Abstract
Bacteriocinogenic cultures can represent a natural way to increase the safety of cheeses made from raw milk, in which a relevant role in ripening and flavor formation is exerted by the nonstarter lactic acid bacteria (NSLAB). Since the latter can be inhibited by bacteriocin producers, this study evaluated to which extent a nisinogenic culture inoculated at low initial levels can affect the growth rate and peptide degradation activity of the nisin-sensitive cheese isolate Lactobacillus plantarum LZ by comparison with its isogenic variant, L. plantarum LZNI, with increased immunity to nisin. A growth delay of the nisin sensitive strain was observed only when its initial number was 100-fold lower than the nisin producer and nisin was added as an inducer of its own production. In this case, the amount of free α-amino groups was significantly different between cultures of L. plantarum LZ and LZNI only at Day 1. Reverse Phase High Performance Liquid Chromatography (RP-HPLC) highlighted a few differences between the peptide profiles of co-cultures L. plantarum LZ and LZNI. However, results showed that the bacteriocin producer did not dramatically influence the behavior of the sensitive NSLAB and that the evaluation of the effects on microbial contaminants in cheese is worthwhile.
1. Introduction
Many Protected Designation of Origin (PDO) cheese productions follow manufacturing guidelines that prescribe use of raw milk. In this way, the diversity of the natural microbiota responsible for sensory distinctness, and, primarily, the starter lactic acid bacteria (SLAB) and nonstarter lactic acid bacteria (NSLAB), is preserved. NSLAB in most cheese varieties are represented by the mesophilic lactobacilli Lactobacillus paracasei and L. plantarum and are found in high numbers during the whole ripening period [1,2,3]. Some reports demonstrated that they exert a positive influence on flavor [4] due in part to their peptide hydrolyzing activity, which increases the amount of short peptides and free amino acids after the proteolysis carried out by rennet and SLAB. In particular, L. plantarum influenced proteolysis and the amounts of individual amino acids in Canestrato Pugliese PDO cheese and predominated from 7–15 days until 90 days of ripening [4]. L. plantarum strains also carry out transamination of amino acids with their conversion to flavor active compounds [5]. Use of raw milk in cheese manufacturing is hygienically hazardous and the good practices of milking and milk storage can reduce but not nullify the safety risk [6]. The possibility of exploiting the antimicrobial potential of bacteriocin producing lactic acid bacteria (LAB) to improve the safety of these products has received little consideration. The effect of bacteriocinogenic cultures on SLAB has been studied with the aim of accelerating ripening by inducing their lysis [7,8]. Moreover, use of a bacteriocin producer was proposed to inhibit adventitious NSLAB in Cheddar cheese from pasteurized milk, thus avoiding quality fluctuations [9,10]. Those investigations utilized a high initial inoculum of the bacteriocinogenic culture, so that use of bacteriocin resistant cultures was necessary to counteract the negative consequences of NSLAB inhibition [10]. No studies are available on the effect of bacteriocin producers on NSLAB at the strain level and on the effectiveness of using low initial numbers of the bacteriocin producer to avoid their strong inhibition.
The lantibiotic nisin A is used worldwide to improve the preservation of food products. However, the application of this natural antimicrobial, legally a food preservative at all effects [11], has not yet been considered for the manufacture of artisanal cheeses. Conversely, use of Lactococcuslactis subsp. lactis nisin producing cultures that have a Qualified Presumption of Safety (QPS) status [12] could be implemented also in these products. Indeed, some studies have demonstrated positive effects of using nisinogenic cultures in cheese manufacturing. Rilla et al., (2003) [13] reported that a nisinogenic culture inhibited Clostridium tyrobutyricum in cheese. Moreover, the addition of a nisin producing L. lactis subsp. lactis strain in Minas cheese made from raw goat milk kept the biogenic amine content at acceptable levels for human consumption and inhibited coagulase-positive cocci [14].
This study aimed at evaluating the feasibility of using low initial levels of a nisinogenic culture in cheese to improve safety without strongly inhibiting the autochthonous microbiota. The model system used was constituted by milk co-cultures of the nisin producer L. lactis subsp. lactis DSM 20729 with the NSLAB strain L. plantarum NI, isolated from Monte Veronese cheese and selected among other NSLAB isolates for its high sensitivity to nisin. The effects on growth and peptidolytic activity were put in evidence by comparison with co-cultures inoculated with a nisin-immune counterpart of L. plantarum LZ, L. plantarum LZNI, expressing the nisin immunity gene nisI and obtained in a previous study [15].
2. Materials and Methods
2.1. Culture Methods
Lactobacillus spp. strains used in this study belong to the species L. paracasei and L. plantarum (Table 1). The cheese isolates were identified in two previous studies [1,15]. These were subcultured in MRS medium (Biolife Italiana Srl., Milan, Italy). L. lactis subsp. lactis DSM 20729, a strain used in Swiss cheese manufacture to suppress gas production by clostridia, was grown in M17 medium (Biolife). All LAB strains were grown aerobically at 28 °C for 24 h–36 h.
L. plantarum counts in the co-cultures were carried out on MRS medium, supplemented with 100 μg/mL vancomycin to inhibit L. lactis subsp. lactis DSM 20729. The latter was enumerated in M17 medium.
