A New ECG Data Processing Approach to Developing an Accurate Driving Fatigue Detection Framework with Heart Rate Variability Analysis and Ensemble Learning
Abstract
1. Introduction
- As shown in Table 1, a number of driving fatigue detection studies using ECG only from 2017 to 2022 achieved a low accuracy, up to 92.5% [10]. Most driving fatigue detection studies have combined a heart rate-related sensor with other physiological sensors to obtain a more accurate classification model. For example, [11] combined ECG, EEG, and driving behavior sensors, resulting in an accuracy of 95.4%. Using more sensors attached to the driver’s body is impractical in real-world driving applications.
- Most driving fatigue detection studies (Table 1) have concentrated on developing feature engineering and classification techniques. Very few studies have focused on developing preprocessing methods. In fact, the preprocessing stage plays the most important part in the classification problem [12,13,14]. Therefore, with the right preprocessing method, driving fatigue detection using ECG could likely increase the model’s performance.
- Most driving fatigue detection studies have focused on developing the classification stage using neural networks and deep learning models to improve model performance. Table 1 shows very good results for the method of [6], which uses this strategy. For example, Huang and Deng proposed a novel classification model for detecting driver fatigue in 2022. They used a combination of neural network models, resulting in a 97.9% accuracy [15]. However, these methods are not perfect. They require a large amount of data and many computational resources, and if the model is overtrained to minimize the error, it may become less generalized [16,17].
- We chose the single-lead ECG method for driving fatigue measurement due to its ease of use. This method is sufficient for measuring heart rate, and heart rate variability is correlated with driver fatigue [18,19]. The ECG recording configuration used for driving fatigue detection in this study is a modified lead-I ECG with two electrodes placed at the second intercostal space.
- In the preprocessing stage, we applied three types of resampling methods—no resampling, resampling only, and resampling with overlapping windows—to obtain and gather more information from the ECG data. Five resampling scenarios were employed in the driving fatigue detection framework (Figure 1) to determine which resampling scenario had the greatest impact on the model’s performance.
- In the feature extraction stage, we applied new feature extraction methods that had not been used in previous driving fatigue detection studies (Table 1). These are Poincare plot analysis and multifractal detrended fluctuation analysis to extract nonlinear properties from ECG data. There are 2 scenarios for the feature extraction method employed in the driving fatigue detection framework: 29 and 54 features are used to evaluate whether the nonlinear analysis method has an effect on the model’s performance. A 29-feature scenario covers the properties of the time domain and frequency domain analysis, while a 54-feature scenario covers the properties of the time domain, frequency domain, and nonlinear analysis.
- In the classification stage, we preferred to use an ensemble learning model to produce a better classification performance than an individual model. We employed four ensemble learning model scenarios (Figure 1), AdaBoost, bagging, gradient boosting, and random forest, to assess which method gave the best model performance. In the proposed driving fatigue detection framework, 40 possible scenarios were employed. A combination of five resampling scenarios, two feature extraction method scenarios, and four ensemble learning model scenarios were considered to determine which scenario produced the best model performance on both the training and testing datasets. In addition, we employed the cross-validation method to evaluate model generalizability and the hyperparameter optimization method to optimize the trained model in the proposed driving fatigue detection framework.
2. Related Works
| No. | Source | Number of Participants | Record. Time | Measurement | Features | Classification | Class | Accuracy |
|---|---|---|---|---|---|---|---|---|
| 1 | [27] | 22 | 80 min | ECG and EEG | 95 | Support vector machine (SVM) | 2 | 80.9 |
| 2 | [28] | 1st: 18; 2nd: 24; 3rd: 44 | 90 min | EEG, ECG, EOG, steering behavior and lane positioning | 54 | Random forest | 2 | 94.1 |
| 3 | [29] | Unknown | 5 min | ECG | 4 | SVM | 2 | 83.9 |
| 4 | [30] | 6 | 67 min | ECG | 12 | SVM | 2 | AUC: 0.95 |
| 5 | [10] | 25 | 80 min | ECG | 24 | Ensemble logistic regression | 2 | 92.5 |
| 6 | [31] | 6 | 60–120 min | ECG | Convolutional neural network (CNN) and recurrence plot | 2 | Accuracy: 70 Precision: 71 Recall: 85 | |
| 7 | [32] | 25 | Unknown | ECG | 32 | SVM | 2 | 87.5 |
| 8 | [33] | 47 | 30 min | ECG signals and vehicle data | 49 | Random forest | 2 | 91.2 |
| 9 | [34] | 23 | 33 min | driving behavior, reaction time and ECG | 13 | Eigenvalue of generation process of driving fatigue (GPDF) | 3 | 72 |
| 10 | [35] | 45 | 45 min | BVP, respiration, skin conductance and skin temperature | 73 | CNN-LSTM | 2 | Recall: 82 Specificity: 71 Sensitivity: 93 AUC: 0.88 |
