Machine Learning in X-ray Diagnosis for Oral Health: A Review of Recent Progress
Abstract
1. Introduction
2. Methods
- Studies between 1 January 2018 to 31 December 2022, since the goal was to access the most recent progress in a rapidly evolving field;
- Studies with a focus on dental/oral imaging techniques based on X-rays, including cone beam computed tomography (CBCT);
- Studies with a focus on diagnostic applications. To our knowledge, this is the first paper that exclusively reviews the application of ML methods in oral health diagnosis.
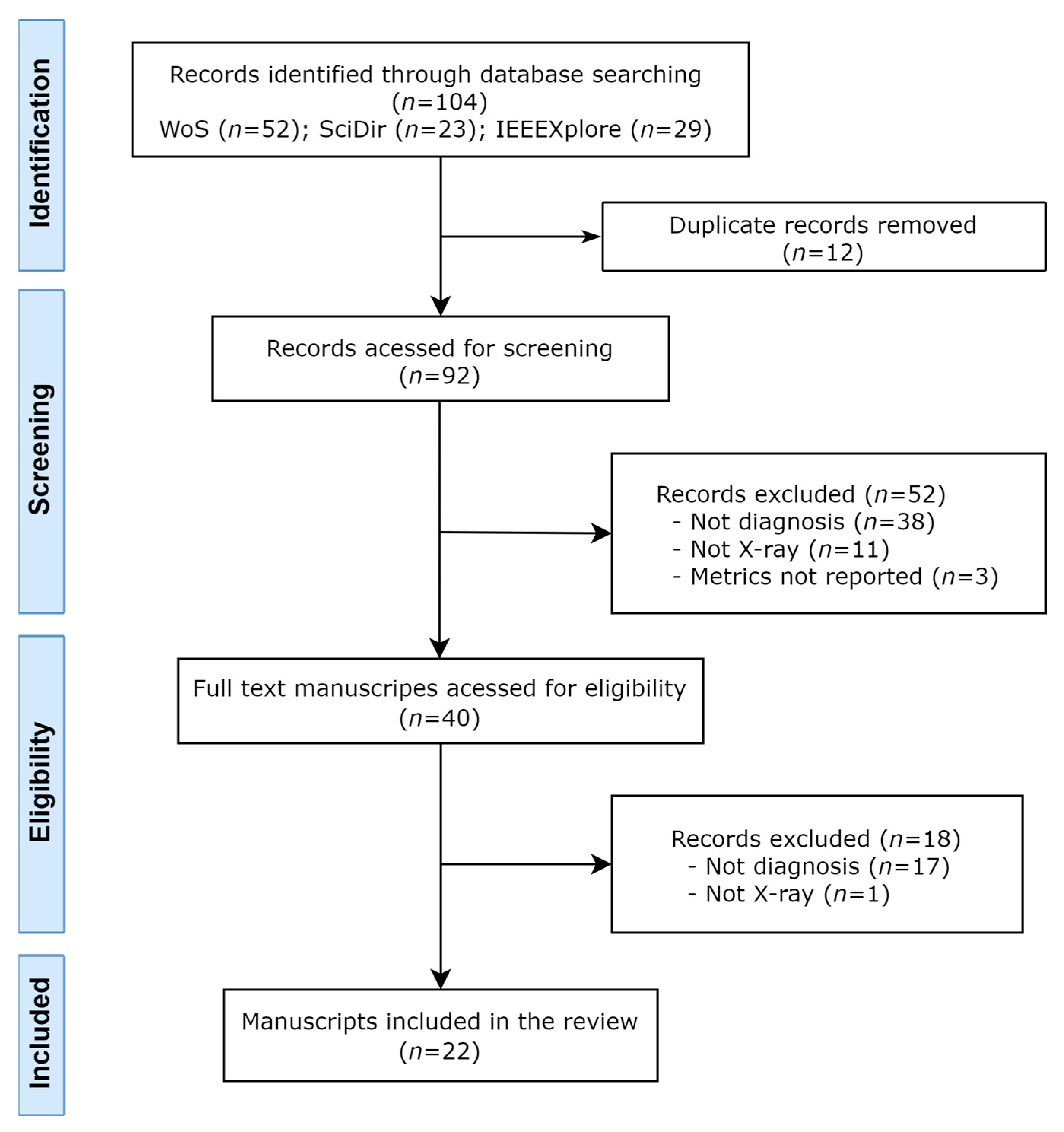
3. Results
3.1. Search and Study Selection
3.2. Included Studies
3.3. Clinical Applications, Image Types, Data Sources and Labeling
3.4. Datasets Size, Partitions, and Data Augmentation
3.5. Machine Learning Tasks and Models
3.6. Outcome Metrics and Model Performance
3.7. Human Comparators
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pitts, N.B.; Zero, D.T.; Marsh, P.D.; Ekstrand, K.; Weintraub, J.A.; Ramos-Gomez, F.; Tagami, J.; Twetman, S.; Tsakos, G.; Ismail, A. Dental Caries. Nat. Rev. Dis. Prim. 2017, 3, 17030. [Google Scholar] [CrossRef]
- Kinane, D.F.; Stathopoulou, P.G.; Papapanou, P.N. Periodontal Diseases. Nat. Rev. Dis. Prim. 2017, 3, 17038. [Google Scholar] [CrossRef]
- The Use of Dental Radiographs: Update and Recommendations. J. Am. Dent. Assoc. 2006, 137, 1304–1312. [CrossRef]
- Ludlow, J.B.; Ivanovic, M. Comparative Dosimetry of Dental CBCT Devices and 64-Slice CT for Oral and Maxillofacial Radiology. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2008, 106, 106–114. [Google Scholar] [CrossRef]