The sensitivity to nisin of NSLAB strains was assessed by the agar spot assay on medium F (4 g/L peptone, 1 g/L yeast extract, 0.5 M K2HPO4, 0.18 M citric acid, 1.5 g/L agar, pH 5.4). Five microliters of a 24 h fresh culture of L. lactis subsp. lactis DSM 20729 were spotted onto F agar plates and grown for 24 h at 28 °C, then the plates were overlaid with 5 mL of F agar soft (F medium plus 0.5 % w/v agar) inoculated with 100 μL of a 24 h NSLAB strain culture. The diameter of the inhibition halo was measured after growth for 48 h at 30 °C. The MICs for nisin of L. plantarum LZ and LZNI were determined in F broth by the microdilution method using a double dilution series of nisin with concentration ranging between 0.5 and 2 × 104 IU mL−1. Nisin was added in the form of a water solution of a 106 IU g−1 commercial preparation (Sigma Aldrich, Milan, Italy) heat treated at 100 °C for 30 min. The initial inoculum of the strain to be tested was of the order of 103 CFU mL−1. Data were recorded after 48 h incubation at 30 °C. The MIC value was defined as the lowest nisin concentration giving a complete inhibition of visible growth in comparison to a non inoculated control well.
2.2. Co-Cultures in Milk Based Medium
L. plantarum LZ and its nisI expressing derivative LZNI were co-cultivated in skim milk (Oxoid, UK) supplemented with 1% (w/v) yeast extract and 1% (w/v) casamino acids (Difco Laboratories, Detroit, MI, USA) with L. lactis subsp. lactis DSM 20729 using two different inoculums ratios (102:102 CFU mL−1 in combinations B and NB and 102:104 CFU mL−1 in combinations A and NA). In the cultures NA and NB 10 IU mL−1 of nisin were added to induce nisin production by L. lactis subsp. lactis DSM 20729 [16]. The growth trials were carried out in triplicate. Cultures were incubated at 28 °C for 28 days. The comparisons between the average data series of growth and pH in co-cultures of L. plantarum LZ and LZNI was done by the Student t-test.
2.3. Quantification of Nisin in the Co-Cultures
The activity of nisin produced in the co-cultures was determined by the agar diffusion assay in F medium, using L. plantarum LZ as indicator, as follows: 500 μL of co-culture were acidified to pH 4.0 with 1 M HCl. Casein precipitates and bacterial cells were removed by centrifugation for 2 min at 8000 rpm. The clear supernatant was heat treated at 100 °C for 30 min. Ten microliters of the supernatants were transferred in wells created in the layer of agarized medium; the diameters of the inhibition haloes were compared to those of nisin solutions of known concentration for approximate quantification of activity.
2.4. Preparation of Peptide Extracts
Procedures recommended for cheese analysis [17] were adapted for peptide extraction from milk samples using a correspondence between mL of culture and g of cheese. Water-soluble nitrogen (WSN) was prepared from 1 mL of culture and following method III, while nitrogen soluble at pH 4.4 (non-casein nitrogen) was prepared from 1.25 mL of culture following method I [18].
2.5. Quantitative Determination of Free Amino Groups
Peptide hydrolysis was quantitatively determined by analysis of free amino groups according to the trinitrobenzenesulphonic acid (TNBS) procedure described by Polychroniadou [19] and using the non-inoculated growth medium as control. Data were expressed as OD at 420 nm and analyzed statistically by the Student t test. Pair wise comparisons between the values in co-cultures of L. plantarum LZ with those of co-cultures of L. plantarum LZNI were done at each time interval.
2.6. RP-HPLC Analysis of Peptide Profiles
The water soluble nitrogen (WSN) and nitrogen soluble at pH 4.4 were analyzed at different time intervals by Reversed-Phase High-Performance Liquid Chromatography (RP-HPLC) as described by Ardö and Gripon, 1995 [20] for pooled cultures B and NA using a Beckman HPLC system (125S Solvent Module coupled with a 166 Detector Module) equipped with a C18Vydac 218TP column (Sigma Aldrich). Chromatograms were acquired and analyzed by the Beckman GoldTM Nouveau ver. 1.7 software.
3. Results
3.1. Sensitivity of NSLAB to Nisin
Data on the sensitivity to nisin of the NSLAB strains tested are reported in Table 1 in terms of diameter of the inhibition halo formed in the deferred agar spot test with L. lactis subsp. lactis DSM 20729. L. plantarum LZ was selected as a suitable test strain, among other strains of L. plantarum and L. paracasei isolated from cheese, for being one of the most sensitive to nisin and for the availability of a food-grade variant with increased immunity to nisin developed in a previous study [15]. The latter was inhibited by 64-fold higher concentrations of nisin compared to the wild type strain, as determined by the broth microdilution test.

Table 1.
