Comparative Analysis of Genetic Structure and Diversity in Five Populations of Yellowtail Kingfish (Seriola aureovittata)
Abstract
1. Introduction
2. Materials and Methods
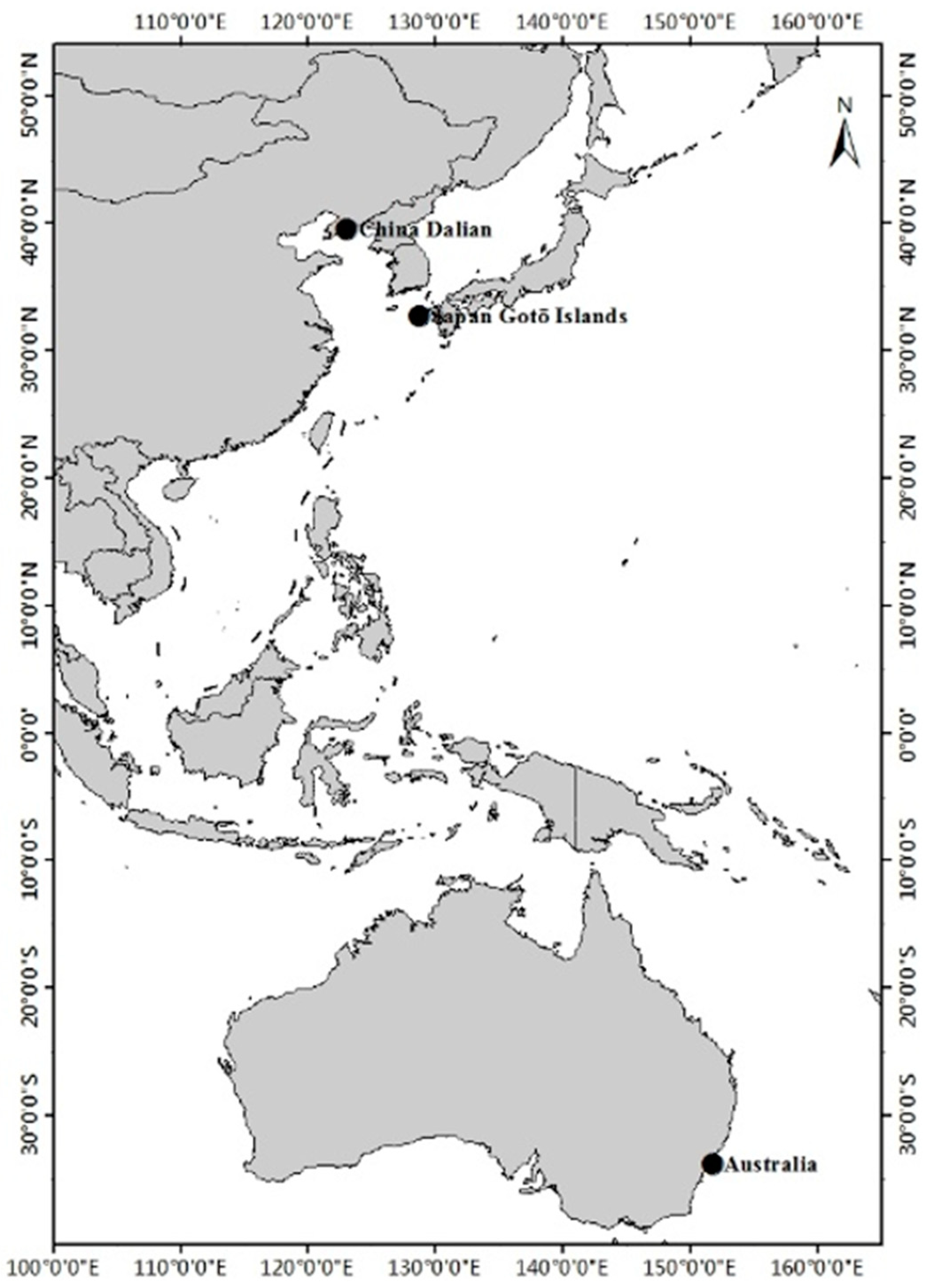
2.1. Fish Sampling
