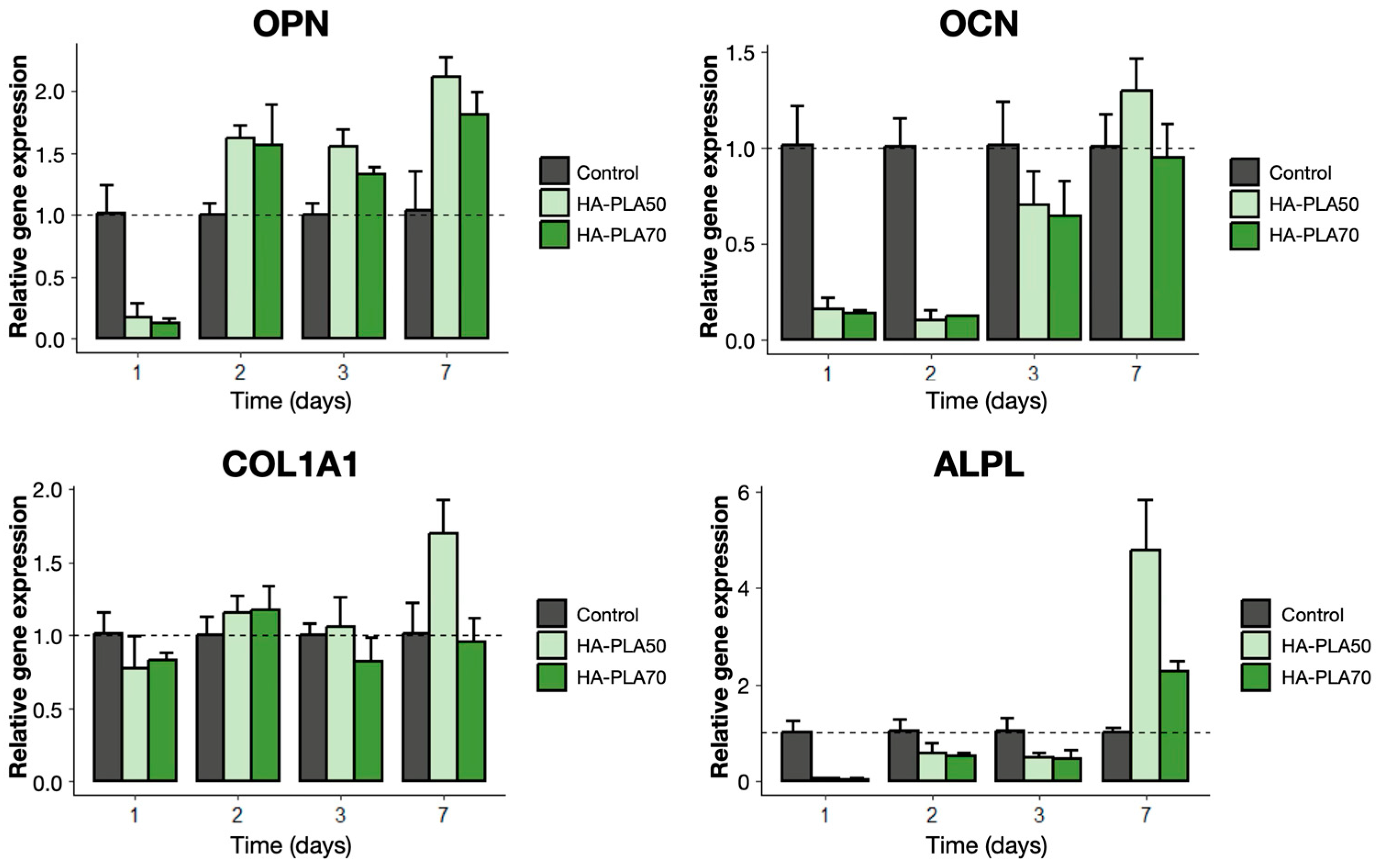
3.6. Gene Expression
3.7. Protein Synthesis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CLSM | Confocal scanning laser microscopy |
| COL1A1 | Collagen type 1 |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| HA | Hydroxyapatite |
| IL | Interleukin |
| M-CSF | Macrophage colony-stimulating factor |
| MMP | Matrix metalloproteinase |
| PLA | Polylactic acid |
| RANKL | Receptor activator of nuclear factor kappa-B ligand |
| RT-qPCR | Reverse transcription-quantitative polymerase chain reaction |
| SEM | Scanning electron microscopy |
| TBP | TATA-box binding protein |
| TGA | Thermogravimetry |
| UV | Ultraviolet |
| WST | Water-soluble tetrazolium |
Appendix A

| Gene | Primer | Sequence |
|---|---|---|
| TBP | Forward | 5′-TGTATCCACAGTGAATCTTGGTTG-3′ |
| Reverse | 5′-GGTTCGTGGCTCTCTTATCCTC-3′ | |
| GAPDH | Forward | 5′-GTCTCCTCTGACTTCAACAGCG-3′ |
| Reverse | 5′-ACCACCCTGTTGCTGTAGCCAA-3′ | |
| OPN | Forward | 5′-CGAGGTGATATAGTGTGGTTTATGG-3′ |
| Reverse | 5′-GCACCATTCAACTCCTCGCTTTC-3′ | |
| OCN | Forward | 5′-CGCTACCTGTATCAATGGCTGG-3′ |
| Reverse | 5′-CTCCTGAAAGCCGATGTGGTCA-3′ | |
| COL1A1 | Forward | 5′-GATTCCCTGGACCTAAAGGTGC-3′ |
| Reverse | 5′-AGCCTCTCCATCTTTGCCAGCA-3′ | |
| ALPL | Forward | 5′-GCTGTAAGGACATCGCCTACCA-3′ |
| Reverse | 5′-CCTGGCTTTCTCGTCACTCTCA-3′ |
| Gene | Infill | N | Relative Gene Expression (Mean (SD)) | p Value (Between Time Points) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 1W | 24 h vs. 48 h | 24 h vs. 72 h | 24 h vs. 1 W | ||||
| ALPL | HA-PLA50 | 3 | 0.05 (0.01) | 0.57 (0.21) | 0.49 (0.08) | 4.77 (1.04) | 0.317 | 0.069 | 0.107 | |
| HA-PLA70 | 3 | 0.04 (0.01) | 0.50 (0.06) | 0.47 (0.17) | 2.28 (0.19) | 0.033 | 0.294 | 0.017 | ||
| CONTROL | 3 | 1.01 (0.22) | 1.02 (0.25) | 1.02 (0.26) | 1.00 (0.08) | 1.000 | 1.000 | 1.000 | ||
| p value (between scaffolds) | HA-PLA50 vs. HA-PLA70 | 0.034 | 1.000 | 1.000 | 0.326 | |||||
| HA-PLA50 vs. Control | 0.324 | 0.681 | 0.414 | 0.161 | ||||||
| HA-PLA70 vs. Control | 0.320 | 0.409 | 0.438 | 0.032 | ||||||
| COL1A1 | HA-PLA50 | 3 | 0.77 (0.21) | 1.16 (0.11) | 1.06 (0.21) | 1.70 (0.22) | 0.528 | 1.000 | <0.001 | |
| HA-PLA70 | 3 | 0.83 (0.05) | 1.17 (0.16) | 0.82 (0.16) | 0.96 (0.16) | 1.000 | 1.000 | 1.000 | ||
| CONTROL | 3 | 1.01 (0.15) | 1.00 (0.12) | 1.00 (0.07) | 1.02 (0.21) | 1.000 | 1.000 | 1.000 | ||
| p value (between scaffolds) | HA-PLA50 vs. HA-PLA70 | 1.000 | 1.000 | 1.000 | <0.001 | |||||