- Tadinada, A. Dental Radiography BT. In Evidence-Based Oral Surgery: A Clinical Guide for the General Dental Practitioner; Ferneini, E.M., Goupil, M.T., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 67–90. ISBN 978-3-319-91361-2. [Google Scholar]
- Shan, T.; Tay, F.R.; Gu, L. Application of Artificial Intelligence in Dentistry. J. Dent. Res. 2021, 100, 232–244. [Google Scholar] [CrossRef]
- Carrillo-Perez, F.; Pecho, O.E.; Morales, J.C.; Paravina, R.D.; Della Bona, A.; Ghinea, R.; Pulgar, R.; Pérez, M.D.M.; Herrera, L.J. Applications of Artificial Intelligence in Dentistry: A Comprehensive Review. J. Esthet. Restor. Dent. 2022, 34, 259–280. [Google Scholar] [CrossRef]
- Mahdi, S.S.; Battineni, G.; Khawaja, M.; Allana, R.; Siddiqui, M.K.; Agha, D. How Does Artificial Intelligence Impact Digital Healthcare Initiatives? A Review of AI Applications in Dental Healthcare. Int. J. Inf. Manag. Data Insights 2023, 3, 100144. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-Based Learning Applied to Document Recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Li, F.-F. ImageNet: A Large-Scale Hierarchical Image Database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009. [Google Scholar]
- Lin, T.-Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common Objects in Context BT. In Computer Vision—ECCV 2014; Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 740–755. [Google Scholar]
- Deng, L. Digit Images for Machine Learning Research. IEEE Signal Process. Mag. 2012, 29, 141–142. [Google Scholar] [CrossRef]
- Krizhevsky, A. Learning Multiple Layers of Features from Tiny Images. 2009. Available online: https://www.cs.toronto.edu/~kriz/learning-features-2009-TR.pdf (accessed on 5 May 2023).
