1. Introduction
Dynamic mode decomposition (DMD) has become increasingly popular in studying complex dynamical systems. Since it was introduced for the first time by Schmid [
1], it has been successfully applied to a wide range of problems, such as video processing [
2], epidemiology [
3], neuroscience [
4], financial trading [
5,
6,
7], robotics [
8], cavity flows [
9,
10], and various jets [
11,
12]. For a review of the DMD literature, we refer the reader to [
13,
14,
15,
16,
17]. For some recent modifications of DMD for non-uniformly sampled data, the higher-order DMD method, parallel implementations of DMD, and some derivative DMD techniques, we recommend [
18,
19,
20,
21,
22,
23,
24,
25,
26,
27,
28,
29].
In this work, we are interested in a recently developed modification of the DMD method called
online dynamic mode decomposition (
online DMD) [
30] (see also [
31,
32]). We introduce an alternative online DMD method that is applicable for both overconstrained and underconstrained datasets.
The outline of this paper is as follows. In the rest of this section, we give a brief summary of the online DMD and alternative online DMD methods. In
Section 2, we introduce and discuss the new approach of the online DMD method. In
Section 3, we present examples demonstrating the new algorithm. The conclusion is in
Section 4.
1.1. Online Dynamic Mode Decomposition (Online DMD)
In this subsection, the Algorithm 1 of so-called online dynamic mode decomposition (online DMD) is introduced, following the description in [
30].
It requires a dataset of snapshot pairs
spaced a fixed time interval apart. Then, the snapshots are stacked into the following pair of matrices:
The following assumptions are made:
The number of snapshots k is large compared with the state dimension n (i.e., we consider the overconstrained case of the dataset, where ).
The matrix has full row rank (i.e., ).
The objective of the online DMD method is to provide an alternative way of computing the DMD operator such that it can be updated incrementally as new snapshots become available. The algorithm of online DMD proceeds as follows [
30]:
| Algorithm 1 Online DMD Method [30] |
- 1.
Collect k snapshot pairs , , where is large enough that , where is given by ( 2). - 2.
- 3.
When a new snapshot pair becomes available, update and :
and
where
|
1.2. Alternative Online DMD
A novel approach was introduced in [
33] to overcome the main shortcomings of online DMD. The new scheme is designed to work with low-rank data, and the condition
has been relaxed.
Consider a time series of data
with
snapshots organized into the following two data matrices:
Then, the truncated SVD is performed, where , , , and is the truncation value. The low-dimensional DMD operator is calculated using the formula .
A single iteration of the modified algorithm can be summarized as follows [
33].
In this study, we aim to improve the alternative online DMD method (Algorithm 2) from a computational perspective. The next section shows how matrix
can be expressed by using a
diagonal-plus-rank-one matrix (DPR1 matrix) instead of matrix
(or
) in Step 1 of Algorithm 2. As a result, the eigendecomposition of a DPR1 matrix can be used instead of the SVD of a matrix of the same dimension.
| Algorithm 2 Alternative Online DMD Method |
Input: Matrices , scalar , last 3 snapshots .
Compute and proceed:
If :
- 1.
Construct the matrix: - 2.
Compute the compact SVD: - 3.
Compute the left singular vectors and singular values of - 4.
If the DMD modes are required, then compute spectral decomposition of
If is the jth eigenvector of , then is jth DMD mode.
Else If :
- 1.
Construct the matrix: - 2.
Compute the truncated SVD: with truncation value . - 3.
Compute the left singular vectors and singular values of :
- 4.
If the DMD modes are required, then compute the spectral decomposition of
where
If is the jth eigenvector of , then is the jth DMD mode.
|
2. Improved Online DMD
Through analogy with the online DMD, let us consider the
k snapshot pairs
for
as in Equation (
1). We form matrices
which both have dimensions of
. Then, the truncated SVD
is performed, where
,
, and
. The truncation value
r is such that
, and it is a predefined value. From the pseudoinverse
, we express the DMD operator
The projected DMD operator has the form
which is of dimension
. We can perform the leading eigendecomposition of
by using the eigendecomposition of
.
Suppose now that we have already computed
(or
) for a given dataset. Whenever a new pair of snapshots becomes available
, we would like to compute matrix
more efficiently than how it is usually carried out (Equation (
6)).
Let us denote the augmented matrices
Suppose that the singular value decomposition of
has the form
From Equation (
8), we express
as
Then, the updated DMD operator has the form
By substituting Equation (
7) into the last expression, we obtain
Using the SVD of
, we obtain
By applying Equation (
5), the last expression yields
Equation (
13) gives a rule for computing
given
, and the new snapshot pair
.
In what follows, we will show that matrices
and
can be expressed from matrices
and
. For this purpose, we consider the covariance matrix
which has the following presentation:
by using Equation (
7) and the SVD of
. It is well known that the leading left singular vectors and singular values of
can be computed by the eigendecomposition of
. We will consider two scenarios in order to obtain an alternative representation of the SVD of
, depending on whether
:
(1) If
, then the expression
is valid, and the covariance matrix
can be presented as
where we define
as a diagonal-plus-rank-one (DPR1) matrix of a dimension
. In this case, the eigendecomposition of
can be reduced to that of a DPR1 matrix
plus two matrix multiplications. Assume that the eigendecomposition of the
matrix
has the form
where
is unitary and
is diagonal. Then, for the left singular vectors and singular values of
, we obtain
Now, from Equation (
13), we can express the matrix
as
(2) If
, then let us define
and
It is straightforward that
By applying the last two expressions in Equation (
15), the formula for
becomes
which is an
hermitian matrix. From Equation (
24) and the definition of
follows the identity
where
is a zero vector and
is the identity matrix.