| HA-PLA50 vs. Control | 1.000 | 1.000 | 1.000 | 0.002 | ||||||
| HA-PLA70 vs. Control | 1.000 | 1.000 | 1.000 | 1.000 | ||||||
| OCN | HA-PLA50 | 3 | 0.15 (0.06) | 0.10 (0.05) | 0.70 (0.17) | 1.29 (0.17) | 0.998 | 0.185 | 0.030 | |
| HA-PLA70 | 3 | 0.13 (0.02) | 0.12 (0.00) | 0.64 (0.18) | 0.95 (0.17) | 0.985 | 0.255 | 0.098 | ||
| CONTROL | 3 | 1.01 (0.20) | 1.00 (0.14) | 1.01 (0.22) | 1.00 (0.16) | 1.000 | 1.000 | 1.000 | ||
| p value (between scaffolds) | HA-PLA50 vs. HA-PLA70 | 1.000 | 1.000 | 1.000 | 0.632 | |||||
| HA-PLA50 vs. Control | 0.100 | 0.040 | 0.843 | 0.841 | ||||||
| HA-PLA70 vs. Control | 0.113 | 0.062 | 0.732 | 1.000 | ||||||
| OPN | HA-PLA50 | 3 | 0.16 (0.11) | 1.62 (0,09) | 1.55 (0.14) | 2.11 (0.16) | <0.001 | <0.001 | <0.001 | |
| HA-PLA70 | 3 | 0.12 (0.03) | 1.57 (0.32) | 1.33 (0.05) | 1.81 (0.19) | <0.001 | <0.001 | <0.001 | ||
| CONTROL | 3 | 1.01 (0.22) | 1.00 (0.09) | 1.00 (0.09) | 1.03 (0.32) | 1.000 | 1.000 | 1.000 | ||
| p value (between scaffolds) | HA-PLA50 vs. HA-PLA70 | 1.000 | 1.000 | 1.000 | 1.000 | |||||
| HA-PLA50 vs. Control | <0.001 | 0.024 | 0.182 | <0.001 | ||||||
| HA-PLA70 vs. Control | <0.001 | 0.057 | 1.000 | 0.002 | ||||||
| Protein Synthesis | Infill | N | Concentration pg/mL (Mean (SD)) | p Value (Between Time Points) | |||||
|---|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 1 W | 24 h vs. 48 h | 24 h vs. 72 h | 24 h vs. 1 W | |||
| IL-6 | HA-PLA50 | 3 | 55.25 (13.32) | 66.09 (1.09) | 80.68 (1.14) | 194.04 (6.72) | 0.954 | 0.461 | 0.007 |
| HA-PLA70 | 3 | 39.33 (9.37) | 77.07 (2.47) | 75.66 (4.28) | 120.65 (5.58) | 0.117 | 0.107 | 0.009 | |
| CONTROL | 3 | 45.40 (6.05) | 46.18 (8.13) | 52.22 (13.42) | 155.57 (15.62) | 1.000 | 1.000 | 0.027 | |
| p value (between scaffolds) | HA-PLA50 vs. HA-PLA70 | 0.916 | 0.077 | 0.813 | 0.003 | ||||
| HA-PLA50 vs. Control | 0.992 | 0.314 | 0.395 | 0.300 | |||||
| HA-PLA70 vs. Control | 1.000 | 0.128 | 0.533 | 0.360 | |||||
| IL-8 | HA-PLA50 | 3 | 408.04 (75.85) | 434.42 (48.83) | 510.72 (48.75) | 1546.95 (81.09) | 1.000 | 1.000 | <0.001 |
| HA-PLA70 | 3 | 293.65 (56.01) | 701.98 (50.19) | 484.16 (95.73) | 1187.54 (90.89) | <0.001 | 0.051 | <0.001 | |
| CONTROL | 3 | 155.10 (20.08) | 188.73 (30.11) | 234.43 (41.37) | 479.21 (29.63) | 1.000 | 1.000 | <0.001 | |
| p value (between scaffolds) | HA-PLA50 vs. HA-PLA70 | 1.000 | <0.001 | 1.000 | <0.001 | ||||
| HA-PLA50 vs. Control | 0.002 | 0.003 | <0.001 | <0.001 | |||||