- Everingham, M.; Eslami, S.M.A.; Van Gool, L.; Williams, C.K.I.; Winn, J.; Zisserman, A. The Pascal Visual Object Classes Challenge: A Retrospective. Int. J. Comput. Vis. 2015, 111, 98–136. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Fix, E.; Hodges, J.L. Discriminatory Analysis. Nonparametric Discrimination: Consistency Properties. Int. Stat. Rev. 1989, 57, 238–247. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Washington DC USA, 14–18 August 2016. [Google Scholar]
- Huang, X.; Lin, J.; Demner-Fushman, D. Evaluation of PICO as a Knowledge Representation for Clinical Questions. AMIA Annu. Symp. Proc. AMIA Symp. 2006, 2006, 359–363. [Google Scholar]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan-a Web and Mobile App for Systematic Reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, D.H.; Jeong, S.N.; Choi, S.H. Detection and Diagnosis of Dental Caries Using a Deep Learning-Based Convolutional Neural Network Algorithm. J. Dent. 2018, 77, 106–111. [Google Scholar] [CrossRef]
- Ekert, T.; Krois, J.; Meinhold, L.; Elhennawy, K.; Emara, R.; Golla, T.; Schwendicke, F. Deep Learning for the Radiographic Detection of Apical Lesions. J. Endod. 2019, 45, 917–922.e5. [Google Scholar] [CrossRef]
- Krois, J.; Ekert, T.; Meinhold, L.; Golla, T.; Kharbot, B.; Wittemeier, A.; Dörfer, C.; Schwendicke, F. Deep Learning for the Radiographic Detection of Periodontal Bone Loss. Sci. Rep. 2019, 9, 8495. [Google Scholar] [CrossRef]
- Geetha, V.; Aprameya, K.S.; Hinduja, D.M. Dental Caries Diagnosis in Digital Radiographs Using Back-Propagation Neural Network. Health Inf. Sci. Syst. 2020, 8, 8. [Google Scholar] [CrossRef]
- Endres, M.G.; Hillen, F.; Salloumis, M.; Sedaghat, A.R.; Niehues, S.M.; Quatela, O.; Hanken, H.; Smeets, R.; Beck-Broichsitter, B.; Rendenbach, C.; et al. Development of a Deep Learning Algorithm for Periapical Disease Detection in Dental Radiographs. Diagnostics 2020, 10, 430. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, D.H.; Jeong, S.N. Diagnosis of Cystic Lesions Using Panoramic and Cone Beam Computed Tomographic Images Based on Deep Learning Neural Network. Oral Dis. 2020, 26, 152–158. [Google Scholar] [CrossRef]
- Hashem, M.; Youssef, A.E. Teeth Infection and Fatigue Prediction Using Optimized Neural Networks and Big Data Analytic Tool. Clust. Comput. 2020, 23, 1669–1682. [Google Scholar] [CrossRef]
- Zheng, Z.; Yan, H.; Setzer, F.C.; Shi, K.J.; Mupparapu, M.; Li, J. Anatomically Constrained Deep Learning for Automating Dental CBCT Segmentation and Lesion Detection. IEEE Trans. Autom. Sci. Eng. 2021, 18, 603–614. [Google Scholar] [CrossRef]
- Lee, D.W.; Kim, S.Y.; Jeong, S.N.; Lee, J.H. Artificial Intelligence in Fractured Dental Implant Detection and Classification: Evaluation Using Dataset from Two Dental Hospitals. Diagnostics 2021, 11, 233. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.H.; Hamamoto, K.; Paing, M.P. Deep Fusion Feature Extraction for Caries Detection on Dental Panoramic Radiographs. Appl. Sci. 2021, 11, 2005. [Google Scholar] [CrossRef]
- Cha, J.Y.; Yoon, H.I.; Yeo, I.S.; Huh, K.H.; Han, J.S. Peri-Implant Bone Loss Measurement Using a Region-Based Convolutional Neural Network on Dental Periapical Radiographs. J. Clin. Med. 2021, 10, 1009. [Google Scholar] [CrossRef]