Bacterial strains tested for sensitivity to nisin in this study with respective halo sizes observed in the deferred agar spot test with the nisin producer L. lactis subsp. lactis DSM 20729.
| Species | Isolation Source | Strain and Halo Size (mm of Diameter) |
|---|---|---|
| L. plantarum | Monte Veronese cheese | LZ (18); R1M2 (16); R2M3 (0); RM2 (10); R12 (9) |
| Reference strains | NCFB 340 (8); ATCC 14917 (10) | |
| L. paracasei | Grana Trentino cheese | RL3 (15); RC4 (14); RL4 (15); RM10 (14); RM133 (16); RM187 (16); RF138 (16); RF102 (15); RF302 (14); RL131 (16); RF135 (15); RM1812 (15); RL188 (15); RR131 (12); RC304 (10); RC303 (18); RC137 (11); RC131 (7); RM1811 (12) |
| Reference strains | ATCC 334 (10); NCFB 151 (11); ATCC 27092 (16); ATCC 25180 (14); DSM 20006 (10); 4114 (11); NCIMB 8001 (8); NCFB 242 (9); NCFB 2743 (11); DSM 20207 (12) |
3.2. Growth and Acidification in Co-Cultures L. plantarum/Nisin Producer
The growth behavior of L. plantarum LZ and LZNI and acidification in co-culture with L. lactis subsp. lactis DSM 20729 are shown in Figure 1 for two combinations of initial inoculums and in presence or absence of 10 IU mL−1 of nisin added as inducer of its own production.
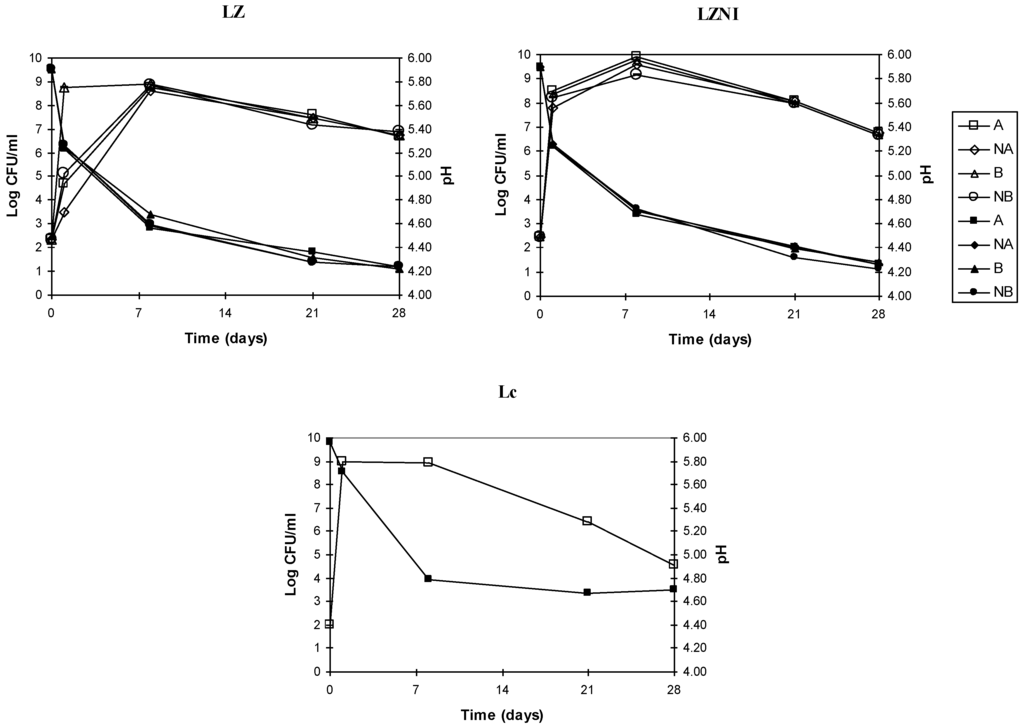
Figure 1.
Growth and pH values in co-cultures of L. plantarum LZ or LZNI with L. lactis subsp. lactis DSM 20729 (Lc):B, combination L. plantarum/nisin producer 102:102 CFU mL−1; NB, combination B with 10 IU/mL of nisin added; A, combinationL. plantarum/nisin producer 102:104 CFU mL−1; NA, combination A with 10 IU/mL of nisin added. Open symbols: Count values; solid symbols: pH values.
In the co-cultures,L. lactis subsp. lactis DSM 20729 reached high numbers at Day 1 (Table 2) but it could not be enumerated later because of the lack of specificity of the M17 medium. Indeed, L. plantarum LZ grew well on this medium not allowing the enumeration of the nisin producer when this was present in lower numbers. Therefore, it can be only stated that, since Day 8, L. lactis subsp. lactis DSM 20729 was present at numbers lower than 108 CFU mL−1 in the co-cultures. The growth behavior of this strain in pure culture indicated that its decline is more rapid than that of L. plantarum LZ and LZNI in the cultivation conditions adopted (Figure 1). Compared to L. plantarum LZNI, the growth of L. plantarum LZ was inhibited in the cultures A, NA and NB during the first day of incubation; however, the series of average CFU mL−1 and pH data of the co-cultures of L. plantarum LZ and LZNI at all the incubation times were not statistically distinct.

Table 2.