| HA-PLA70 vs. Control | 0.655 | <0.001 | 0.002 | <0.001 | |||||
| M-CSF | HA-PLA50 | 3 | 492.79 (84.50) | 755.61 (59.26) | 1055.86 (6.84) | 3054.39 (396.82) | 0.183 | 0.050 | 0.046 |
| HA-PLA70 | 3 | 333.43 (80.04) | 918.58 (84.30) | 1137.25 (55.17) | 3756.68 (175.99) | 0.016 | 0.004 | 0.001 | |
| CONTROL | 3 | 582.21 (74.77) | 821.08 (100.85) | 1065.47 (31.64) | 3870.45 (477.06) | 0.371 | 0.031 | 0.043 | |
| p value (between scaffolds) | HA-PLA50 vs. HA-PLA70 | 0.673 | 0.539 | 0.640 | 0.542 | ||||
| HA-PLA50 vs. Control | 0.981 | 0.999 | 1.000 | 0.710 | |||||
| HA-PLA70 vs. Control | 0.229 | 0.989 | 0.827 | 1.000 | |||||
| MMP-1 | HA-PLA50 | 3 | 893.41 (140.99) | 1146.76 (143.42) | 1333.12 (145.00) | 2823.84 (513.31) | 0.749 | 0.257 | 0.133 |
| HA-PLA70 | 3 | 919.62 (56.51) | 1242.56 (200.89) | 1519.94 (140.59) | 3212.64 (114.59) | 0.592 | 0.089 | 0.001 | |
| CONTROL | 3 | 307.15 (17.50) | 525.09 (30.18) | 646.04 (101.22) | 1451.68 (138.29) | 0.016 | 0.178 | 0.031 | |
| p value (between scaffolds) | HA-PLA50 vs. HA-PLA70 | 1.000 | 1.000 | 0.944 | 0.976 | ||||
| HA-PLA50 vs. Control | 0.121 | 0.105 | 0.052 | 0.262 | |||||
| HA-PLA70 vs. Control | 0.012 | 0.159 | 0.021 | 0.002 | |||||
References
- Schropp, L.; Wenzel, A.; Kostopoulos, L.; Karring, T. Bone healing and soft tissue contour changes following single-tooth extraction: A clinical and radiographic 12-month prospective study. Int. J. Periodontics Restor. Dent. 2003, 23, 313–323. [Google Scholar]
- Wang, H.L.; Boyapati, L. "PASS" principles for predictable bone regeneration. Implant. Dent. 2006, 15, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Sanchez, I.; Ortiz-Vigon, A.; Sanz-Martin, I.; Figuero, E.; Sanz, M. Effectiveness of Lateral Bone Augmentation on the Alveolar Crest Dimension: A Systematic Review and Meta-analysis. J. Dent. Res. 2015, 94, 128S–142S. [Google Scholar] [CrossRef] [PubMed]
- Urban, I.A.; Montero, E.; Amerio, E.; Palombo, D.; Monje, A. Techniques on vertical ridge augmentation: Indications and effectiveness. Periodontol 2000 2023, 93, 153–182. [Google Scholar] [CrossRef] [PubMed]
- Rogers, G.F.; Greene, A.K. Autogenous bone graft: Basic science and clinical implications. J. Craniofac. Surg. 2012, 23, 323–327. [Google Scholar] [CrossRef]
- Mir-Mari, J.; Wui, H.; Jung, R.E.; Hammerle, C.H.; Benic, G.I. Influence of blinded wound closure on the volume stability of different GBR materials: An in vitro cone-beam computed tomographic examination. Clin. Oral Implant. Res. 2016, 27, 258–265. [Google Scholar] [CrossRef]