- Kearney, V.P.; Yansane, A.I.M.; Brandon, R.G.; Vaderhobli, R.; Lin, G.H.; Hekmatian, H.; Deng, W.; Joshi, N.; Bhandari, H.; Sadat, A.S.; et al. A Generative Adversarial Inpainting Network to Enhance Prediction of Periodontal Clinical Attachment Level. J. Dent. 2022, 123, 104211. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, J.; Zhou, Z.; Zhou, Z.; Wu, X.; Li, Y.; Wang, S.; Liao, W.; Ying, S.; Zhao, Z. Artificial Intelligence for Caries and Periapical Periodontitis Detection. J. Dent. 2022, 122, 104107. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Y.; Li, S.; Ying, S.; Zheng, L.; Zhao, Z. Artificial Intelligence-Aided Detection of Ectopic Eruption of Maxillary First Molars Based on Panoramic Radiographs. J. Dent. 2022, 125, 104239. [Google Scholar] [CrossRef]
- Aljabri, M.; Aljameel, S.S.; Min-Allah, N.; Alhuthayfi, J.; Alghamdi, L.; Alduhailan, N.; Alfehaid, R.; Alqarawi, R.; Alhareky, M.; Shahin, S.Y.; et al. Canine Impaction Classification from Panoramic Dental Radiographic Images Using Deep Learning Models. Inform. Med. Unlocked 2022, 30, 100918. [Google Scholar] [CrossRef]
- Ying, S.; Wang, B.; Zhu, H.; Liu, W.; Huang, F. Caries Segmentation on Tooth X-Ray Images with a Deep Network. J. Dent. 2022, 119, 104076. [Google Scholar] [CrossRef] [PubMed]
- Imak, A.; Celebi, A.; Siddique, K.; Turkoglu, M.; Sengur, A.; Salam, I. Dental Caries Detection Using Score-Based Multi-Input Deep Convolutional Neural Network. IEEE Access 2022, 10, 18320–18329. [Google Scholar] [CrossRef]
- Tajima, S.; Okamoto, Y.; Kobayashi, T.; Kiwaki, M.; Sonoda, C.; Tomie, K.; Saito, H.; Ishikawa, Y.; Takayoshi, S. Development of an Automatic Detection Model Using Artificial Intelligence for the Detection of Cyst-like Radiolucent Lesions of the Jaws on Panoramic Radiographs with Small Training Datasets. J. Oral Maxillofac. Surg. Med. Pathol. 2022, 34, 553–560. [Google Scholar] [CrossRef]
- Feher, B.; Krois, J. Emulating Clinical Diagnostic Reasoning for Jaw Cysts with Machine Learning. Diagnostics 2022, 12, 1968. [Google Scholar] [CrossRef]
- Tsoromokos, N.; Parinussa, S.; Claessen, F.; Moin, D.A.; Loos, B.G. Estimation of Alveolar Bone Loss in Periodontitis Using Machine Learning. Int. Dent. J. 2022, 72, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Xu, T.; Peng, L.; Cao, Y.; Zhao, X.; Li, S.; Zhao, Y.; Meng, F.; Ding, J.; Liang, S. Faster-RCNN Based Intelligent Detection and Localization of Dental Caries. Displays 2022, 74, 102201. [Google Scholar] [CrossRef]
- Muhammed Sunnetci, K.; Ulukaya, S.; Alkan, A. Periodontal Bone Loss Detection Based on Hybrid Deep Learning and Machine Learning Models with a User-Friendly Application. Biomed. Signal Process. Control 2022, 77, 103844. [Google Scholar] [CrossRef]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 386–397. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Schwendicke, F.; Golla, T.; Dreher, M.; Krois, J. Convolutional Neural Networks for Dental Image Diagnostics: A Scoping Review. J. Dent. 2019, 91, 103226. [Google Scholar] [CrossRef]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G.M. Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD): The TRIPOD Statement. BMC Med. 2015, 13, 1. [Google Scholar] [CrossRef]
- Wolff, R.F.; Moons, K.G.M.; Riley, R.D.; Whiting, P.F.; Westwood, M.; Collins, G.S.; Reitsma, J.B.; Kleijnen, J.; Mallett, S. PROBAST: A Tool to Assess the Risk of Bias and Applicability of Prediction Model Studies. Ann. Intern. Med. 2019, 170, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.S.; Dhiman, P.; Navarro, C.L.A.; Ma, J.; Hooft, L.; Reitsma, J.B.; Logullo, P.; Beam, A.L.; Peng, L.; Van Calster, B.; et al. Protocol for Development of a Reporting Guideline (TRIPOD-AI) and Risk of Bias Tool (PROBAST-AI) for Diagnostic and Prognostic Prediction Model Studies Based on Artificial Intelligence. BMJ Open 2021, 11, e048008. [Google Scholar] [CrossRef] [PubMed]