| 11 | [11] | 16 | 30 min | EEG, ECG, driving behavior | 80 | Majority voting classifier (kNN, LR, SVM) | 2 | 95.4 |
| 12 | [36] | 16 | Unknown | ECG | Multiple-objective genetic algorithm (MOGA) optimized deep multiple kernel learning support vector machine (D-MKL-SVM) + cross-correlation coefficient | 2 | AUC: 0.97 | |
| 13 | [37] | 35 | 30 min | ECG, EEG, EOG | 13 | Artificial neural networks (ANNs) | 2 | 83.5 |
| 14 | [15] | 9 | >10 min | EDA, RESP, and PPG | 15 | ANN, backpropagation neural network, and cascade forward neural network | 2 | 97.9 |
| 15 | [38] | 20 | 20 min | EEG and ECG | Product fuzzy convolutional network (PFCN) | 2 | 94.19 | |
3. Materials and Methods
3.1. Dataset
3.2. Driving Fatigue Detection Framework
3.3. Data Splitting and Labelling
3.4. Resampling Methods
3.5. Feature Extraction
| No. | Type of Analysis | Feature Name | Feature Description | |
|---|---|---|---|---|
| 1 | Time Domain | Statistical analysis [47,66] | MeanNN | Mean of the NN intervals of time series data |
| 2 | SDNN | Standard deviation of the NN intervals of time series data | ||
| 3 | SDANN | Standard deviation of the average NN intervals of each 5-minute segment of time series data | ||
| 4 | SDNNI | Mean of the standard deviations of NN intervals of each 5-minute segment of time series data | ||
| 5 | RMSSD | Square root of the mean of the sum of successive differences between adjacent NN intervals | ||
| 6 | SDSD | Standard deviation of the successive differences between NN intervals of time series data | ||
| 7 | CVNN | Ratio of SDNN to MeanNN | ||
| 8 | CVSD | Ratio of RMSSD and MeanNN | ||
| 9 | MedianNN | Median of the absolute values of the successive differences between NN intervals of time series data | ||
| 10 | MadNN | Median absolute deviation of the NN intervals of time series data | ||
| 11 | HCVNN | Ratio of MadNN to Median | ||
| 12 | IQRNN | Interquartile range (IQR) of the NN intervals | ||
| 13 | Prc20NN | The 20th percentile of the NN intervals | ||
| 14 | Prc80NN | The 80th percentile of the NN intervals | ||
| 15 | pNN50 | The proportion of NN intervals greater than 50 ms out of the total number of NN intervals of time series data | ||
| 16 | pNN20 | The proportion of NN intervals greater than 20 ms out of the total number of NN intervals of time series data | ||
| 17 | MinNN | Minimum of the NN intervals of time series data | ||
| 18 | MaxNN | Maximum of the NN intervals of time series data | ||
| 19 | Geometrical analysis [47,66] | TINN | Width of the baseline of the distribution of the NN interval obtained by triangular interpolation | |
| 20 | HTI | HRV triangular index | ||
| 21 | Frequency Domain | Spectral analysis [47,66] | ULF | Power in the ultralow frequency range |
| 22 | VLF | Power in the very low-frequency range | ||
| 23 | LF | Power in the low-frequency range | ||
| 24 | HF | Power in the high-frequency range | ||
| 25 | VHF | Power in the very high-frequency range | ||
| 26 | LFHF | Ratio of LF to HF | ||
| 27 | LFn | Normalized power in the low-frequency range | ||
| 28 | HFn | Normalized power in the high-frequency range | ||
| 29 | LnHF | Natural logarithm of power in the high frequency range |
| No | Type of Analysis | Feature Name | Feature Description |
|---|---|---|---|
| 1 | Poincare analysis [66,71,79] | SD1 | Standard deviation perpendicular to the line of identity |
| 2 | SD2 | Standard deviation along the identity line | |
| 3 | SD1/SD2 | Ratio of SD1 to SD2 | |
| 4 | S | Area of the ellipse described by SD1 and SD2 | |
| 5 | CSI | Cardiac Sympathetic Index | |
| 6 | CVI | Cardiac Vagal Index | |
| 7 | CSI modified | Modified CSI | |
| 8 | Detrended fluctuation analysis (DFA) [66,78,80] | DFA α1 | Detrended fluctuation analysis |
| 9 | MFDFA α1—Width | Multifractal DFA α1—width parameter | |
| 10 | MFDFA α1—Peak | Multifractal DFA α1—peak parameter | |
| 11 | MFDFA α1—Mean | Multifractal DFA α1—mean parameter | |
| 12 | MFDFA α1—Max | Multifractal DFA α1—maximum parameter | |
| 13 | MFDFA α1—Delta | Multifractal DFA α1—delta parameter | |
| 14 | MFDFA α1—Asymmetry | Multifractal DFA α1—asymmetry parameter | |
| 15 | MMFDFA α1—Fluctuation | Multifractal DFA α1—fluctuation parameter | |
| 16 | MFDFA α1—Increment | Multifractal DFA—increment parameter | |
| 17 | DFA α2 | Detrended fluctuation analysis | |
| 18 | MFDFA α2—Width | Multifractal DFA α2—width parameter | |
| 19 | MFDFA α2—Peak | Multifractal DFA α2—peak parameter | |
| 20 | MFDFA α2—Mean | Multifractal DFA α2—mean parameter | |
| 21 | MFDFA α2—Max | Multifractal DFA α2—maximum parameter | |
| 22 | MFDFA α2—Delta | Multifractal DFA α2—delta parameter | |
| 23 | MFDFA α2—Asymmetry | Multifractal DFA α2—asymmetry parameter | |
| 24 | MFDFA α2—Fluctuation | Multifractal DFA α2—fluctuation parameter | |
| 25 | MFDFA α2—Increment | Multifractal DFA α2—increment parameter |
3.6. Classification Model
3.7. Cross-Validation and Hyperparameter Optimization
4. Results and Discussion
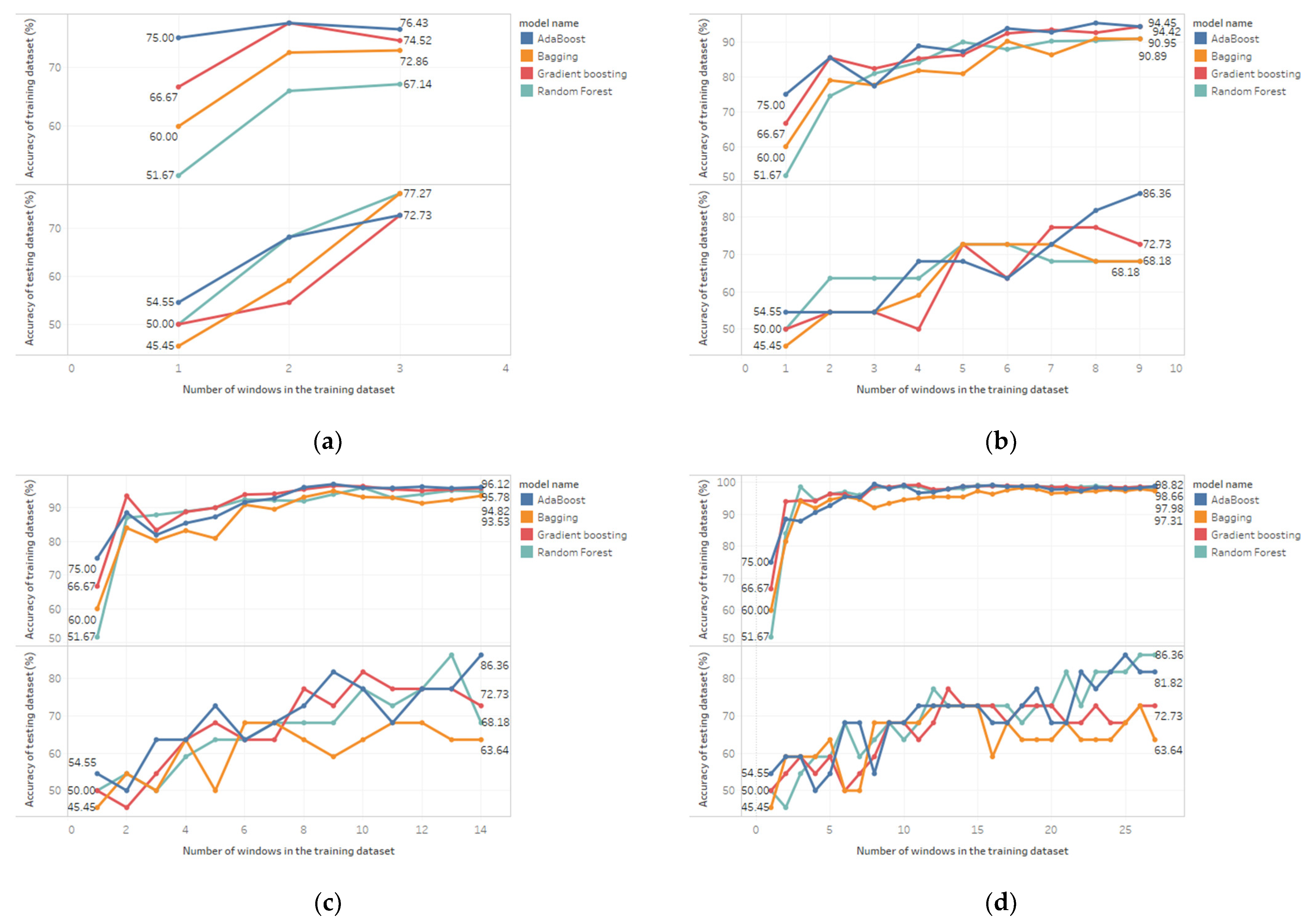
4.1. The Effect of Various Resampling Methods on the Model’s Performance
4.2. The Effect of 29 and 54 Features on the Model’s Performance
4.3. Model Selection Considerations
4.4. Future Work Developments
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Road Traffic Injuries. 2022. Available online: https://www.who.int/news-room/fact-sheets/detail/road-traffic-injuries (accessed on 22 September 2022).