- Ivanovski, S.; Breik, O.; Carluccio, D.; Alayan, J.; Staples, R.; Vaquette, C. 3D printing for bone regeneration: Challenges and opportunities for achieving predictability. Periodontol 2000 2023, 93, 358–384. [Google Scholar] [CrossRef]
- Wang, C.; Huang, W.; Zhou, Y.; He, L.; He, Z.; Chen, Z.; He, X.; Tian, S.; Liao, J.; Lu, B.; et al. 3D printing of bone tissue engineering scaffolds. Bioact. Mater. 2020, 5, 82–91. [Google Scholar] [CrossRef]
- Obregon, F.; Vaquette, C.; Ivanovski, S.; Hutmacher, D.W.; Bertassoni, L.E. Three-Dimensional Bioprinting for Regenerative Dentistry and Craniofacial Tissue Engineering. J. Dent. Res. 2015, 94, 143S–152S. [Google Scholar] [CrossRef]
- Persson, M.; Lorite, G.S.; Kokkonen, H.E.; Cho, S.W.; Lehenkari, P.P.; Skrifvars, M.; Tuukkanen, J. Effect of bioactive extruded PLA/HA composite films on focal adhesion formation of preosteoblastic cells. Colloids Surf. B Biointerfaces 2014, 121, 409–416. [Google Scholar] [CrossRef]
- Chocholata, P.; Kulda, V.; Babuska, V. Fabrication of Scaffolds for Bone-Tissue Regeneration. Materials 2019, 12, 568. [Google Scholar] [CrossRef]
- Cao, S.S.; Li, S.Y.; Geng, Y.M.; Kapat, K.; Liu, S.B.; Perera, F.H.; Li, Q.; Terheyden, H.; Wu, G.; Che, Y.J.; et al. Prefabricated 3D-Printed Tissue-Engineered Bone for Mandibular Reconstruction: A Preclinical Translational Study in Primate. ACS Biomater. Sci. Eng. 2021, 7, 5727–5738. [Google Scholar] [CrossRef]
- Huang, J.; Xiong, J.; Liu, J.; Zhu, W.; Chen, J.; Duan, L.; Zhang, J.; Wang, D. Evaluation of the novel three-dimensional porous poly (L-lactic acid)/nano-hydroxyapatite composite scaffold. Biomed. Mater. Eng. 2015, 26 (Suppl. S1), S197–S205. [Google Scholar] [CrossRef]
- Ferrández-Montero, A.; Ortega-Columbrans, P.; Eguiluz, A.; Sanchez-Herencia, A.J.; Detsch, R.; Boccaccini, A.R.; Ferrari, B. Biocompatible colloidal feedstock for material extrusion processing of bioceramic-based scaffolds. Polym. Compos. 2024, 45, 7237–7255. [Google Scholar] [CrossRef]
- Kinoshita, Y.; Maeda, H. Recent developments of functional scaffolds for craniomaxillofacial bone tissue engineering applications. Sci. J. 2013, 2013, 863157. [Google Scholar] [CrossRef]
- Bartnikowski, M.; Klein, T.J.; Melchels, F.P.; Woodruff, M.A. Effects of scaffold architecture on mechanical characteristics and osteoblast response to static and perfusion bioreactor cultures. Biotechnol. Bioeng. 2014, 111, 1440–1451. [Google Scholar] [CrossRef]