Average and SD of counts of L. lactis subsp. lactis DSM 20729 in the co-cultures with L. plantarum LZ and LZNI at time 0 and day 1.
| Day | LZA | LZNA | LZB | LZNB | LZNIA | LZNINA | LZNIB | LZNINB |
|---|---|---|---|---|---|---|---|---|
| 0 | 4.31±0.01 | 4.31±0.01 | 2.31±0.01 | 2.31±0.01 | 4.31±0.01 | 4.31±0.01 | 2.31±0.01 | 2.31±0.01 |
| 1 | 9.6±0.02 | 9.77±0.05 | 9.77±0.01 | 9.77±0.03 | 9.77±0.05 | 9.77±0.02 | 9.3±0.01 | 9.6±0.02 |
3.3. Nisin Activity and Peptide Hydrolysis in Co-Cultures
The activity of nisin in the cultures, expressed as diameter of the halo formed by filtered aliquots of the growth medium in lawns of L. plantarum LZ, and the concentration of free amino groups in terms of OD at 420 nm are reported in Figure 2. In all the co-cultures, except the B combinations at Day 1, nisin was detected during the whole incubation period of 28 days and could be roughly estimated, by referring to the calibration curve reported in Figure 3, as ranging between lower than 1 × 10 and about 3 × 102 IU mL−1, though a non-linear relationship between halo size and nisin activity was found for the lowest levels of nisin tested. During incubation nisin activity exhibited a fluctuation, since it increased until Day 8, decreased until Day 21 and increased again later.
Stimulation of nisin production was observed at all incubation times in the co-cultures NB, A and NA (Figure 2). The nisin concentration in the cultures was approximately estimated by plotting the diameter of haloes formed in lawns of L. plantarum LZ by known amounts of nisin expressed as IU mL−1 (Figure 3).
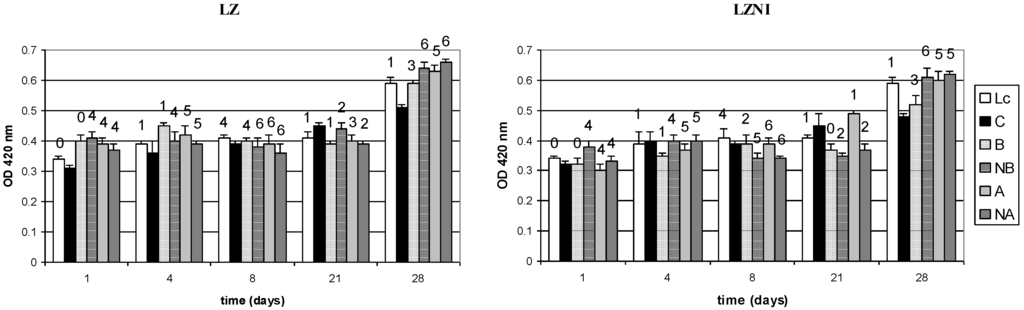
Figure 2.
Values of free amino groups, expressed as OD at 420nm and determined by the trinitrobenzenesulphonic acid (TNBS) method, and halo diameter (mm) formed by the nisin produced in the cultures in lawns of L. plantarum LZ. Lc, pure culture of L. lactis subsp. lactis DSM 20729; C, pure culture of L. plantarum LZ or LZNI; B, combination L. plantarum/nisin producer 102:102 CFU mL−1; NB, combination B with 10 IU/mL of nisin added; A, combination L. plantarum/nisin producer 102:104 CFU mL−1; NA, combinations A with 10 IU/mL of nisin added. Numerals above histograms represent the average diameter of the inhibition haloes formed by 10 μL aliquots of the co-cultures.
The amounts of free amino groups in the cultures ranged between 50 mm and 115 mm, according to a calibration curve obtained by plotting the OD at 420 nm of TNBS preparations obtained from known concentrations of leucine (Figure 4). The OD 420 nm data series of co-cultures with L. plantarum LZ or LZNI were statistically different at Day 1 (p = 0.01) but not later on.
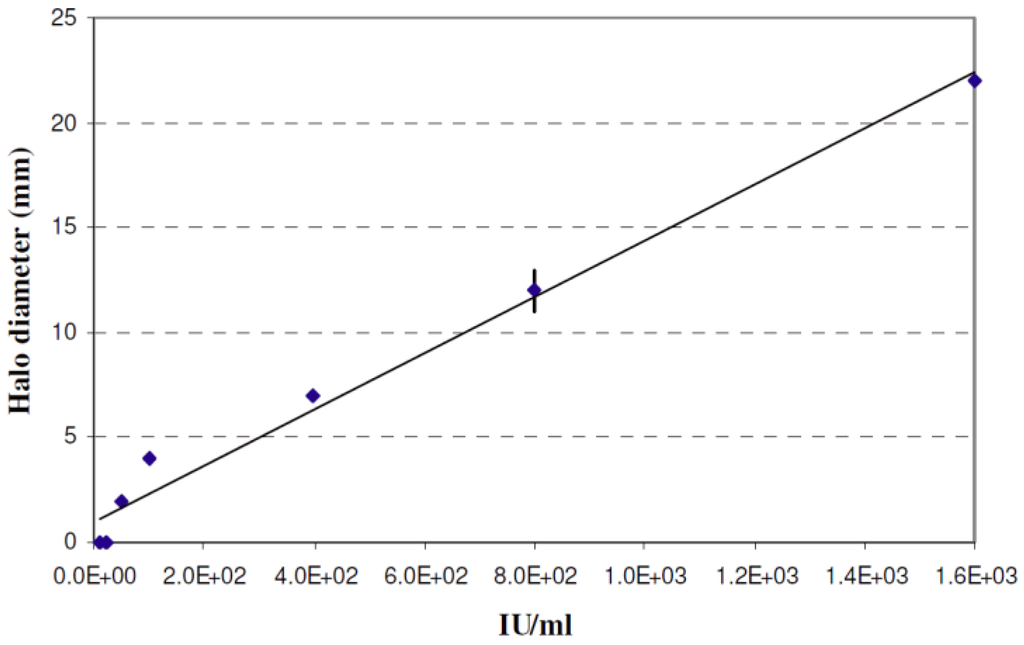
Figure 3.