- Schwendicke, F.; Singh, T.; Lee, J.H.; Gaudin, R.; Chaurasia, A.; Wiegand, T.; Uribe, S.; Krois, J. Artificial Intelligence in Dental Research: Checklist for Authors, Reviewers, Readers. J. Dent. 2021, 107, 103610. [Google Scholar] [CrossRef] [PubMed]
- Norgeot, B.; Quer, G.; Beaulieu-Jones, B.K.; Torkamani, A.; Dias, R.; Gianfrancesco, M.; Arnaout, R.; Kohane, I.S.; Saria, S.; Topol, E.; et al. Minimum Information about Clinical Artificial Intelligence Modeling: The MI-CLAIM Checklist. Nat. Med. 2020, 26, 1320–1324. [Google Scholar] [CrossRef]
- Bonfanti, S.; Gargantini, A.; Mashkoor, A. A Systematic Literature Review of the Use of Formal Methods in Medical Software Systems. J. Softw. Evol. Process 2018, 30, e1943. [Google Scholar] [CrossRef]



| Study Question | |
|---|---|
| Population | Oral X-ray diagnostic images of patients (radiography, CBCT) |
| Intervention | Artificial intelligence-based forms of diagnosis |
| Control | Oral health |
| Outcome | Quality of the predictive models |
| Time | Last five years |
| Name | Acronym | URL |
|---|---|---|
| IEEE Xplore | IEEEXplore | https://ieeexplore.ieee.org/Xplore/home.jsp (accessed on 6 March 2023) |
| Science Direct | SciDir | https://www.sciencedirect.com/ (accessed on 6 March 2023) |
| Web of Science | WoS | https://www.webofscience.com/wos/ (accessed on 6 March 2023) |
| Study | Country, Year | Diagnosis of | ImageType | Data Source | Dataset Size | Machine Learning Task | Metrics | Models |
|---|---|---|---|---|---|---|---|---|
| [25] | South Korea, 2018 | Dental caries | Periapical | Hospital | 24,600 | Classification | Acc, Sens, Spec, PPV, NPV, ROC-AUC | GoogLeNet |
| [26] | Germany, 2019 | Apical lesions | Panoramic | University | 2877 | Classification | ROC-AUC, Sens, Spec, PPV, NPV | Proprietary CNN |
| [27] | Germany, 2019 | Periodontal diseases | Panoramic | University | 2538 | Classification | Acc, ROC-AUC, F1, Sens, Spec, PPV, NPV | Proprietary CNN |
| [28] | India, 2020 | Dental caries | Periapical | University | 105 | Classification | Acc, FPR, PRC, MCC | BPNN |
| [29] | Germany, 2020 | Apical lesions | Panoramic | University | 3099 | Classification | PPV, Sens, F1, Prec, TPR | U-Net |
| [30] | South Korea, 2020 | Oral lesions | CBCT, Panoramic | University | 170,525 | Classification | ROC-AUC, Sens, Specificity | GoogLeNet |
| [31] | Saudi Arabia, 2020 | Apical lesions, dental caries, periodontal diseases | Periapical | Database | 120 | Classification | Acc, Spec, Prec, Rec, F1 | Proprietary CNN |
| [32] | USA, 2021 | Oral lesions | CBCT | University | 100 | Classification | Prec, Rec, Dice, Acc | Proprietary CNN |
| [33] | South Korea, 2020 | Implant defects | Periapical, Panoramic | Hospital | 1,292,360 | Classification | ROC-AUC, Sens, Spec, YI | VGG, GoogLeNet, Proprietary CNN |
| [34] | Japan, 2021 | Dental caries | Panoramic | Hospital | 533 | Classification | Acc, Sens, Spec, PPV, NPV, F1 | Alexnet, GoogLeNet, VGG, ResNet, Xception, SVM, KNN, DT, NB, RF |
| [35] | South Korea, 2021 | Periodontal diseases | Periapical | University | 708 | Classification | Prec, Rec, mOKS | Mask R-CNN, ResNet |
| [36] | USA, 2022 | Periodontal diseases | Bitewing, Periapical | Private clinic | 133,304 | Generative; Regression | MAE, MBE | Proprietary CNN, DeepLabV3, DETR |
| [37] | China, 2022 | Periodontal diseases, Dental caries | Periapical | Hospital | 7924 | Classification | Sens, Spec, PPV, NPV, F1, ROC-AUC | Modified ResNet-18 |
| [38] | China, 2022 | Ectopic eruption | Panoramic | Hospital | 3160 | Classification | Sens, Spec, PPV, NPV, ROC-AUC, F1 | Proprietary CNN |