- Abbasi, N.; Ivanovski, S.; Gulati, K.; Love, R.M.; Hamlet, S. Role of offset and gradient architectures of 3-D melt electrowritten scaffold on differentiation and mineralization of osteoblasts. Biomater. Res. 2020, 24, 2. [Google Scholar] [CrossRef]
- Bartnikowski, M.; Vaquette, C.; Ivanovski, S. Workflow for highly porous resorbable custom 3D printed scaffolds using medical grade polymer for large volume alveolar bone regeneration. Clin. Oral Implant. Res. 2020, 31, 431–441. [Google Scholar] [CrossRef]
- Yu, N.; Nguyen, T.; Cho, Y.D.; Kavanagh, N.M.; Ghassib, I.; Giannobile, W.V. Personalized scaffolding technologies for alveolar bone regenerative medicine. Orthod. Craniofac. Res. 2019, 22 (Suppl. S1), 69–75. [Google Scholar] [CrossRef]
- Giannitelli, S.M.; Accoto, D.; Trombetta, M.; Rainer, A. Current trends in the design of scaffolds for computer-aided tissue engineering. Acta Biomater. 2014, 10, 580–594. [Google Scholar] [CrossRef]
- Abbasi, N.; Abdal-Hay, A.; Hamlet, S.; Graham, E.; Ivanovski, S. Effects of Gradient and Offset Architectures on the Mechanical and Biological Properties of 3-D Melt Electrowritten (MEW) Scaffolds. ACS Biomater. Sci. Eng. 2019, 5, 3448–3461. [Google Scholar] [CrossRef]
- Smith, J.A.; Mele, E. Electrospinning and Additive Manufacturing: Adding Three-Dimensionality to Electrospun Scaffolds for Tissue Engineering. Front. Bioeng. Biotechnol. 2021, 9, 674738. [Google Scholar] [CrossRef]
- Ferrandez-Montero, A.; Lieblich, M.; Benavente, R.; González-Carrasco, J.L.; Ferrari, B. New approach to improve polymer-Mg interface in biodegradable PLA/Mg composites through particle surface modification. Surf. Coat. Technol. 2020, 383, 125285. [Google Scholar] [CrossRef]
- Oberbek, P.; Bolek, T.; Chlanda, A.; Hirano, S.; Kusnieruk, S.; Rogowska-Tylman, J.; Nechyporenko, G.; Zinchenko, V.; Swieszkowski, W.; Puzyn, T. Characterization and influence of hydroxyapatite nanopowders on living cells. Beilstein J. Nanotechnol. 2018, 9, 3079–3094. [Google Scholar] [CrossRef]
- Ribeiro, M.M.; Neumann, V.A.; Padoveze, M.C.; Graziano, K.U. Efficacy and effectiveness of alcohol in the disinfection of semi-critical materials: A systematic review. Rev. Lat. Am. Enferm. 2015, 23, 741–752. [Google Scholar] [CrossRef]
- Jin, P.; Zhao, Y.; Ngalame, Y.; Panelli, M.C.; Nagorsen, D.; Monsurró, V.; Smith, K.; Hu, N.; Su, H.; Taylor, P.R.; et al. Selection and validation of endogenous reference genes using a high throughput approach. BMC Genom. 2004, 5, 55. [Google Scholar] [CrossRef]