- Chand, A.; Jayesh, S.; Bhasi, A. Road traffic accidents: An overview of data sources, analysis techniques and contributing factors. Mater. Today Proc. 2021, 47, 5135–5141. [Google Scholar] [CrossRef]
- Razzaghi, A.; Soori, H.; Kavousi, A.; Abadi, A.; Khosravi, A.K.; Alipour, A. Risk factors of deaths related to road traffic crashes in World Health Organization regions: A systematic review. Arch. Trauma Res. 2019, 8, 57–86. [Google Scholar]
- Bucsuházy, K.; Matuchová, E.; Zůvala, R.; Moravcová, P.; Kostíková, M.; Mikulec, R. Human factors contributing to the road traffic accident occurrence. Transp. Res. Procedia 2020, 45, 555–561. [Google Scholar] [CrossRef]
- Smith, A.P. A UK survey of driving behaviour, fatigue, risk taking and road traffic accidents. BMJ Open 2016, 6, e011461. [Google Scholar] [CrossRef]
- Albadawi, Y.; Takruri, M.; Awad, M. A review of recent developments in driver drowsiness detection systems. Sensors 2022, 22, 2069. [Google Scholar] [CrossRef]
- Khunpisuth, O.; Chotchinasri, T.; Koschakosai, V.; Hnoohom, N. Driver drowsiness detection using eye-closeness detection. In Proceedings of the 2016 12th International Conference on Signal-Image Technology & Internet-Based Systems (SITIS), Naples, Italy, 28 November–1 December 2016; pp. 661–668. [Google Scholar]
- Khare, S.K.; Bajaj, V. Entropy-Based Drowsiness Detection Using Adaptive Variational Mode Decomposition. IEEE Sens. J. 2021, 21, 6421–6428. [Google Scholar] [CrossRef]
- Khushaba, R.N.; Kodagoda, S.; Lal, S.; Dissanayake, G. Driver drowsiness classification using fuzzy wavelet-packet-based feature-extraction algorithm. IEEE Trans. Biomed. Eng. 2010, 58, 121–131. [Google Scholar] [CrossRef]
- Babaeian, M.; Amal Francis, K.; Dajani, K.; Mozumdar, M. Real-time driver drowsiness detection using wavelet transform and ensemble logistic regression. Int. J. Intell. Transp. Syst. Res. 2019, 17, 212–222. [Google Scholar] [CrossRef]
- Gwak, J.; Hirao, A.; Shino, M. An investigation of early detection of driver drowsiness using ensemble machine learning based on hybrid sensing. Appl. Sci. 2020, 10, 2890. [Google Scholar] [CrossRef]
- Singhal, S.; Jena, M. A study on WEKA tool for data preprocessing, classification and clustering. Int. J. Innov. Technol. Explor. Eng. (IJITEE) 2013, 2, 250–253. [Google Scholar]
- Benhar, H.; Idri, A.; Fernández-Alemán, J. Data preprocessing for heart disease classification: A systematic literature review. Comput. Methods Programs Biomed. 2020, 195, 105635. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekar, P.; Qian, K. The impact of data preprocessing on the performance of a naive bayes classifier. In Proceedings of the 2016 IEEE 40th Annual Computer Software and Applications Conference (COMPSAC), Atlanta, GA, USA, 10–14 June 2016; pp. 618–619. [Google Scholar]
- Huang, Y.; Deng, Y. A Hybrid Model Utilizing Principal Component Analysis and Artificial Neural Networks for Driving Drowsiness Detection. Appl. Sci. 2022, 12, 6007. [Google Scholar] [CrossRef]
- Shi, Z.; He, L.; Suzuki, K.; Nakamura, T.; Itoh, H. Survey on neural networks used for medical image processing. Int. J. Comput. Sci. 2009, 3, 86. [Google Scholar] [PubMed]
- Noguerol, T.M.; Paulano-Godino, F.; Martín-Valdivia, M.T.; Menias, C.O.; Luna, A. Strengths, weaknesses, opportunities, and threats analysis of artificial intelligence and machine learning applications in radiology. J. Am. Coll. Radiol. 2019, 16, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.; Nguyen, T.; Jang, H.; Kim, J.G.; Jun, S.C. Exploring neuro-physiological correlates of drivers’ mental fatigue caused by sleep deprivation using simultaneous EEG, ECG, and fNIRS data. Front. Hum. Neurosci. 2016, 10, 219. [Google Scholar] [CrossRef]
- Bier, L.; Wolf, P.; Hilsenbek, H.; Abendroth, B. How to measure monotony-related fatigue? A systematic review of fatigue measurement methods for use on driving tests. Theor. Issues Ergon. Sci. 2020, 21, 22–55. [Google Scholar] [CrossRef]
- May, J.F.; Baldwin, C.L. Driver fatigue: The importance of identifying causal factors of fatigue when considering detection and countermeasure technologies. Transp. Res. Part F Traffic Psychol. Behav. 2009, 12, 218–224. [Google Scholar] [CrossRef]
- Johns, M.W. A new method for measuring daytime sleepiness: The Epworth sleepiness scale. Sleep 1991, 14, 540–545. [Google Scholar] [CrossRef]
- Samn, S.W.; Perelli, L.P. Estimating Aircrew Fatigue: A Technique with Application to Airlift Operations; School of Aerospace Medicine: Brooks AFB, TX, USA, 1982. [Google Scholar]
- Shahid, A.; Wilkinson, K.; Marcu, S.; Shapiro, C.M. Stanford sleepiness scale (SSS). In Stop, That and One Hundred Other Sleep Scales; Springer: Berlin/Heidelberg, Germany, 2011; pp. 369–370. [Google Scholar]
- Åkerstedt, T.; Gillberg, M. Subjective and objective sleepiness in the active individual. Int. J. Neurosci. 1990, 52, 29–37. [Google Scholar] [CrossRef]
- Cella, M.; Chalder, T. Measuring fatigue in clinical and community settings. J. Psychosom. Res. 2010, 69, 17–22. [Google Scholar] [CrossRef]