Halo diameters (mm) formed by nisin suspensions of different concentration (IU mL−1) in lawns of L. plantarum LZ used as indicator.
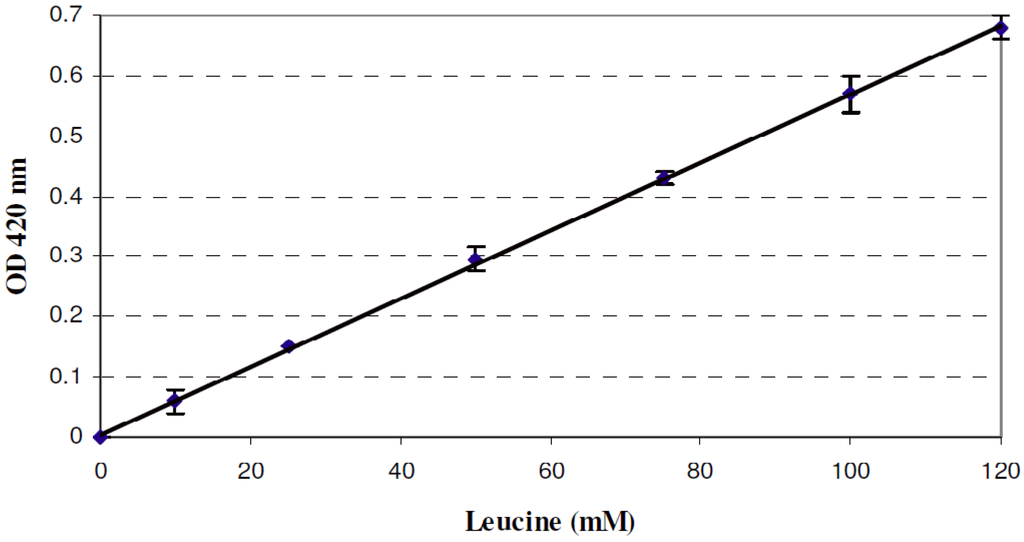
Figure 4.
Correlation between OD at 420 nm and leucine concentration determined by the TNBS method.
3.4. Reversed-Phase High Performance Liquid Chromatography (RP-HPLC) Analysis
The Reversed-Phase High Performance Liquid Chromatography (RP-HPLC) profiles of the water soluble nitrogen (WSN) and of nitrogen soluble at pH 4.4 of pure cultures of L. lactis subsp. lactis DSM 20972 and of its co-cultures B and NA with L. plantarum LZ and LZNI are shown in Figure 5. It appeared that main peaks eluting at 35 and 45 min were found in all cultures and in the not inoculated medium. Very little variations were observed for the pure cultures of L. lactis subsp. lactis DSM 20972 at different times, thus indicating a low peptidolytic activity of this strain. The WSN profiles varied mostly for the L. plantarum LZ co-cultures, showing the increase of a group of peaks eluting at about 20 min at Day 21 that were later degraded. At 28 days, the cultures NA of L. plantarum LZ showed higher peaks compared to the L. plantarum LZNI co-cultures, thus demonstrating the presence of higher amounts of some peptide hydrolysis products. On the other hand, the co-cultures with L. plantarum LZNI presented a high, early eluting peak at Day 8 and hydrophobic peptides eluting after 45 min. Peaks corresponding to hydrophobic peptides were also evident in the pure cultures of L. lactis subsp. lactis DSM 20729, but were not observed in the co-cultures with L. plantarum LZ, thus indicating their degradation.
The profiles of nitrogen soluble at pH 4.4 were very similar for all cultures and inoculum ratios. The main changes observed were the formation of hydrophobic peaks in the profiles of the L. plantarum LZ co-culture B at eight and 21 days.