By substituting Equation (
26) into Equation (
25), we obtain
where
is a diagonal-plus-rank-one (DPR1) matrix of the dimensions
. Assume that the eigendecomposition of
is
where
is unitary and
is diagonal. Then, for the left singular vectors of
(or the eigenvectors of
) and singular values of
, we obtain
Furthermore, we can compute the reduced-order approximation of
:
which is an
matrix. An equivalent representation of
using the expression in Equation (
26) and the definition of
is
where the following expression holds:
In the particular case of
for
, from the orthogonality of
and
, it follows that
Therefore, we have
where
is the last row of matrix
. From the SVD of
, it follows that
which yields the last column of
:
Then, the expression in Equation (
32) takes the form
where
The next paragraph summarizes the obtained results as an improved variant of Algorithm 2.
Algorithm for Improved Alternative Online DMD
Let us consider a time series of data
with
snapshots organized into the following two data matrices:
Then, we compute the truncated SVD of
as in Equation (
4), where
is the truncation value, and compute the matrix
as in Equation (
6).
For each new data point , the first task is to determine whether the basis contained in should be expanded. To accomplish this, the residual is computed, and if is greater than some pre-specified tolerance , then we expand by appending .
A single iteration of the updated algorithm can be summarized as follows.
The main advantages of the presented algorithm over Algorithm 2 are in Step 1 and Step 2. Algorithm 3 uses a matrix with a simpler structure in Step 1 (in both parts), namely the DPR1 matrix
(or
, respectively). In addition, at Step 2 in Algorithm 3, spectral decomposition of a DPR1 matrix is used instead of SVD of a regular matrix.
| Algorithm 3 Improved Online DMD Method |
Input: Matrices , scalar , last 3 snapshots .
Output: Matrices .
Compute and proceed:
If :
- 1.
Construct the DPR1 matrix:
- 2.
Compute the spectral decomposition:
- 3.
Compute the left singular vectors and singular values of - 4.
If the DMD modes are required, then compute the spectral decomposition of
If is the jth eigenvector of , then is the jth DMD mode.
Else If :
- 1.
Construct the DPR1 matrix:
- 2.
Compute the truncated SVD:
with truncation value: . - 3.
Compute the left singular vectors and singular values of :
where
- 4.
If the DMD modes are required, then compute the spectral decomposition of
where
If is the jth eigenvector of , then is the jth DMD mode.
|
The main advantages of Algorithm 3 (as well as Algorithm 2) over Algorithm 1 are that it does not require full rank data matrix, and unlike Algorithm 1, Algorithm 3 (and Algorithm 2) does not require the number of snapshots to be greater than the state dimension.
In the next section, we present the framework with which Algorithm 3 (and Algorithm 2) can be modified to a weighted alternative.
3. Weighted Modifications to Online DMD
The algorithms described above are suitable for time-varying systems. They allow the DMD matrix
to be updated in real time. In such cases, we may want to give more weight to recent snapshots and less weight to older snapshots. This can be achieved by using a modified cost function. As is known in the standard DMD method, for the given data matrices
and
in Equation (
3), the DMD operator
is found by minimizing the following cost function:
where
denotes the Euclidean norm on the vectors and
denotes the Frobenius norm on the matrices.
In cases where the system changes over time to give more weight to newer snapshots than older snapshots, we can use the modified cost function
for some constant
where
. In fact, for
, the two formulas match, and for
, errors in past snapshots are discounted.
We will show below that Algorithm 3 can easily be modified to use this weighting scheme (i.e., to minimize a cost function of the form in Equation (
36) instead of the original cost function in Equation (
35)). For convenience, let us denote
, where
, and write the cost function (2) as
Let us define the matrices based on scaled versions of the snapshots as
Then, the cost function in Equation (
37) can be written as
The DMD operator
, which is a unique solution that minimizes this cost function, is given by
where the truncated SVD
is used. Then, the reduced-order DMD operator will have the form
On the next (
th) step, we want to compute
. Let us denote
and
By analogy with
Section 2, we can repeat the expressions from Equation (
8) to Equation (
12) to obtain
for the updated DMD matrix, where
and
are the singular vectors and value matrices of
, respectively.
In a similar way to that in Equations (
14) and (
15), we can extract the singular vectors and singular values of
through the eigendecomposition of
Matrix
differs from
in Equation (
15) only by a factor of
. Again, we will have two scenarios, depending on whether
. At each step, we have to factorize one of the following two matrices:
or
where
,
, and
. The matrices
and
differ only by a factor of
from the corresponding matrices
and
in Equations (
18) and (
28), respectively. Then, the singular vectors and values of
are given by
where
and
are from the eigendecomposition of
or
.
The reduced order DMD matrix is then given by
or
for the two considered scenarios. Observe that the updates in Equations (
45) and (
46) for the reduced order approximation
are similar to the updates in Equations (31) and (32). The only difference is the factor of
.
Therefore, the algorithm for weighted alternative online DMD follows the same steps as in Algorithm 3. The only difference is the calculation of matrices
(or
) and
, where Equations (
42), (
43) and (
45), (
46) are used, respectively. We have to note that the choice of the weighting factor
matters. Therefore, for example, choosing
will ensure that snapshots have a
half-life of
m samples [
30]. In practice, the choice of
depends on the rate of change of the dynamics. Choosing a smaller
value will result in faster tracking while making the model more sensitive to noise in the data.
4. Numerical Illustrations
The new algorithm (Algorithm 3), introduced in
Section 2, offers the advantage of being more cost-effective than the alternative algorithm (Algorithm 2). Considering that Algorithms 2 and 3 produce identical results, in this part, we will mainly present the results of the application of Algorithm 3 and its comparison with standard methods.
Example 1. An illustrative example of a toy.
We consider a simple example of incrementally updated DMD in a toy problem. In this example, we have an arbitrary
n-dimensional dynamical system with two characteristic frequencies. A total of
m snapshot pairs are measured sequentially and are subject to additive zero-mean Gaussian noise with covariance:
where
are random state directions,
are the characteristic frequencies,
is the time sampling, and
is the independent identically distributed zero-mean Gaussian noise with covariance
.