- Ramzan, M.; Khan, H.U.; Awan, S.M.; Ismail, A.; Ilyas, M.; Mahmood, A. A survey on state-of-the-art drowsiness detection techniques. IEEE Access 2019, 7, 61904–61919. [Google Scholar] [CrossRef]
- Awais, M.; Badruddin, N.; Drieberg, M. A hybrid approach to detect driver drowsiness utilizing physiological signals to improve system performance and wearability. Sensors 2017, 17, 1991. [Google Scholar] [CrossRef] [PubMed]
- Mårtensson, H.; Keelan, O.; Ahlström, C. Driver sleepiness classification based on physiological data and driving performance from real road driving. IEEE Trans. Intell. Transp. Syst. 2018, 20, 421–430. [Google Scholar] [CrossRef]
- Lei, J.; Liu, F.; Han, Q.; Tang, Y.; Zeng, L.; Chen, M.; Ye, L.; Jin, L. Study on driving fatigue evaluation system based on short time period ECG signal. In Proceedings of the 2018 21st International Conference on Intelligent Transportation Systems (ITSC), Maui, HI, USA, 4–7 November 2018; pp. 2466–2470. [Google Scholar]
- Kim, J.; Shin, M. Utilizing HRV-derived respiration measures for driver drowsiness detection. Electronics 2019, 8, 669. [Google Scholar] [CrossRef]
- Lee, H.; Lee, J.; Shin, M. Using wearable ECG/PPG sensors for driver drowsiness detection based on distinguishable pattern of recurrence plots. Electronics 2019, 8, 192. [Google Scholar] [CrossRef]
- Babaeian, M.; Mozumdar, M. Driver drowsiness detection algorithms using electrocardiogram data analysis. In Proceedings of the 2019 IEEE 9th Annual Computing and Communication Workshop and Conference (CCWC), Las Vegas, NV, USA, 7–9 January 2019; pp. 0001–0006. [Google Scholar]
- Arefnezhad, S.; Eichberger, A.; Frühwirth, M.; Kaufmann, C.; Moser, M. Driver Drowsiness Classification Using Data Fusion of Vehicle-based Measures and ECG Signals. In Proceedings of the 2020 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Toronto, ON, Canada, 11–14 October 2020; pp. 451–456. [Google Scholar]
- Peng, Z.; Rong, J.; Wu, Y.; Zhou, C.; Yuan, Y.; Shao, X. Exploring the different patterns for generation process of driving fatigue based on individual driving behavior parameters. Transp. Res. Rec. 2021, 2675, 408–421. [Google Scholar] [CrossRef]
- Papakostas, M.; Das, K.; Abouelenien, M.; Mihalcea, R.; Burzo, M. Distracted and drowsy driving modeling using deep physiological representations and multitask learning. Appl. Sci. 2020, 11, 88. [Google Scholar] [CrossRef]
- Chui, K.T.; Lytras, M.D.; Liu, R.W. A generic design of driver drowsiness and stress recognition using MOGA optimized deep MKL-SVM. Sensors 2020, 20, 1474. [Google Scholar] [CrossRef]
- Hasan, M.M.; Watling, C.N.; Larue, G.S. Physiological signal-based drowsiness detection using machine learning: Singular and hybrid signal approaches. J. Saf. Res. 2022, 80, 215–225. [Google Scholar] [CrossRef]
- Du, G.; Long, S.; Li, C.; Wang, Z.; Liu, P.X. A Product Fuzzy Convolutional Network for Detecting Driving Fatigue. IEEE Trans. Cybern. 2022, 1–14. [Google Scholar] [CrossRef]
- Rather, A.A.; Sofi, T.A.; Mukhtar, N. A Survey on Fatigue and Drowsiness Detection Techniques in Driving. In Proceedings of the 2021 International Conference on Computing, Communication, and Intelligent Systems (ICCCIS), Greater Noida, India, 19–20 February 2021; pp. 239–244. [Google Scholar]
- Fujiwara, K.; Abe, E.; Kamata, K.; Nakayama, C.; Suzuki, Y.; Yamakawa, T.; Hiraoka, T.; Kano, M.; Sumi, Y.; Masuda, F. Heart rate variability-based driver drowsiness detection and its validation with EEG. IEEE Trans. Biomed. Eng. 2018, 66, 1769–1778. [Google Scholar] [CrossRef] [PubMed]
- Hendra, M.; Kurniawan, D.; Chrismiantari, R.V.; Utomo, T.P.; Nuryani, N. Drowsiness detection using heart rate variability analysis based on microcontroller unit. Proc. J. Phys. Conf. Ser. 2019, 1153, 012047. [Google Scholar] [CrossRef]
- Halomoan, J.; Ramli, K.; Sudiana, D. Statistical analysis to determine the ground truth of fatigue driving state using ECG Recording and subjective reporting. In Proceedings of the 2020 1st International Conference on Information Technology, Advanced Mechanical and Electrical Engineering (ICITAMEE), Yogyakarta, Indonesia, 13–14 October 2020; pp. 244–248. [Google Scholar]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [PubMed]
- Terzano, M.G.; Parrino, L.; Sherieri, A.; Chervin, R.; Chokroverty, S.; Guilleminault, C.; Hirshkowitz, M.; Mahowald, M.; Moldofsky, H.; Rosa, A. Atlas, rules, and recording techniques for the scoring of cyclic alternating pattern (CAP) in human sleep. Sleep Med. 2001, 2, 537–554. [Google Scholar] [CrossRef]
- Sahayadhas, A.; Sundaraj, K.; Murugappan, M. Detecting driver drowsiness based on sensors: A review. Sensors 2012, 12, 16937–16953. [Google Scholar] [CrossRef]
- Kwon, O.; Jeong, J.; Kim, H.B.; Kwon, I.H.; Park, S.Y.; Kim, J.E.; Choi, Y. Electrocardiogram sampling frequency range acceptable for heart rate variability analysis. Healthc. Inform. Res. 2018, 24, 198–206. [Google Scholar] [CrossRef]