- Dai, Z.; Ronholm, J.; Tian, Y.; Sethi, B.; Cao, X. Sterilization techniques for biodegradable scaffolds in tissue engineering applications. J. Tissue Eng. 2016, 7, 2041731416648810. [Google Scholar] [CrossRef]
- Rodovalho, A.J.R.L.; Barbosa, W.T.; Vieira, J.L.; Oliva, C.A.d.; Gonçalves, A.P.B.; Cardoso, P.d.S.M.; Modolon, H.B.; Montedo, O.R.K.; Arcaro, S.; Hodel, K.V.S.; et al. Influence of size and crystallinity of nanohydroxyapatite (nHA) particles on the properties of Polylactic Acid/nHA nanocomposite scaffolds produced by 3D printing. J. Mater. Res. Technol. 2024, 30, 3101–3111. [Google Scholar] [CrossRef]
- Chen, X.; Gao, C.; Jiang, J.; Wu, Y.; Zhu, P.; Chen, G. 3D printed porous PLA/nHA composite scaffolds with enhanced osteogenesis and osteoconductivity in vivo for bone regeneration. Biomed. Mater. 2019, 14, 065003. [Google Scholar] [CrossRef]
- Gregor, A.; Filova, E.; Novak, M.; Kronek, J.; Chlup, H.; Buzgo, M.; Blahnova, V.; Lukasova, V.; Bartos, M.; Necas, A.; et al. Designing of PLA scaffolds for bone tissue replacement fabricated by ordinary commercial 3D printer. J. Biol. Eng. 2017, 11, 31. [Google Scholar] [CrossRef]
- Karanth, D.; Puleo, D.; Dawson, D.; Holliday, L.S.; Sharab, L. Characterization of 3D printed biodegradable piezoelectric scaffolds for bone regeneration. Clin. Exp. Dent. Res. 2023, 9, 398–408. [Google Scholar] [CrossRef]
- Pamula, E.; Filova, E.; Bacakova, L.; Lisa, V.; Adamczyk, D. Resorbable polymeric scaffolds for bone tissue engineering: The influence of their microstructure on the growth of human osteoblast-like MG 63 cells. J. Biomed. Mater. Res. A 2009, 89, 432–443. [Google Scholar] [CrossRef]
- Bahraminasab, M.; Arab, S.; Ghaffari, S. Osteoblastic cell response to Al2O3-Ti composites as bone implant materials. Bioimpacts 2022, 12, 247–259. [Google Scholar] [CrossRef]
- Vaquette, C.; Mitchell, J.; Fernandez-Medina, T.; Kumar, S.; Ivanovski, S. Resorbable additively manufactured scaffold imparts dimensional stability to extraskeletally regenerated bone. Biomaterials 2021, 269, 120671. [Google Scholar] [CrossRef]
- Ivanovski, S.; Staples, R.; Arora, H.; Vaquette, C.; Alayan, J. Alveolar bone regeneration using a 3D-printed patient-specific resorbable scaffold for dental implant placement: A case report. Clin. Oral Implant. Res. 2024, 35, 1655–1668. [Google Scholar] [CrossRef]