For the simulation, we set the parameters as follows:
,
,
,
,
, and
. By using the built-in function
in Matlab, we defined the vectors
and
via the following syntax:
The 3D visualization of the dynamics is illustrated in
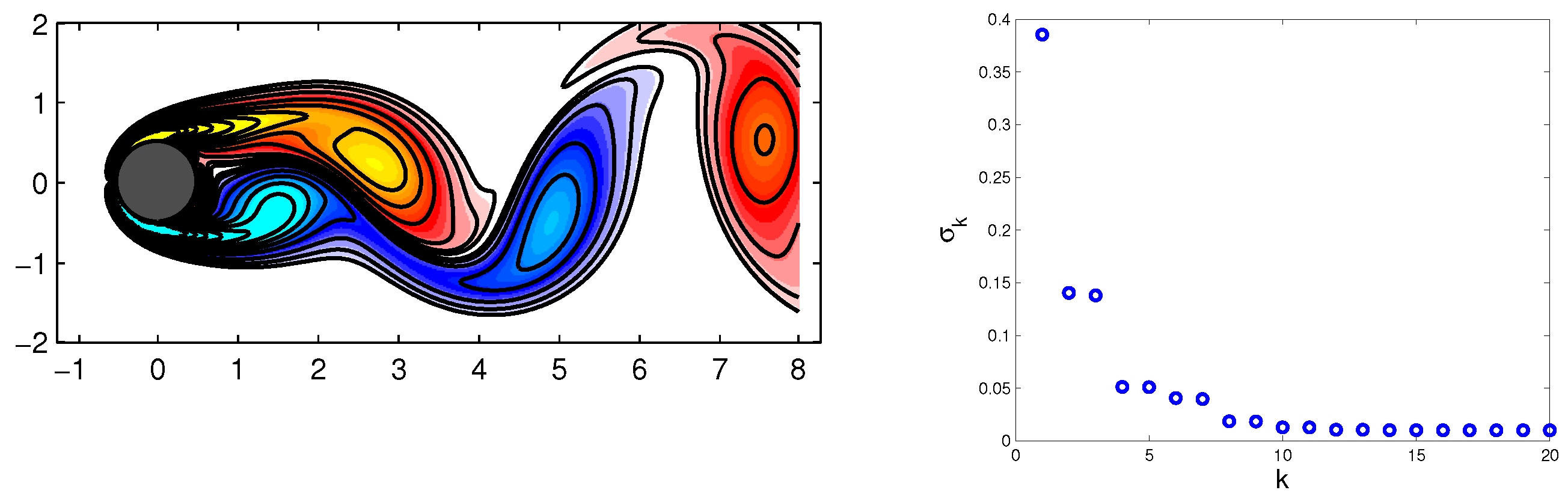
Figure 1. The singular values of data matrix
X, illustrated in
Figure 1, show that the data can be adequately represented by the rank-four (
) approximation.
The standard online DMD method (Algorithm 1) is not applicable in this case because . Therefore, here we will compare the standard DMD method and improved online DMD method (Algorithm 3). We initialized the improved online DMD algorithm with the first three snapshots () (i.e., the initial matrices and have the form and , respectively).
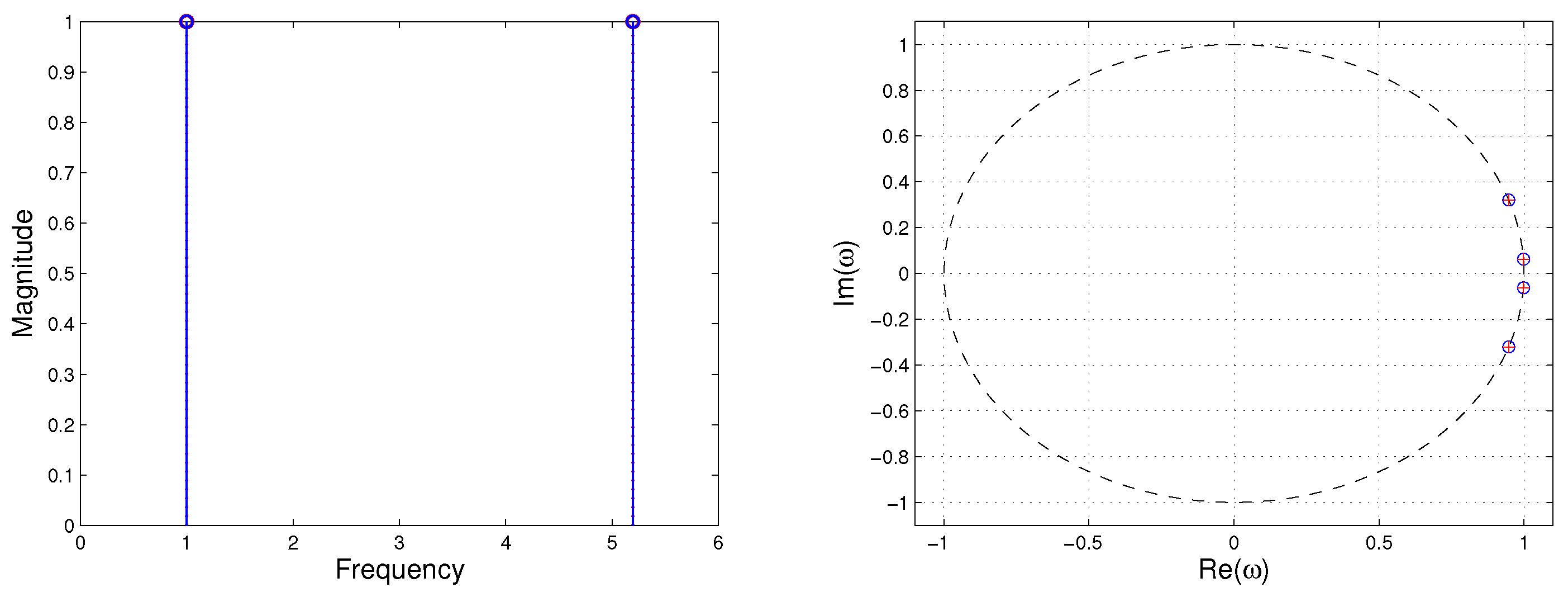
We performed both methods with rank reduction of
. The two algorithms, standard DMD and Algorithm 3, were applied to different numbers of snapshots in the interval from 50 to 100. All runs of both algorithms yielded the same DMD eigenvalues shown in
Figure 2.