- Shaffer, F.; Ginsberg, J.P. An overview of heart rate variability metrics and norms. Front. Public Health 2017, 5, 258. [Google Scholar] [CrossRef]
- Oweis, R.J.; Al-Tabbaa, B.O. QRS detection and heart rate variability analysis: A survey. Biomed. Sci. Eng. 2014, 2, 13–34. [Google Scholar]
- Fariha, M.; Ikeura, R.; Hayakawa, S.; Tsutsumi, S. Analysis of Pan-Tompkins algorithm performance with noisy ECG signals. Proc. J. Phys. Conf. Ser. 2020, 1532, 012022. [Google Scholar] [CrossRef]
- Pan, J.; Tompkins, W.J. A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 1985, BME-32, 230–236. [Google Scholar] [CrossRef]
- Wang, L.; Lin, Y.; Wang, J. A RR interval based automated apnea detection approach using residual network. Comput. Methods Programs Biomed. 2019, 176, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Van, R.G.; Drake, F. Python 3 reference manual. Scotts Val. CA Creat. 2009, 10, 1593511. [Google Scholar]
- McKinney, W. Pandas: A foundational Python library for data analysis and statistics. Python High Perform. Sci. Comput. 2011, 14, 1–9. [Google Scholar]
- Van Der Walt, S.; Colbert, S.C.; Varoquaux, G. The NumPy array: A structure for efficient numerical computation. Comput. Sci. Eng. 2011, 13, 22–30. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Raybaut, P. Spyder-Documentation. 2009. Available online: https://www.spyder-ide.org/ (accessed on 22 September 2022).
- Arlot, S.; Celisse, A. A survey of cross-validation procedures for model selection. Stat. Surv. 2010, 4, 40–79. [Google Scholar] [CrossRef]
- Meng, Z.; McCreadie, R.; Macdonald, C.; Ounis, I. Exploring data splitting strategies for the evaluation of recommendation models. In Proceedings of the Fourteenth ACM Conference on Recommender Systems, Online, 22–26 September 2020; pp. 681–686. [Google Scholar]
- Vilette, C.; Bonnell, T.; Henzi, P.; Barrett, L. Comparing dominance hierarchy methods using a data-splitting approach with real-world data. Behav. Ecol. 2020, 31, 1379–1390. [Google Scholar] [CrossRef]
- Malik, M. Heart rate variability: Standards of measurement, physiological interpretation, and clinical use: Task force of the European Society of Cardiology and the North American Society for Pacing and Electrophysiology. Ann. Noninvasive Electrocardiol. 1996, 1, 151–181. [Google Scholar] [CrossRef]
- El-Amir, H.; Hamdy, M. Data Resampling. In Deep Learning Pipeline; Springer: Berlin/Heidelberg, Germany, 2020; pp. 207–231. [Google Scholar]
- Zhou, Z.-H. Ensemble learning. In Machine Learning; Springer: Berlin/Heidelberg, Germany, 2021; pp. 181–210. [Google Scholar]
- Sagi, O.; Rokach, L. Ensemble learning: A survey. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2018, 8, e1249. [Google Scholar] [CrossRef]
- Rahi, P.K.; Mehra, R. Analysis of power spectrum estimation using welch method for various window techniques. Int. J. Emerg. Technol. Eng. 2014, 2, 106–109. [Google Scholar]
- Fell, J.; Röschke, J.; Mann, K.; Schäffner, C. Discrimination of sleep stages: A comparison between spectral and nonlinear EEG measures. Electroencephalogr. Clin. Neurophysiol. 1996, 98, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Makowski, D.; Pham, T.; Lau, Z.J.; Brammer, J.C.; Lespinasse, F.; Pham, H.; Schölzel, C.; Chen, S. NeuroKit2: A Python toolbox for neurophysiological signal processing. Behav. Res. Methods 2021, 53, 1689–1696. [Google Scholar] [CrossRef]
- Mourot, L.; Bouhaddi, M.; Perrey, S.; Cappelle, S.; Henriet, M.T.; Wolf, J.P.; Rouillon, J.D.; Regnard, J. Decrease in heart rate variability with overtraining: Assessment by the Poincare plot analysis. Clin. Physiol. Funct. Imaging 2004, 24, 10–18. [Google Scholar] [CrossRef]
- Zeng, C.; Wang, W.; Chen, C.; Zhang, C.; Cheng, B. Poincaré plot indices of heart rate variability for monitoring driving fatigue. In Proceedings of the 19th COTA International Conference of Transportation Professionals (CICTP), Nanjing, China, 6–8 July 2019; pp. 652–660. [Google Scholar]
- Guo, W.; Xu, C.; Tan, J.; Li, Y. Review and implementation of driving fatigue evaluation methods based on RR interval. In Proceedings of the International Conference on Green Intelligent Transportation System and Safety, Changchun, China, 1–2 July 2017; pp. 833–843. [Google Scholar]
- Hsu, C.-H.; Tsai, M.-Y.; Huang, G.-S.; Lin, T.-C.; Chen, K.-P.; Ho, S.-T.; Shyu, L.-Y.; Li, C.-Y. Poincaré plot indexes of heart rate variability detect dynamic autonomic modulation during general anesthesia induction. Acta Anaesthesiol. Taiwanica 2012, 50, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Tulppo, M.P.; Makikallio, T.H.; Takala, T.; Seppanen, T.; Huikuri, H.V. Quantitative beat-to-beat analysis of heart rate dynamics during exercise. Am. J. Physiol.-Heart Circ. Physiol. 1996, 271, H244–H252. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Venkatasubramanian, K. EKG/ECG based driver alert system for long haul drive. Indian J. Sci. Technol. 2015, 8, 8–13. [Google Scholar] [CrossRef]