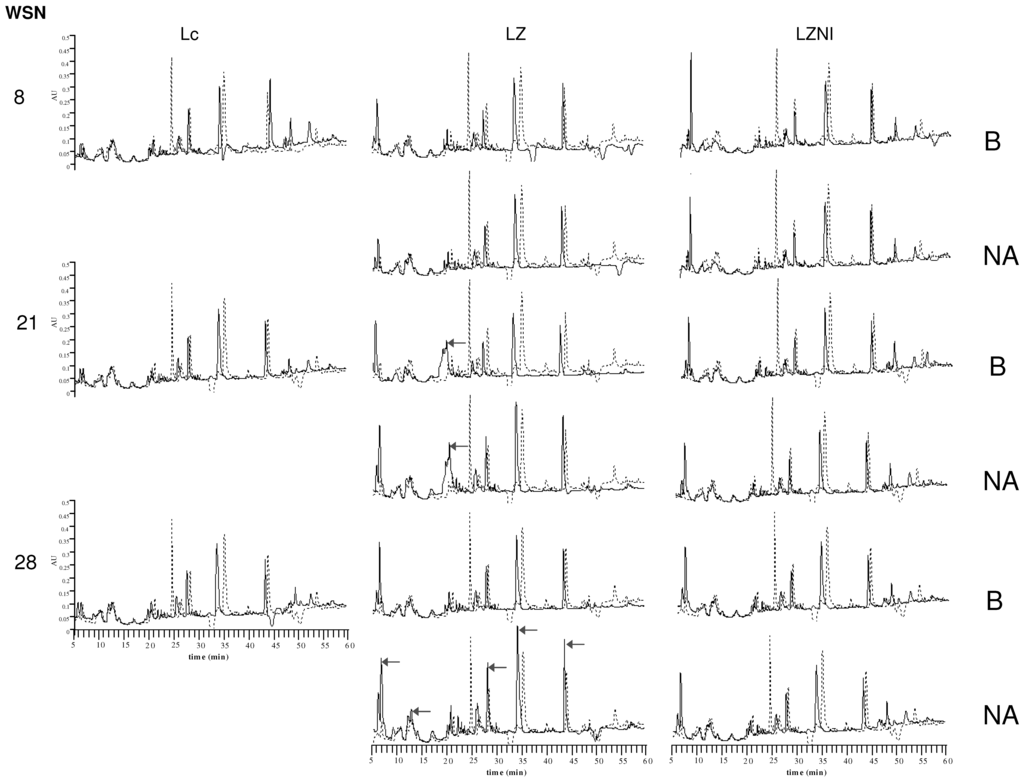
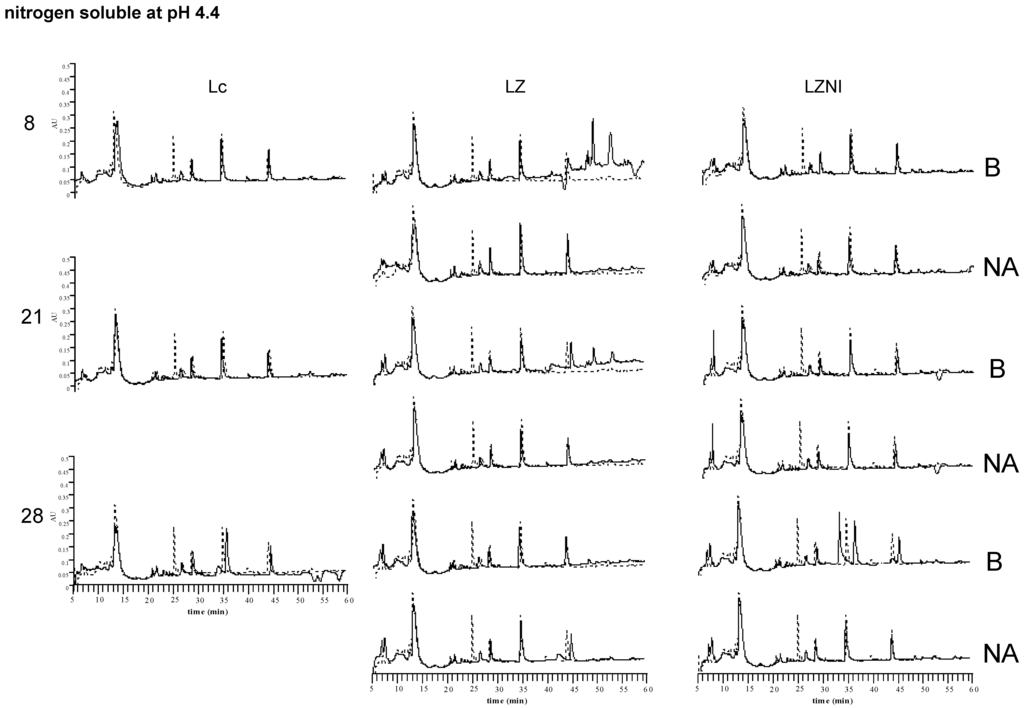
Figure 5.
RP-HPLC profiles of Water Soluble Nitrogen (WSN) and pH 4.4 soluble nitrogen in co-cultures of L. plantarum LZ or LZNI with the nisinogenic L. lactis subsp. lactis DSM 20729 (Lc). The time (days) of analysis is indicated on the left. Arrows indicate the peaks that better discriminated between co-cultures of L. plantarum LZ and L. plantarum LZNI. The dashed profile is that of the not inoculated skim milk. B, combination L. plantarum/nisin producer 102:102 CFU mL−1; NA, combination L. plantarum/nisin producer 102:104 CFU mL−1 with 10 IU/mL of nisin added.