To make quantitative comparisons among alternative online DMD and standard DMD, we computed the
norm of the difference between the spatial modes extracted by the two algorithms. We computed the residual errors as follows:
for
, where
and
denote the DMD modes computed by both algorithms. The error values for two cases at 50 and 100 snapshots are shown in
Table 1.
Example 2. Flow around a cylinder wake ().
In this example, we consider a dataset representing a time series of fluid vorticity fields for the wake behind a circular cylinder at Reynolds number
. The data for this example are publicly available at
www.siam.org/books/ot149/flowdata (see [
15]). The collected data consisted of
snapshots at regular intervals in time
, where
, sampling five periods of vortex shedding.
Each vorticity field snapshot from the finest computational domain was reshaped into a large vector
, and these vectors comprised columns of matrices
X and
Y, as described in Equation (
34). A snapshot example of the cylinder wake data is shown in
Figure 3.
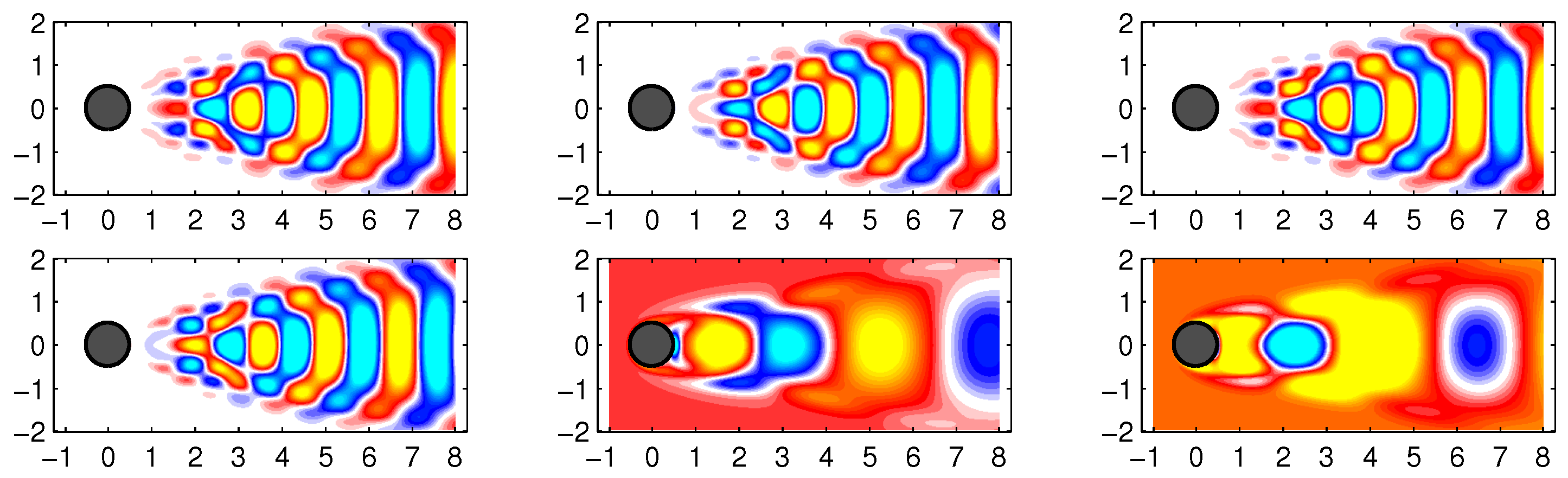
Here, we compare the standard DMD method and the improved online DMD method, since online DMD (Algorithm 1) is not applicable. In order to capture the low-dimensional dynamic, we performed rank-seven truncation. We used Algorithm 3 to obtain the DMD reconstruction of the data. The two algorithms reproduced the same DMD eigenvalues and modes. We compared the results with the standard DMD algorithm for the same rank-seven truncation. The DMD eigenvalues computed by the improved alternative online DMD and standard DMD algorithms are shown in
Figure 4.