- Mohanavelu, K.; Lamshe, R.; Poonguzhali, S.; Adalarasu, K.; Jagannath, M. Assessment of human fatigue during physical performance using physiological signals: A review. Biomed. Pharmacol. J. 2017, 10, 1887–1896. [Google Scholar] [CrossRef]
- Talebinejad, M.; Chan, A.D.; Miri, A. Fatigue estimation using a novel multi-fractal detrended fluctuation analysis-based approach. J. Electromyogr. Kinesiol. 2010, 20, 433–439. [Google Scholar] [CrossRef]
- Wang, F.; Wang, H.; Zhou, X.; Fu, R. Study on the effect of judgment excitation mode to relieve driving fatigue based on MF-DFA. Brain Sci. 2022, 12, 1199. [Google Scholar] [CrossRef]
- Rogers, B.; Mourot, L.; Doucende, G.; Gronwald, T. Fractal correlation properties of heart rate variability as a biomarker of endurance exercise fatigue in ultramarathon runners. Physiol. Rep. 2021, 9, e14956. [Google Scholar] [CrossRef]
- Wang, F.; Wang, H.; Zhou, X.; Fu, R. A Driving Fatigue Feature Detection Method Based on Multifractal Theory. IEEE Sens. J. 2022, 22, 19046–19059. [Google Scholar] [CrossRef]
- Kantelhardt, J.W.; Zschiegner, S.A.; Koscielny-Bunde, E.; Havlin, S.; Bunde, A.; Stanley, H.E. Multifractal detrended fluctuation analysis of nonstationary time series. Phys. A Stat. Mech. Appl. 2002, 316, 87–114. [Google Scholar] [CrossRef]
- Jeppesen, J.; Beniczky, S.; Johansen, P.; Sidenius, P.; Fuglsang-Frederiksen, A. Detection of epileptic seizures with a modified heart rate variability algorithm based on Lorenz plot. Seizure 2015, 24, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ihlen, E.A. Introduction to multifractal detrended fluctuation analysis in Matlab. Front. Physiol. 2012, 3, 141. [Google Scholar] [CrossRef]
- Gholami, R.; Fakhari, N. Support vector machine: Principles, parameters, and applications. In Handbook of Neural Computation; Elsevier: Amsterdam, The Netherlands, 2017; pp. 515–535. [Google Scholar]
- Ansari, A.; Bakar, A.A. A comparative study of three artificial intelligence techniques: Genetic algorithm, neural network, and fuzzy logic, on scheduling problem. In Proceedings of the 2014 4th International Conference on Artificial Intelligence with Applications in Engineering and Technology, Sabah, Malaysia, 3–5 December 2014; pp. 31–36. [Google Scholar]
- Emanet, N.; Öz, H.R.; Bayram, N.; Delen, D. A comparative analysis of machine learning methods for classification type decision problems in healthcare. Decis. Anal. 2014, 1, 6. [Google Scholar] [CrossRef]
- González, S.; García, S.; Del Ser, J.; Rokach, L.; Herrera, F. A practical tutorial on bagging and boosting based ensembles for machine learning: Algorithms, software tools, performance study, practical perspectives and opportunities. Inf. Fusion 2020, 64, 205–237. [Google Scholar] [CrossRef]
- Reitermanova, Z. Data splitting. In Proceedings of the WDS, Prague, Czech Republic, 1–4 June 2010; pp. 31–36. [Google Scholar]
- Xu, Y.; Goodacre, R. On splitting training and validation set: A comparative study of cross-validation, bootstrap and systematic sampling for estimating the generalization performance of supervised learning. J. Anal. Test. 2018, 2, 249–262. [Google Scholar] [CrossRef]
- Refaeilzadeh, P.; Tang, L.; Liu, H. Cross-validation. Encycl. Database Syst. 2009, 5, 532–538. [Google Scholar]
- Yang, L.; Shami, A. On hyperparameter optimization of machine learning algorithms: Theory and practice. Neurocomputing 2020, 415, 295–316. [Google Scholar] [CrossRef]
- Andonie, R. Hyperparameter optimization in learning systems. J. Membr. Comput. 2019, 1, 279–291. [Google Scholar] [CrossRef]
- Kotsiantis, S. Feature selection for machine learning classification problems: A recent overview. Artif. Intell. Rev. 2011, 42, 157–176. [Google Scholar] [CrossRef]
- Rodrigues, J.; Liu, H.; Folgado, D.; Belo, D.; Schultz, T.; Gamboa, H. Feature-Based Information Retrieval of Multimodal Biosignals with a Self-Similarity Matrix: Focus on Automatic Segmentation. Biosensors 2022, 12, 1182. [Google Scholar] [CrossRef] [PubMed]
- do Vale Madeiro, J.P.; Marques, J.A.L.; Han, T.; Pedrosa, R.C. Evaluation of mathematical models for QRS feature extraction and QRS morphology classification in ECG signals. Measurement 2020, 156, 107580. [Google Scholar] [CrossRef]
- Khan, T.T.; Sultana, N.; Reza, R.B.; Mostafa, R. ECG feature extraction in temporal domain and detection of various heart conditions. In Proceedings of the 2015 International Conference on Electrical Engineering and Information Communication Technology (ICEEICT), Savar, Dhaka, Bangladesh, 21–23 May 2015; pp. 1–6. [Google Scholar]


| Participant | Sleep-Good (SG)/Alert | Sleep-Bad (SB)/Fatigue | ||||
|---|---|---|---|---|---|---|
| ECG Data | Total NN Interval () in Msec | ECG Data | Total NN Interval () in Msec | |||
| in Mins | in Msec | in Mins | in Msec | |||
| 1 | 30.05 | 1,803,000 | 1,800,267 | 30.05 | 1,803,000 | 1,800,945 |
| 2 | 30.05 | 1,803,000 | 1,800,827 | 30.05 | 1,803,000 | 1,801,166 |
| 3 | 30.05 | 1,803,000 | 1,801,157 | 30.05 | 1,803,000 | 1,800,932 |
| 4 | 53.88 | 3,232,750 | 3,229,156 | 30.05 | 1,803,000 | 1,801,279 |