4. Discussion
Use of low inoculums of a bacteriocin producer was tested in this study to avoid a strong impact on the development and metabolic activity of the NSLAB strains. This condition better satisfies the need of not altering the microbial complexity in traditional cheeses. Moreover, it could prevent the formation of unwanted sensory notes, like bitterness, that can arise when high levels of a bacteriocinogenic culture are added [20]. The initial level of the NSLAB strain was set to values close to those most likely found in raw milk for cheese manufacturing.
The comparison between isogenic variants of L. plantarum LZ, differing only for the level of immunity to nisin, was a distinctive aspect of this study that permitted an insight in the extent of growth inhibition and modification of the peptide degrading activity of NSLAB at the single strain level.
Results showed that a strong inhibition of the nisin-sensitive L. plantarum only occurred in the earliest phase of the experiment of co-cultivation with the nisin producer, while this strain reached and maintained high numbers during the remaining incubation time. This finding suggests that the application of initial low levels of a nisin producer in cheese would not cause the inhibition of NSLAB populations, as a consequence of the relatively low activity of nisin found in the cultures, notwithstanding the high numbers reached by the producer strain. Probably, the effect of stimulation of nisin production observed in the co-cultures NB, A and NA was not due to the leakage of compounds that stimulated the growth of L. lactis subsp. lactis DSM 20729 from damaged cells of L. plantarum, since the numbers of the nisin producer were comparable in all the co-cultures, at least at Day 1. This aspect should be better elucidated in order to potentiate the antimicrobial effect of the bacteriocinogenic culture in cheese. Though the amount of nisin formed appeared to be rather low, it would be opportune to test the behavior of nisin sensitive foodborne pathogens or spoiling agents in the same conditions. Indeed, according to available literature data, strains of Bacillus spp., Clostridium spp., Listeria spp. and Staphylococcus spp., among the microbial contaminants of relevance for dairy products, have relatively low MIC values for nisin [21,22]. Moreover, nisin concentrations of the order of 102 IU mL−1 were bactericidal or bacteriostatic for L. monocytogenes [23] and reduced the counts of Staphylococcus aureus in curd and whey for Minas Frescal cheese made from highly contaminated milk, keeping this pathogen at levels lower than the control also during storage at 4 °C [24]. Therefore, the amount of nisin formed already at Day 4 in the A and NA co-cultures and also at Day 8 in the NB co-cultures of the nisin-sensitive L. plantarum LZ might be effective against the pathogens mentioned above.
Nisin activity fluctuated during incubation, possibly because it was degraded and produced again later. The possibility of enzymatic degradation of nisin by bacterial proteases was demonstrated in a study with ruminal mixed cultures [25]. The evidences of its new formation during incubation indicated that using a producing culture instead of preparations of the bacteriocin can ensure the maintenance of the antimicrobial effect of the bacteriocin for longer time in a fermented food system.
The differences in the peptide profiles between cultures of L. plantarum LZ and LZNI indicated that the formation of peptides that might influence flavor by the nisin sensitive NSLAB strain was enhanced by co-cultivation with the nisin producer, though only small effects on its growth capacity were observed.
5. Conclusions
In conclusion, the results of this study suggested that use of low initial levels of bacteriocinogenic cultures should be evaluated for its effect on safety of traditional cheeses made from raw milk. Moreover, trials in a traditional cheese might also elucidate the effects of adding a nisinogenic culture on the sensorial profile and on the nature of flavor-active peptides that are formed.
Author Contributions
Franca Rossi and Gianluca Veneri equally contributed to the research plan, execution and writing of the manuscript.
Conflicts of Interest
The authors declare that no conflict of interest regards this study.