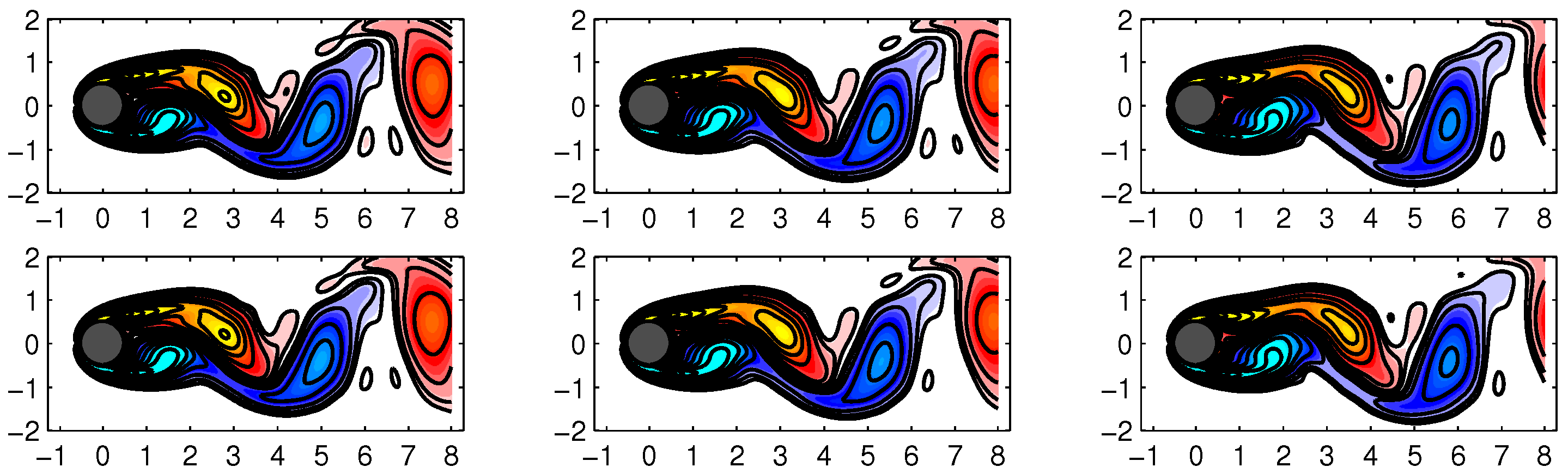
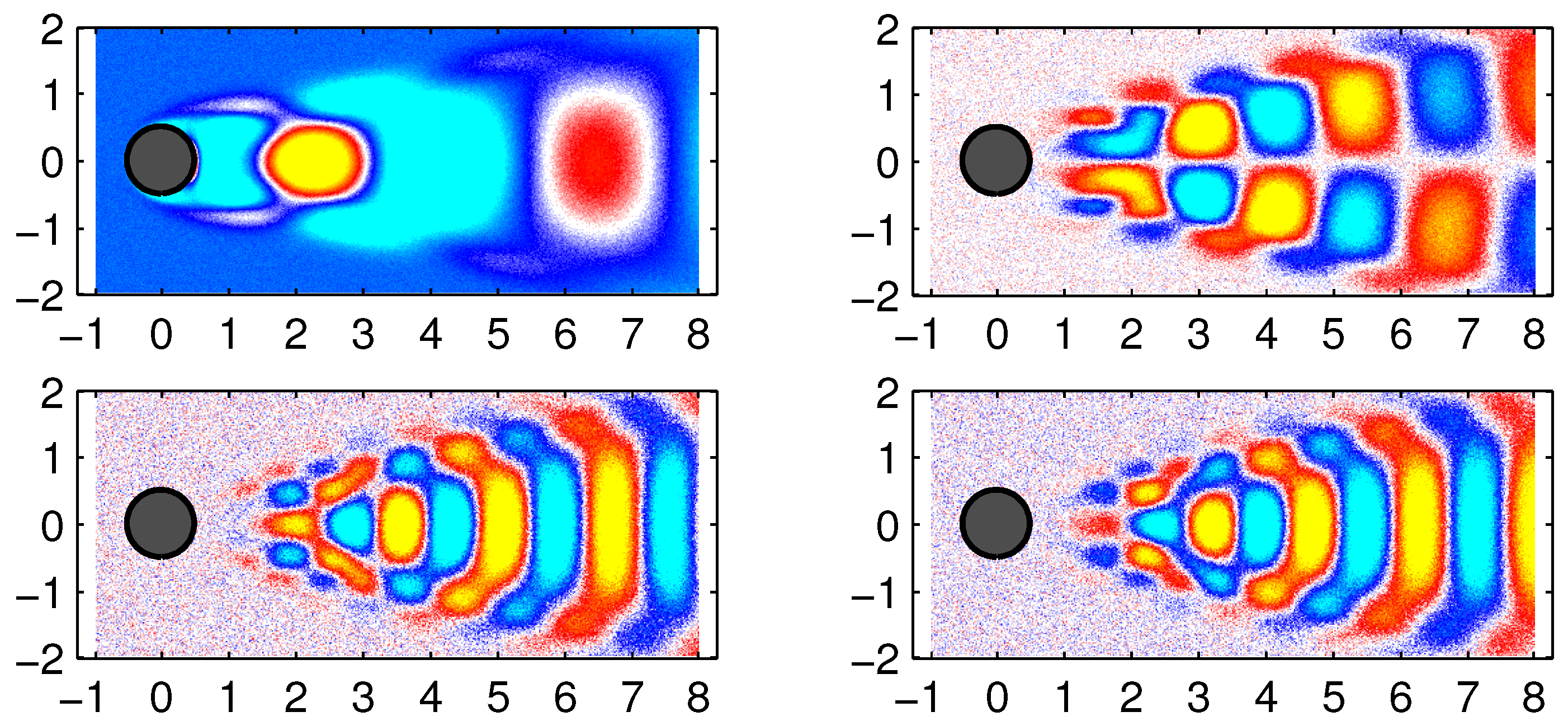
The first 6 DMD modes computed by the improved online DMD and standard DMD algorithms are shown in
Figure 5 and
Figure 6, respectively.