| 5 | 51.53 | 3,091,500 | 3,088,254 | 31.45 | 1,887,250 | 1,884,670 |
| 6 | 30.86 | 1,851,500 | 1,849,644 | 30.05 | 1,803,000 | 1,800,605 |
| 7 | 40.13 | 2,407,750 | 2,404,552 | 30.05 | 1,803,000 | 1,800,367 |
| 8 | 44.52 | 2,671,250 | 2,668,485 | 33.74 | 2,024,250 | 2,022,111 |
| 9 | 35.1 | 2,106,250 | 2,103,198 | 30.05 | 1,803,000 | 1,800,723 |
| 10 | 36.1 | 2,166,250 | 2,163,819 | 30.05 | 1,803,000 | 1,800,733 |
| 11 | 23.59 | 1,415,482 | 1,413,316 | 30.05 | 1,803,000 | 1,800,578 |
| Min | 23.59 | 1,415,482 | 1,413,316 | 30.05 | 1,803,000 | 1,800,367 |
| Max | 53.88 | 3,232,750 | 3,229,156 | 33.74 | 2,024,250 | 2,022,111 |
| St. Dev. | 9.64 | 578,288 | 577,807 | 1.15 | 68,967 | 68,949 |
| Average | 36.90 | 2,213,794 | 2,211,152 | 30.51 | 1,830,773 | 1,828,555 |
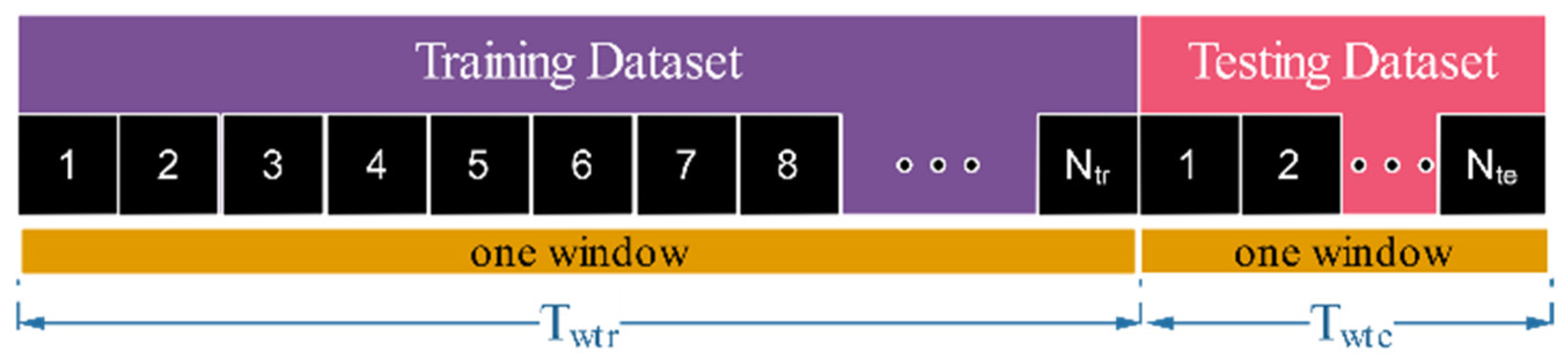
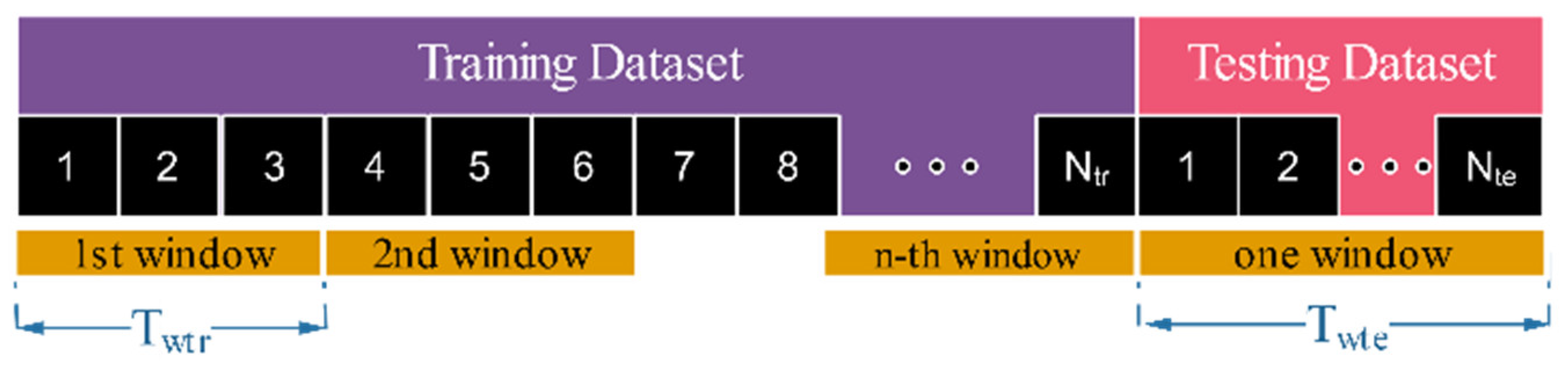
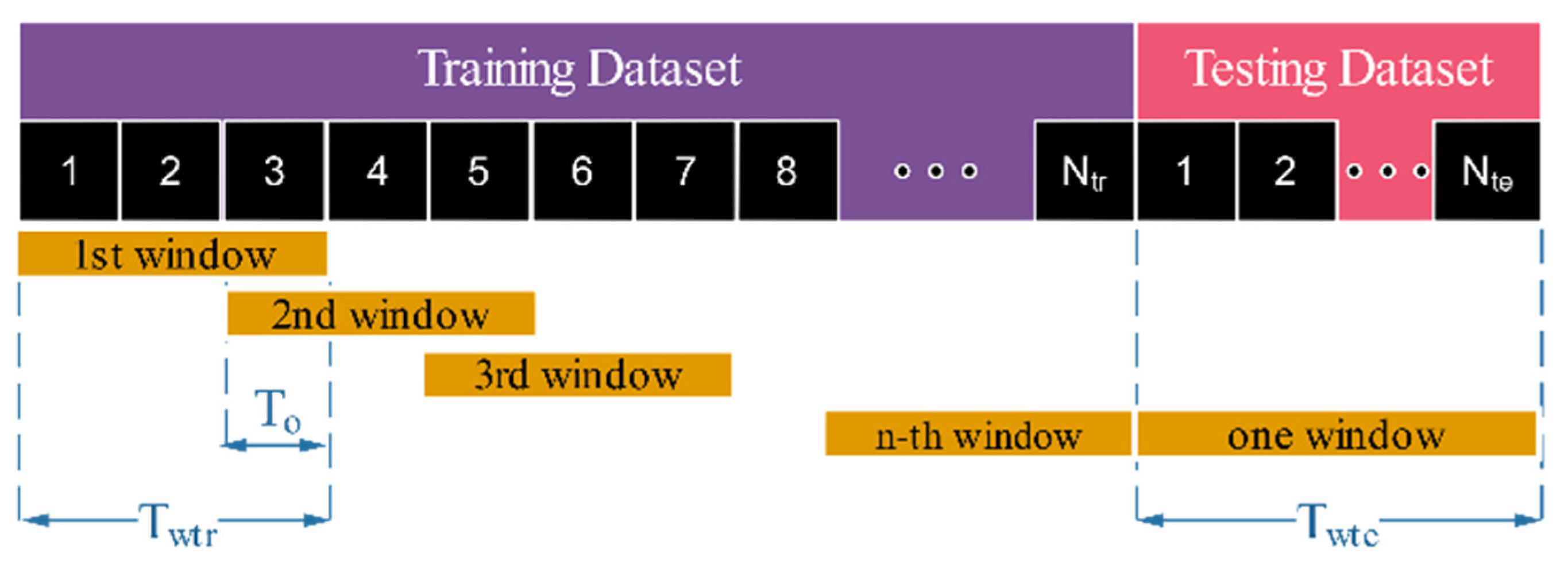
| Resampling Method | Training Dataset (78%) | Testing Dataset (22%) | Term | ||
|---|---|---|---|---|---|
| Duration | Number of Windows () | Duration | Number of Windows () | ||
| No resampling | 18 min or 1080 s | 1 | 300 s | 1 | NoR |
| Resampling only | 300 s 0 s | 3 | RO | ||
| Resampling with overlapping windows | 300 s 210 s | 9 | ROW210 | ||
| 300 s 240 s | 14 | ROW240 | |||
| 300 s 270 s | 27 | ROW270 | |||
| Model | Hyperparameter | Description | Range |
|---|---|---|---|
| AdaBoost | n_estimators | The maximum number of estimators | [10, 20, 50, 100, 500] |
| learning_rate | The weight that is assigned to each weak learner in the model | [0.0001, 0.001, 0.01, 0.1, 1.0] | |
| Bagging | n_estimators | The number of base estimators in the ensemble | [10, 20, 50, 100] |
| Gradient boosting | n_estimators | The number of boosting stages to perform | [10, 100, 500, 1000] |
| learning_rate | The step size that controls the model weight update at each iteration | [0.001, 0.01, 0.1] | |
| Subsample | A random subset used for fitting the individual base learners | [0.5, 0.7, 1.0] | |
| max_depth | The maximum number of levels in a decision tree | [3, 7, 9] | |
| Random forest | n_estimators | The number of trees in the forest | [10, 20, 50, 100] |
| max_features | The number of features to consider when looking for the best split | [‘sqrt’, ‘log2’] |
| Data Resampling Scenarios | Feature Extraction Scenarios | Classification Scenarios | |
|---|---|---|---|
| Term | Description | ||
| NoR | No Resampling |
|
|
| RO | Resampling only ( = 0 s) | ||
| ROW210 | Resampling with overlapping window = 210 s | ||
| ROW240 | Resampling with overlapping window = 240 s | ||
| ROW270 | Resampling with overlapping window = 270 s | ||
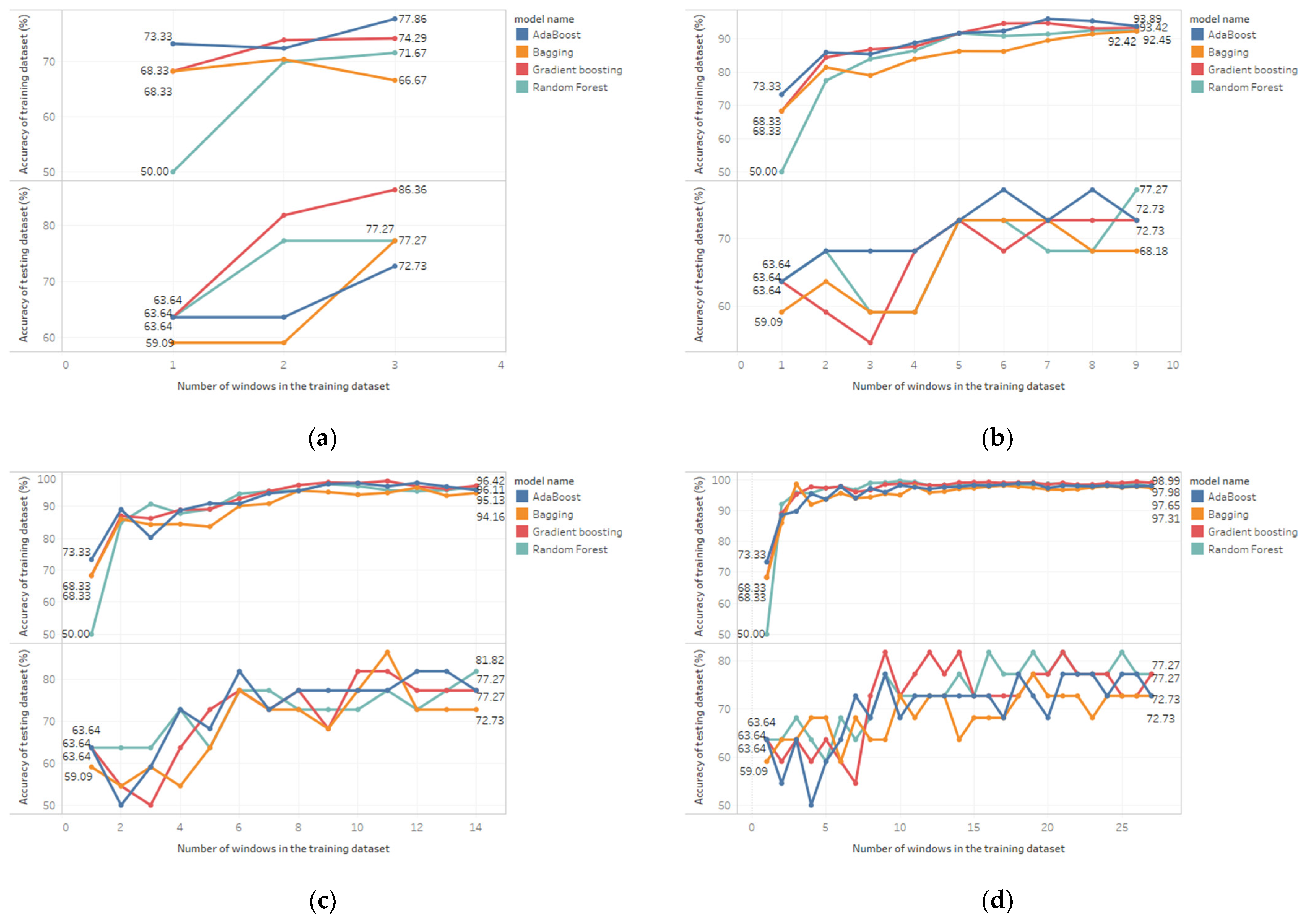
| Classifier | Features | Resampling Scenario | Accuracy on the Training Dataset (%) | Accuracy on the Testing Dataset (%) | ||||
|---|---|---|---|---|---|---|---|---|
| 1 Window | All Windows | Increase | 1 Window | All Windows | Increase | |||
| AdaBoost | 29 | RO | 73.33 | 77.86 | 4.53 | 63.64 | 72.73 | 9.09 |
| ROW210 | 73.33 | 93.89 | 20.56 | 63.64 | 72.73 | 9.09 | ||
| ROW240 | 73.33 | 95.13 | 21.8 | 63.64 | 77.27 | 13.63 | ||
| ROW270 | 73.33 | 97.98 | 24.65 | 63.64 | 72.73 | 9.09 | ||
| 54 | RO | 75 | 76.43 | 1.43 | 54.55 | 72.73 | 18.18 | |
| ROW210 | 75 | 94.45 | 19.45 | 54.55 | 86.36 | 31.81 | ||
| ROW240 | 75 | 96.12 | 21.12 | 54.55 | 86.36 | 31.81 | ||
| ROW270 | 75 | 98.82 | 23.82 | 54.55 | 81.82 | 27.27 | ||
| Bagging | 29 | RO | 68.33 | 66.67 | - | 59.09 | 77.27 | 18.18 |
| ROW210 | 68.33 | 92.42 | 24.09 | 59.09 | 68.18 | 9.09 | ||
| ROW240 | 68.33 | 94.16 | 25.83 | 59.09 | 72.73 | 13.64 | ||
| ROW270 | 68.33 | 97.31 | 28.98 | 59.09 | 72.73 | 13.64 | ||
| 54 | RO | 60 | 72.86 | 12.86 | 45.45 | 77.27 | 31.82 | |
| ROW210 | 60 | 90.89 | 30.89 | 45.45 | 68.18 | 22.73 | ||
| ROW240 | 60 | 93.53 | 33.53 | 45.45 | 63.64 | 18.19 | ||
| ROW270 | 60 | 97.31 | 37.31 | 45.45 | 63.64 | 18.19 | ||
| Gradient boosting | 29 | RO | 68.33 | 74.29 | 5.96 | 63.64 | 86.36 | 22.72 |
| ROW210 | 68.33 | 93.42 | 25.09 | 63.64 | 72.73 | 9.09 | ||
| ROW240 | 68.33 | 96.42 | 28.09 | 63.64 | 77.27 | 13.63 | ||
| ROW270 | 68.33 | 98.99 | 30.66 | 63.64 | 77.27 | 13.63 | ||
| 54 | RO | 66.67 | 74.52 | 7.85 | 50 | 72.73 | 22.73 | |
| ROW210 | 66.67 | 94.42 | 27.75 | 50 | 72.73 | 22.73 | ||
| ROW240 | 66.67 | 95.78 | 29.11 | 50 | 72.73 | 22.73 | ||
| ROW270 | 66.67 | 98.66 | 31.99 | 50 | 72.73 | 22.73 | ||
| Random forest | 29 | RO | 50 | 71.67 | 21.67 | 63.64 | 77.27 | 13.63 |
| ROW210 | 50 | 92.45 | 42.45 | 63.64 | 77.27 | 13.63 | ||
| ROW240 | 50 | 96.11 | 46.11 | 63.64 | 81.82 | 18.18 | ||
| ROW270 | 50 | 97.65 | 47.65 | 63.64 | 77.27 | 13.63 | ||
| 54 | RO | 51.67 | 67.14 | 15.47 | 50 | 77.27 | 27.27 | |
| ROW210 | 51.67 | 90.95 | 39.28 | 50 | 68.18 | 18.18 | ||
| ROW240 | 51.67 | 94.82 | 43.15 | 50 | 68.18 | 18.18 | ||
| ROW270 | 51.67 | 97.98 | 46.31 | 50 | 86.36 | 36.36 | ||
| Classifier | Features | Resampling Scenario | Performance Metrics (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Training Dataset | Testing Dataset | ||||||||