| Infill | N | Absorbance (Mean (SD)) | p Value (Between Time Points) | |||||
|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 1 W | 24 h vs. 48 h | 24 h vs. 72 h | 24 h vs. 1 W | ||
| HA-PLA50 | 6 | 0.36 (0.04) | 0.62 (0.10) | 0.96 (0.19) | 3.01 (0.28) | 0.014 | 0.006 | <0.001 |
| HA-PLA70 | 6 | 0.33 (0.09) | 0.57 (0.09) | 1.01 (0.19) | 3.04 (0.39) | 0.020 | 0.001 | <0.001 |
| p value (between scaffolds) | 1.000 | 1.000 | 1.000 | 1.000 | ||||
| Infill | N | Absorbance (Mean (SD)) | p Value (Between Time Points) | |||||
|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 1 W | 24 h vs. 48 h | 24 h vs. 72 h | 24 h vs. 1 W | ||
| HA-PLA50 | 6 | 1.94 (0.28) | 1.94 (0.20) | 2.41 (0.51) | 2.21 (0.11) | 1.000 | 0.716 | 0.666 |
| HA-PLA70 | 6 | 2.56 (0.19) | 1.66 (0.50) | 2.40 (0.37) | 2.12 (0.17) | 0.080 | 1.000 | 0.177 |
| p value (between scaffolds) | 0.071 | 0.974 | 1.000 | 0.999 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shan, E.; Chamorro, C.; Ferrández-Montero, A.; Martin-Rodriguez, R.M.; Ferrari, B.; Sanchez-Herencia, A.J.; Virto, L.; Marín, M.J.; Figuero, E.; Sanz, M. In Vitro Biological Properties Assessment of 3D-Printed Hydroxyapatite–Polylactic Acid Scaffolds Intended for Bone Regeneration. J. Funct. Biomater. 2025, 16, 218. https://doi.org/10.3390/jfb16060218
Shan E, Chamorro C, Ferrández-Montero A, Martin-Rodriguez RM, Ferrari B, Sanchez-Herencia AJ, Virto L, Marín MJ, Figuero E, Sanz M. In Vitro Biological Properties Assessment of 3D-Printed Hydroxyapatite–Polylactic Acid Scaffolds Intended for Bone Regeneration. Journal of Functional Biomaterials. 2025; 16(6):218. https://doi.org/10.3390/jfb16060218
Chicago/Turabian StyleShan, Eddy, Cristina Chamorro, Ana Ferrández-Montero, Rosa M. Martin-Rodriguez, Begoña Ferrari, Antonio Javier Sanchez-Herencia, Leire Virto, María José Marín, Elena Figuero, and Mariano Sanz. 2025. "In Vitro Biological Properties Assessment of 3D-Printed Hydroxyapatite–Polylactic Acid Scaffolds Intended for Bone Regeneration" Journal of Functional Biomaterials 16, no. 6: 218. https://doi.org/10.3390/jfb16060218
APA StyleShan, E., Chamorro, C., Ferrández-Montero, A., Martin-Rodriguez, R. M., Ferrari, B., Sanchez-Herencia, A. J., Virto, L., Marín, M. J., Figuero, E., & Sanz, M. (2025). In Vitro Biological Properties Assessment of 3D-Printed Hydroxyapatite–Polylactic Acid Scaffolds Intended for Bone Regeneration. Journal of Functional Biomaterials, 16(6), 218. https://doi.org/10.3390/jfb16060218