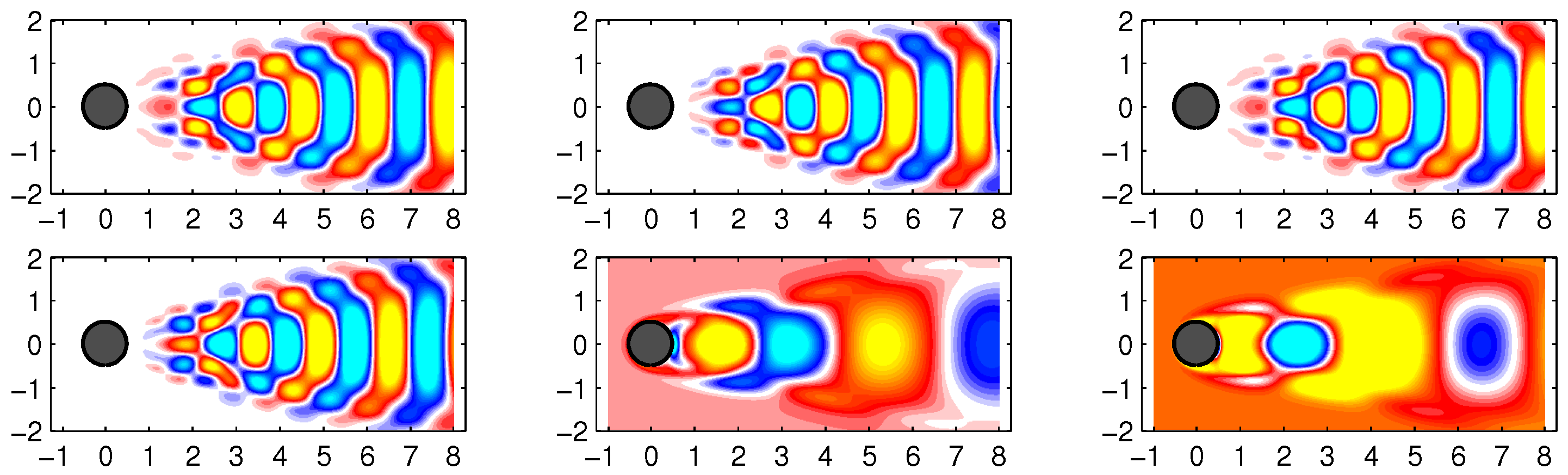
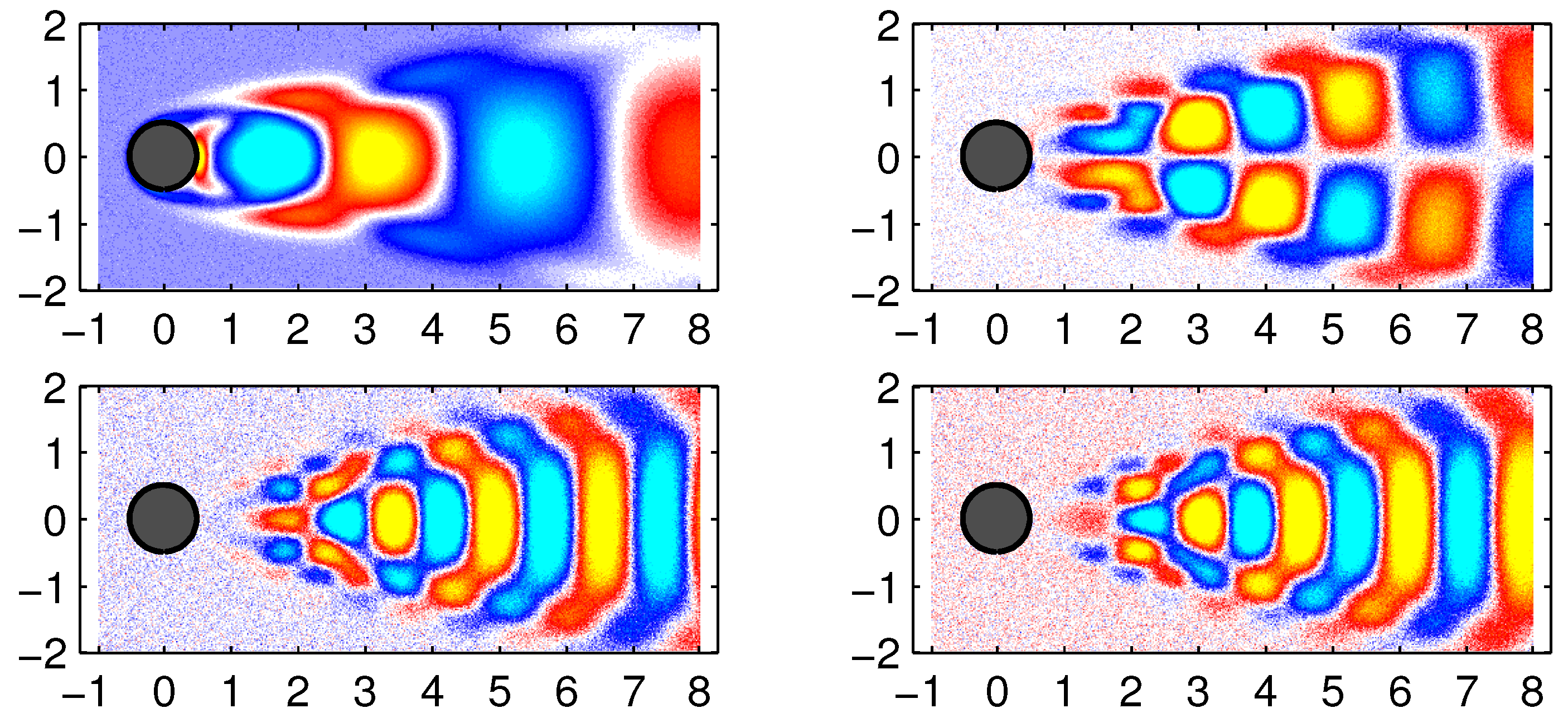
In
Figure 7, some examples of data reconstruction by the standard DMD and improved online DMD methods are shown.
There was no distinction between the DMD modes and eigenvalues generated by the standard DMD algorithm (Algorithm 1) and Algorithm 3.
Example 3. Flow around a cylinder wake with added noise.
It is well known that the DMD method’s performance depends on the cleanliness of the data collected. The noise in the collected data can affect the discovered model’s accuracy. In this example, we consider the same fluid vorticity fields for the wake behind a circular cylinder at Reynolds number , as in Example 2. Measurement noise was generated by adding Gaussian white noise to the data matrix.
As in Example 2, the data-set consisted of snapshots , where each snapshot had coordinates (i.e., the data matrix ).
The updated snapshots were generated by
where
is a zero-mean Gaussian white noise process with unit standard deviation. In this example, we kept the noise variance fixed at
.
Here, we compare the standard DMD method and the improved online DMD method. As in Example 2, we performed rank-seven truncation. We used Algorithm 3 to obtain the DMD reconstruction of the data. The two algorithms reproduced the same DMD eigenvalues and modes.
Figure 8 shows some examples of data reconstruction by the standard DMD and improved online DMD methods for the noisy flow data.