| Acc | Acc | F1 Score | Precision | Sensitivity | Specificity | AUC | |||
| AdaBoost | 29 | NoR | 71.67 | 59.09 | 64 | 57.14 | 72.73 | 45.45 | 0.71 |
| RO | 77.86 | 72.73 | 76.92 | 66.67 | 90.91 | 54.55 | 0.77 | ||
| ROW210 | 93.89 | 72.73 | 75 | 69.23 | 81.82 | 63.64 | 0.88 | ||
| ROW240 | 95.13 | 77.27 | 78.26 | 75 | 81.82 | 72.73 | 0.9 | ||
| ROW270 | 97.98 | 72.73 | 75 | 69.23 | 81.82 | 63.64 | 0.88 | ||
| 54 | NoR | 56.67 | 68.18 | 63.16 | 75 | 54.55 | 81.82 | 0.68 | |
| RO | 76.43 | 72.73 | 76.92 | 66.67 | 90.91 | 54.55 | 0.81 | ||
| ROW210 | 94.45 | 86.36 | 86.96 | 83.33 | 90.91 | 81.82 | 0.9 | ||
| ROW240 | 96.12 | 86.36 | 85.71 | 90 | 81.82 | 90.91 | 0.89 | ||
| ROW270 | 98.82 | 81.82 | 81.82 | 81.82 | 81.82 | 81.82 | 0.9 | ||
| Bagging | 29 | NoR | 56.67 | 72.73 | 75 | 69.23 | 81.82 | 63.64 | 0.83 |
| RO | 66.67 | 77.27 | 80 | 71.43 | 90.91 | 63.64 | 0.86 | ||
| ROW210 | 92.42 | 68.18 | 69.57 | 66.67 | 72.73 | 63.64 | 0.85 | ||
| ROW240 | 94.16 | 72.73 | 72.73 | 72.73 | 72.73 | 72.73 | 0.86 | ||
| ROW270 | 97.31 | 72.73 | 72.73 | 72.73 | 72.73 | 72.73 | 0.86 | ||
| 54 | NoR | 61.67 | 68.18 | 69.57 | 66.67 | 72.73 | 63.64 | 0.76 | |
| RO | 72.86 | 77.27 | 80 | 71.43 | 90.91 | 63.64 | 0.76 | ||
| ROW210 | 90.89 | 68.18 | 69.57 | 66.67 | 72.73 | 63.64 | 0.81 | ||
| ROW240 | 93.53 | 63.64 | 63.64 | 63.64 | 63.64 | 63.64 | 0.84 | ||
| ROW270 | 97.31 | 63.64 | 63.64 | 63.64 | 63.64 | 63.64 | 0.84 | ||
| Gradient boosting | 29 | NoR | 66.67 | 63.64 | 66.67 | 61.54 | 72.73 | 54.55 | 0.64 |
| RO | 74.29 | 86.36 | 86.96 | 83.33 | 90.91 | 81.82 | 0.91 | ||
| ROW210 | 93.42 | 72.73 | 75 | 69.23 | 81.82 | 63.64 | 0.9 | ||
| ROW240 | 96.42 | 77.27 | 78.26 | 75 | 81.82 | 72.73 | 0.83 | ||
| ROW270 | 98.99 | 77.27 | 78.26 | 75 | 81.82 | 72.73 | 0.85 | ||
| 54 | NoR | 61.67 | 68.18 | 66.67 | 70 | 63.64 | 72.73 | 0.84 | |
| RO | 74.52 | 72.73 | 76.92 | 66.67 | 90.91 | 54.55 | 0.72 | ||
| ROW210 | 94.42 | 72.73 | 75 | 69.23 | 81.82 | 63.64 | 0.86 | ||
| ROW240 | 95.78 | 72.73 | 75 | 69.23 | 81.82 | 63.64 | 0.88 | ||
| ROW270 | 98.66 | 72.73 | 75 | 69.23 | 81.82 | 63.64 | 0.87 | ||
| Random forest | 29 | NoR | 55 | 72.73 | 72.73 | 72.73 | 72.73 | 72.73 | 0.78 |
| RO | 71.67 | 77.27 | 80 | 71.43 | 90.91 | 63.64 | 0.84 | ||
| ROW210 | 92.45 | 77.27 | 80 | 71.43 | 90.91 | 63.64 | 0.87 | ||
| ROW240 | 96.11 | 81.82 | 83.33 | 76.92 | 90.91 | 72.73 | 0.9 | ||
| ROW270 | 97.65 | 77.27 | 78.26 | 75 | 81.82 | 72.73 | 0.9 | ||
| 54 | NoR | 55 | 68.18 | 66.67 | 70 | 63.64 | 72.73 | 0.81 | |
| RO | 67.14 | 77.27 | 78.26 | 75 | 81.82 | 72.73 | 0.88 | ||
| ROW210 | 90.95 | 68.18 | 72 | 64.29 | 81.82 | 54.55 | 0.86 | ||
| ROW240 | 94.82 | 68.18 | 66.67 | 70 | 63.64 | 72.73 | 0.82 | ||
| ROW270 | 97.98 | 86.36 | 86.96 | 83.33 | 90.91 | 81.82 | 0.96 | ||
| Source | Number of Participants | Record. Time | Measurement | Features | Classification | Accuracy 1 |
|---|---|---|---|---|---|---|
| [28] | 1st:18; 2nd:24; 3rd:44 | 90 min | EEG, ECG, EOG, and vehicle data | 54 | Random forest | 94.1 |
| [30] | 6 | 67 min | ECG | 12 | SVM | 0.95 (AUC) |
| [32] | 25 | 80 min | ECG | 24 | Ensemble logistic regression | 92.5 |
| [33] | 47 | 30 min | ECG and vehicle data | 49 | Random forest | 91.2 |
| [11] | 16 | 30 min | EEG, ECG, and vehicle data | 80 | Random forest | 95.4 |
| [15] | 9 | >10 min | EDA, RESP, and PPG | 15 | ANN, backpropagation neural network (BPNN), cascade forward neural network (CFNN) | 97.9 |
| [38] | 20 | 20 min | EEG and ECG | Product fuzzy convolutional network (PFCN) | 94.19 | |
| Ours | 11 | 30 min | ECG | 54 features, resampling with overlapping windows () | Random forest | 97.98 1 86.36 2 |
| AdaBoost | 98.82 1 81.82 2 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Halomoan, J.; Ramli, K.; Sudiana, D.; Gunawan, T.S.; Salman, M. A New ECG Data Processing Approach to Developing an Accurate Driving Fatigue Detection Framework with Heart Rate Variability Analysis and Ensemble Learning. Information 2023, 14, 210. https://doi.org/10.3390/info14040210
Halomoan J, Ramli K, Sudiana D, Gunawan TS, Salman M. A New ECG Data Processing Approach to Developing an Accurate Driving Fatigue Detection Framework with Heart Rate Variability Analysis and Ensemble Learning. Information. 2023; 14(4):210. https://doi.org/10.3390/info14040210
Chicago/Turabian StyleHalomoan, Junartho, Kalamullah Ramli, Dodi Sudiana, Teddy Surya Gunawan, and Muhammad Salman. 2023. "A New ECG Data Processing Approach to Developing an Accurate Driving Fatigue Detection Framework with Heart Rate Variability Analysis and Ensemble Learning" Information 14, no. 4: 210. https://doi.org/10.3390/info14040210
APA StyleHalomoan, J., Ramli, K., Sudiana, D., Gunawan, T. S., & Salman, M. (2023). A New ECG Data Processing Approach to Developing an Accurate Driving Fatigue Detection Framework with Heart Rate Variability Analysis and Ensemble Learning. Information, 14(4), 210. https://doi.org/10.3390/info14040210