References
- Rossi, F.; Gatto, V.; Sabattini, G.; Torriani, S. An assessment of factors characterising the microbiology of Grana Trentino cheese, a Grana-type cheese. Int. J. Dairy Technol. 2012, 65, 401–409. [Google Scholar] [CrossRef]
- Licitra, G.; Carpino, S. The microfloras and sensory profiles of selected protected designation of origin Italian cheeses. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef]
- Aydemir, O.; Harth, H.; Weckx, S.; Dervişoğlu, M.; De Vuyst, L. Microbial communities involved in Kaşar cheese ripening. Food Microbiol. 2015, 46, 587–595. [Google Scholar] [CrossRef] [PubMed]
- De Pasquale, I.; Calasso, M.; Mancini, L.; Ercolini, D.; La Storia, A.; De Angelis, M.; Di Cagno, R.; Gobbetti, M. Causal relationship between microbial ecology dynamics and proteolysis during manufacture and ripening of protected designation of origin (PDO) cheese Canestrato Pugliese. Appl. Environ. Microbiol. 2014, 80, 4085–4094. [Google Scholar] [CrossRef] [PubMed]
- Amarita, F.; Requena, T.; Taborda, G.; Amigo, L.; Pelaez, C. Lactobacillus casei and Lactobacillus plantarum initiate catabolism of methionine by transamination. J. Appl. Microbiol. 2001, 90, 971–978. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Poli, A.; Guglielmini, E.; Sembeni, S.; Spiazzi, M.; Dellaglio, F.; Rossi, F.; Torriani, S. Detection of Staphylococcus aureus and enterotoxin genotype diversity in Monte Veronese, a protected designation of origin Italian cheese. Lett. Appl. Microbiol. 2007, 45, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Garde, S.; Tomillo, J.; Gaya, P.; Medina, M.; Nuñez, M. Proteolysis in Hispánico cheese manufactured using a mesophilic starter, a thermophilic starter and bacteriocin-producing Lactococcus lactis subsp. lactis INIA 415 adjunct culture. J. Agric. Food Chem. 2002, 50, 3479–3485. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cuesta, M.C.; Requena, T.; Peláez, C. Use of a bacteriocin-producing transconjugant as starter in acceleration of cheese ripening. Int. J. Food Microbiol. 2001, 70, 79–88. [Google Scholar] [CrossRef]
- Ryan, M.P.; Rea, M.C.; Hill, C.; Ross, R.P. An application in Cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl. Environ. Microbiol. 1996, 62, 612–619. [Google Scholar] [PubMed]
- Ryan, M.P.; Ross, R.P.; Hill, C. Strategy for manipulation of cheese flora using combinations of lacticin-producing and -resistant cultures. Appl. Environ. Microbiol. 2001, 67, 2699–2704. [Google Scholar] [CrossRef] [PubMed][Green Version]
- European Parliament and Council Directive No. 95/2/EC on Food Additives other than Colours and Sweeteners. Available online: http://ec.europa.eu/food/fs/sfp/addit_flavor/flav11_en.pdf (accessed on 14 February 2016).
- EFSA Panel on Biological Hazards (BIOHAZ). Scientific opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2013 update). EFSA J. 2013, 11, 3449–3555. [Google Scholar]
- Rilla, N.; Martínez, B.; Delgado, T.; Rodríguez, A. Inhibition of Clostridium tyrobutyricum in Vidiago cheese by Lactococcus lactis ssp. lactis IPLA 729, a nisin Z producer. Int. J. Food Microbiol. 2003, 85, 23–33. [Google Scholar] [CrossRef]
- Perin, L.M.; Dal Bello, B.; Belviso, S.; Zeppa, G.; Carvalho, A.F.; Cocolin, L.; Nero, L.A. Microbiota of Minas cheese as influenced by the nisin producer Lactococcus lactis subsp. lactis GLc05. Int. J. Food Microbiol. 2015, 214, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Capodaglio, A.; Dellaglio, F. Genetic modification of Lactobacillus plantarum by heterologous gene integration in a not functional region of the chromosome. Appl. Microbiol. Biotechnol. 2008, 80, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Kuipers, O.P.; Beerthuyzen, M.M.; De Ruyter, P.G.; Luesink, E.J.; De Vos, W.M. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 1995, 270, 27299–27304. [Google Scholar] [CrossRef] [PubMed]
- Ardö, Y.; Polychroniadou, A. Laboratory Manual for Chemical Analysis of Cheese; COST: Brussels, Belgium, 1999; pp. 35–36. [Google Scholar]
- Polychroniadou, A. A simple procedure using trinitrobenzenesulphonic acid for monitoring proteolysis in cheese. J. Dairy Res. 1988, 55, 585–596. [Google Scholar] [CrossRef]
- Ardö, Y.; Gripon, J.C. Comparative study of peptidolysis in some semi-hard round-eyed cheese varieties with different fat contents. J. Dairy Res. 1995, 62, 543–547. [Google Scholar] [CrossRef]
- Benech, R.-O.; Kheadr, E.E.; Lacroix, C.; Fliss, I. Impact of nisin producing culture and liposome-encapsulated nisin on ripening of Lactobacillus added-Cheddar cheese. J. Dairy Sci. 2003, 86, 1895–1909. [Google Scholar] [CrossRef]
- Mota-Meira, M.; LaPointe, G.; Lacroix, C.; Lavoie, M.C. MICs of mutacin B-Ny266, nisin A, vancomycin, and oxacillin against bacterial pathogens. Antimicrob. Agents Chemother. 2000, 44, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Avila, M.; Gómez-Torres, N.; Hernández, M.; Garde, S. Inhibitory activity of reuterin, nisin, lysozyme and nitrite against vegetative cells and spores of dairy-related Clostridium species. Int. J. Food Microbiol. 2014, 172, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, M.P.; Daroit, D.J.; Brandelli, A. Inhibition of Listeria monocytogenes in Minas Frescal cheese by free and nanovesicle-encapsulated nisin. Braz. J. Microbiol. 2012, 43, 1414–1418. [Google Scholar]
- Felicio, B.A.; Pinto, M.S.; Oliveira, F.S.; Lempk, M.W.; Pires, A.C.; Lelis, C.A. Effects of nisin on Staphylococcus aureus count and physicochemical properties of Minas Frescal cheese. J. Dairy Sci. 2015, 98, 4364–4369. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.S.; Mantovani, H.C.; Russell, J.B. The binding and degradation of nisin by mixed ruminal bacteria. FEMS Microbiol. Ecol. 2002, 42, 339–345. [Google Scholar] [CrossRef]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).