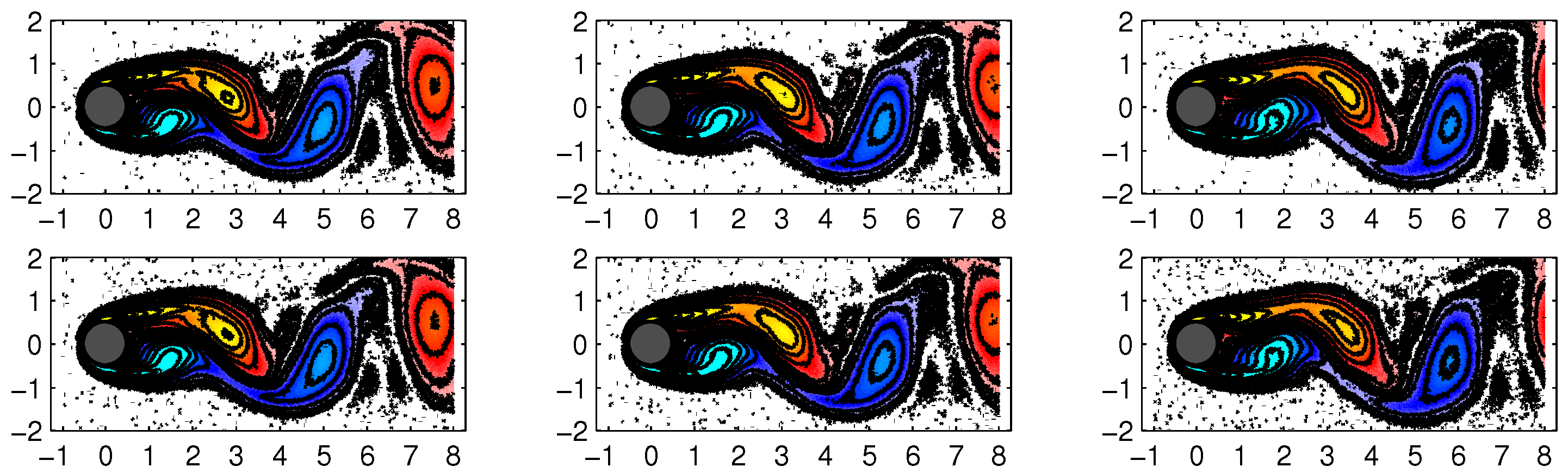
The first four DMD modes computed by the improved online DMD and standard DMD algorithms are shown in
Figure 9 and
Figure 10, respectively.
Note again that there was no distinction between the modes generated by the standard DMD algorithm and Algorithm 3. The visualizations of the DMD eigenvalues calculated by the improved alternative online DMD method and the standard DMD method were omitted as they matched those shown in
Figure 4.
Example 4. Linear time-varying system.
We now illustrate the improved alternative online DMD algorithm in a simple linear system that slowly varies over time:
where
, and the time-varying matrix
A(
t) is given by
where
(see [
30]). The eigenvalues of
are
, and
is constant in
t. We simulated the system for
from the initial condition
, and the snapshots were taken with a time step of
, as shown in
Figure 11.
With the snapshots as input, we applied standard DMD, online DMD (Algorithm 1), and improved alternative online DMD (Algorithm 3) and compared the results. The three methods used the first
snapshot pairs to initialize and start iterating from time
. The discrete-time eigenvalues
computed by the algorithms were converted to continuous-time DMD eigenvalues
by the formula
where
is the time spacing between snapshot pairs. Observe from
Figure 12 that the eigenvalues computed by the alternative online DMD algorithm agreed with those identified by the standard DMD and online DMD algorithms. The true eigenvalues are also shown for comparison. The DMD modes calculated by the three algorithms were also identical, as can be seen from
Figure 12.
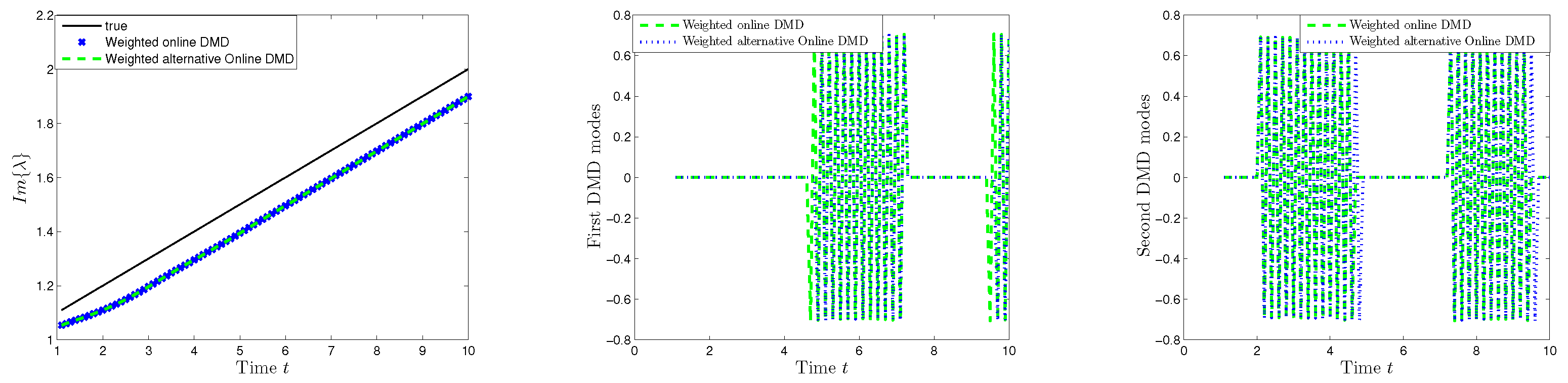
The alternative weighted online DMD algorithm was applied under the same conditions. A comparison was made between the results of the weighted variant of Algorithm 3 and the weighted online DMD algorithm introduced in [
30].
As before, the results show identical DMD modes and DMD eigenvalues for both algorithms.