| [39] | Saudi Arabia, 2022 | Impacted tooth | Panoramic | University | 416 | Classification | Acc, Prec, Rec, Spec, F1 | DenseNet, VGG, Inception V3, ResNet-50 |
| [40] | China, 2022 | Dental caries | Periapical | University | 840 | Classification | DICE, Prec, Sens, Spec | Proprietary CNN |
| [41] | Turkey, 2022 | Dental caries | Periapical | Private clinic | 340 | Classification | Acc, ROC-AUC, CM | Proprietary CNN, VGG, SqueezeNet, GoogleNet, ResNet, ShuffleNet, Xception, MobileNet, DarkNet |
| [42] | Japan, 2022 | Oral lesions | Panoramic | Hospital | 7260 | Classification | Acc, Sens, Spec, Prec, Rec, F1 | YOLO v3 |
| [43] | Germany, 2022 | Oral lesions | Panoramic | University | 1239 | Classification | Prec, Rec, NPV, Spec, F1 | ResNet, RF |
| [44] | Netherlands, 2022 | Periodontal diseases | Periapical | University | 1546 | Regression | MSE | Proprietary CNN |
| [45] | China, 2022 | Dental caries | Periapical | University | 800 | Classification | Prec, F1 | Proprietary CNN |
| [46] | Turkey, 2022 | Periodontal diseases | X-ray, type not defined | Database | 1432 | Classification | Acc, Sens, Spec, Prec, F1 | AlexNet, SqueezeNet, EfficientNet, DT, KNN, NB, RUSBoost, SVM, |
| Journal | n | % |
|---|---|---|
| Journal of Dentistry | 5 | 23% |
| Diagnostics | 3 | 14% |
| Biomedical Signal Processing and Control | 1 | 5% |
| Scientific Reports | 1 | 5% |
| Journal of Oral and Maxillofacial Surgery, Medicine, and Pathology | 1 | 5% |
| Informatics in Medicine Unlocked | 1 | 5% |
| Cluster Computing | 1 | 5% |
| International Dental Journal | 1 | 5% |
| Journal of Clinical Medicine | 1 | 5% |
| Displays | 1 | 5% |
| Journal of Endodontics | 1 | 5% |
| Health Information Science and Systems | 1 | 5% |
| Oral Diseases | 1 | 5% |
| IEEE Access | 1 | 5% |
| Applied Sciences | 1 | 5% |
| IEEE Transactions on Automation Science and Engineering | 1 | 5% |
| Dataset Size | Number of Datasets |
|---|---|
| <500 | 5 |
| 500–1000 | 4 |
| 1000–1500 | 2 |
| 1500–2000 | 1 |
| 2000–5000 | 4 |
| 5000–10,000 | 2 |
| 10,000–50,000 | 1 |
| 50,000–100,000 | 0 |
| 10,000–500,000 | 2 |
| 500,000–1,000,000 | 0 |
| >1,000,000 | 1 |
| Metric | n | Average | Minimum | Maximum |
|---|---|---|---|---|
| Recall | 17 | 0.84 | 0.51 | 0.96 |
| Precision | 16 | 0.81 | 0.67 | 0.99 |
| Specificity | 14 | 0.85 | 0.51 | 1.00 |
| F1 score | 13 | 0.81 | 0.58 | 0.97 |
| Accuracy | 9 | 0.92 | 0.81 | 0.98 |
| ROC-AUC * | 8 | 0.93 | 0.85 | 0.98 |
| NPV ** | 7 | 0.83 | 0.68 | 0.95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martins, M.V.; Baptista, L.; Luís, H.; Assunção, V.; Araújo, M.-R.; Realinho, V. Machine Learning in X-ray Diagnosis for Oral Health: A Review of Recent Progress. Computation 2023, 11, 115. https://doi.org/10.3390/computation11060115
Martins MV, Baptista L, Luís H, Assunção V, Araújo M-R, Realinho V. Machine Learning in X-ray Diagnosis for Oral Health: A Review of Recent Progress. Computation. 2023; 11(6):115. https://doi.org/10.3390/computation11060115
Chicago/Turabian StyleMartins, Mónica Vieira, Luís Baptista, Henrique Luís, Victor Assunção, Mário-Rui Araújo, and Valentim Realinho. 2023. "Machine Learning in X-ray Diagnosis for Oral Health: A Review of Recent Progress" Computation 11, no. 6: 115. https://doi.org/10.3390/computation11060115
APA StyleMartins, M. V., Baptista, L., Luís, H., Assunção, V., Araújo, M.-R., & Realinho, V. (2023). Machine Learning in X-ray Diagnosis for Oral Health: A Review of Recent Progress. Computation, 11(6), 115. https://doi.org/10.3390/computation11060115