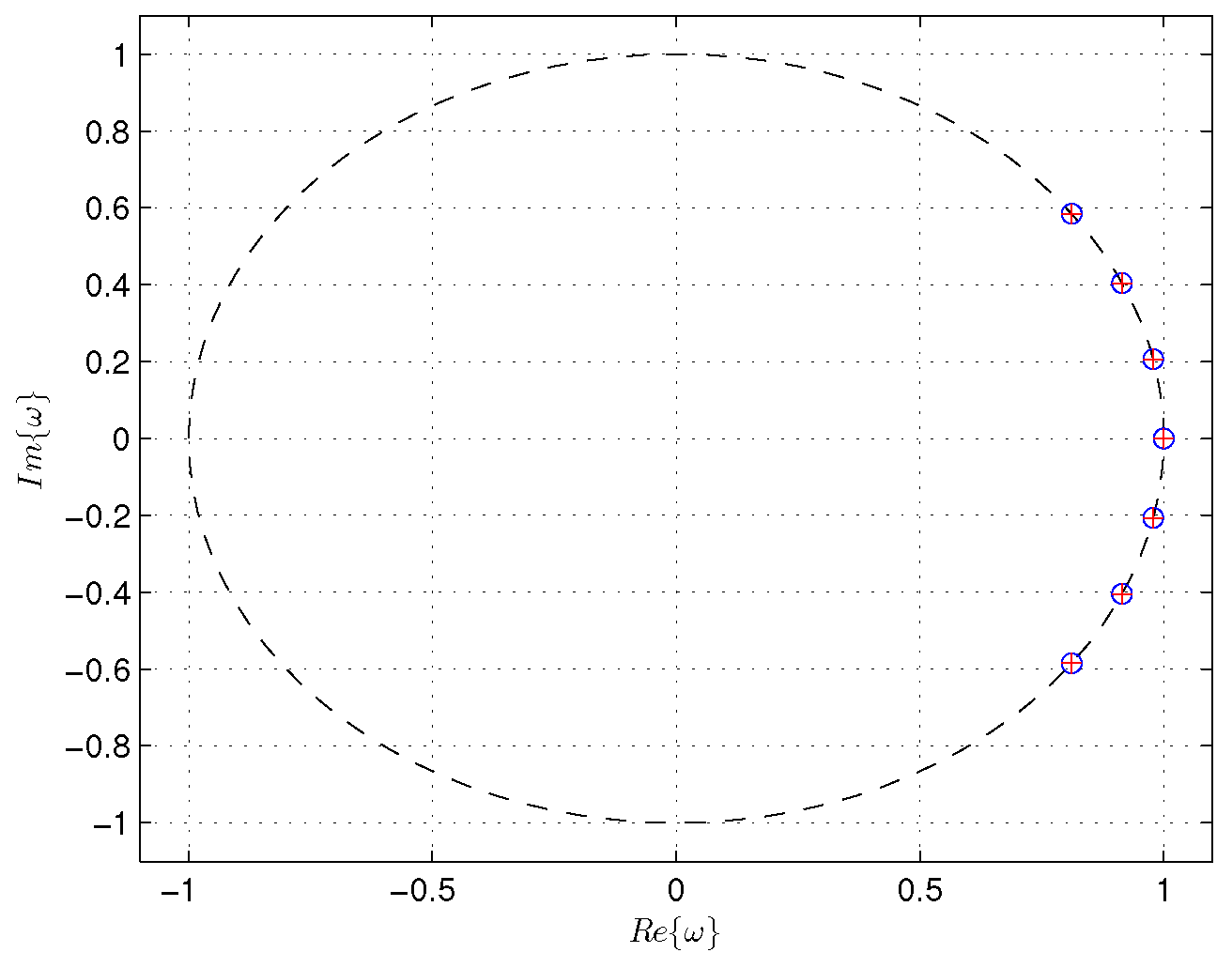
Figure 13 shows the eigenvalues and modes computed by the weighted online DMD and alternative weighted online DMD methods with a factor
.
5. Discussion
The introduced variants of online dynamic mode decomposition approximate the Koopman operator as well as its eigenvalues and modes by processing continuous data streams. Online DMD has applications in various fields, including control systems, sensor networks, weather forecasting, and monitoring dynamic systems in real time. By providing up-to-date decomposition of the data, it allows for the identification of evolving patterns, prediction of future behavior, and monitoring system health in real time.
Online DMD offers several advantages over the standard DMD method when analyzing streaming or real-time data:
Computational efficiency and reduced memory requirements: By processing data incrementally, online DMD avoids redundant computations of previously analyzed data, which results in improved efficiency. It only requires storage for the most recent data rather than the entire dataset, which reduces the memory requirements.
Continuous monitoring and prediction: Online DMD allows for continuous monitoring of evolving systems and the ability to make predictions based on the estimated modes. This is valuable for applications where forecasting or early detection of anomalies is essential, such as in weather forecasting or fault detection.
Flexibility in data input: Online DMD can handle data streams with missing or irregularly spaced samples, accommodating the challenges often encountered in real-time data acquisition and processing.
Adaptability to changing dynamics: Streaming data often exhibit time-varying or non-stationary behavior. Online DMD and the weighted online DMD modifications continuously update the decomposition using the most recent data, allowing them to capture and adapt to changes in the underlying dynamics.
In this paper, we introduced extended modifications of the standard online DMD method introduced by Zhang et al. in [
30]. The main advantages of the proposed modifications over the standard online DMD scheme are that they are applicable regardless of whether the data system is full-rank or not. In addition, the requirement that the number of snapshots be larger than the state dimension is dropped.
The ability of the introduced techniques to handle streaming data and provide real-time insights make them valuable in various scientific domains where continuous monitoring, prediction, and analysis of dynamic systems are essential. Such areas of application include structural health monitoring, biomedical research, weather forecasting, and robotics and control systems. Additionally, these techniques can be useful in scenarios where there are large snapshots and real-time analysis is required. Such areas of application include image and video processing, computational biology and genomics, financial markets, and social media and web analytics. Some related papers include [
34,
35,
36,
37].
We believe that there is a scope for further research and applications of the presented methods for online DMD. Potential directions for further research and development include adaptive and data-driven parameter selection, online DMD for multi-modal and heterogeneous data, and integration of online DMD with machine learning techniques. Some related papers include [
38,
39,
40,
41].