2.2. DNA Extraction, SNP Library Construction, and SNP Marker Typing
2.3. Population Genetic Diversity
2.4. Population Genetic Structure
3. Results
3.1. Yellowtail Kingfish Population Genetics
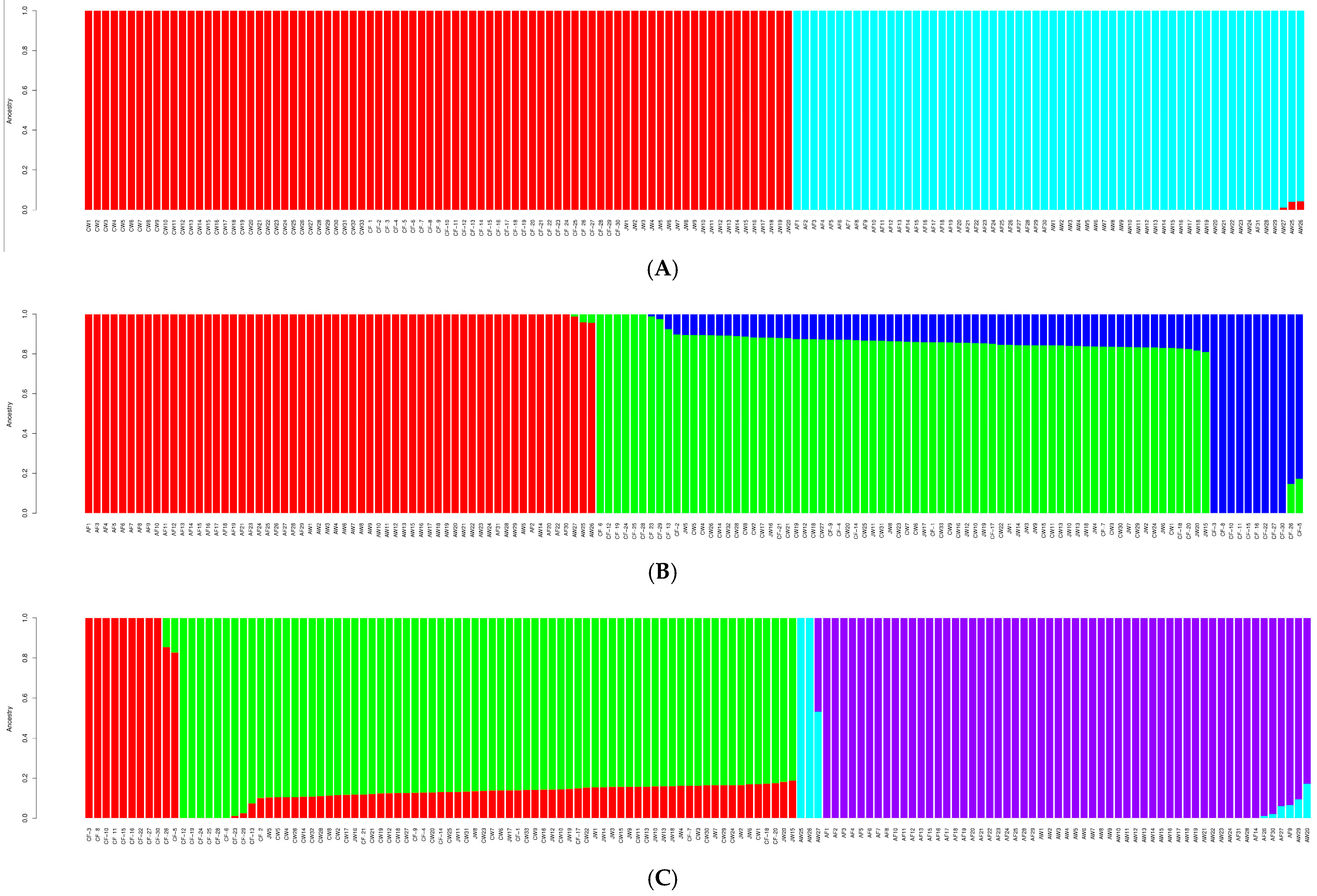
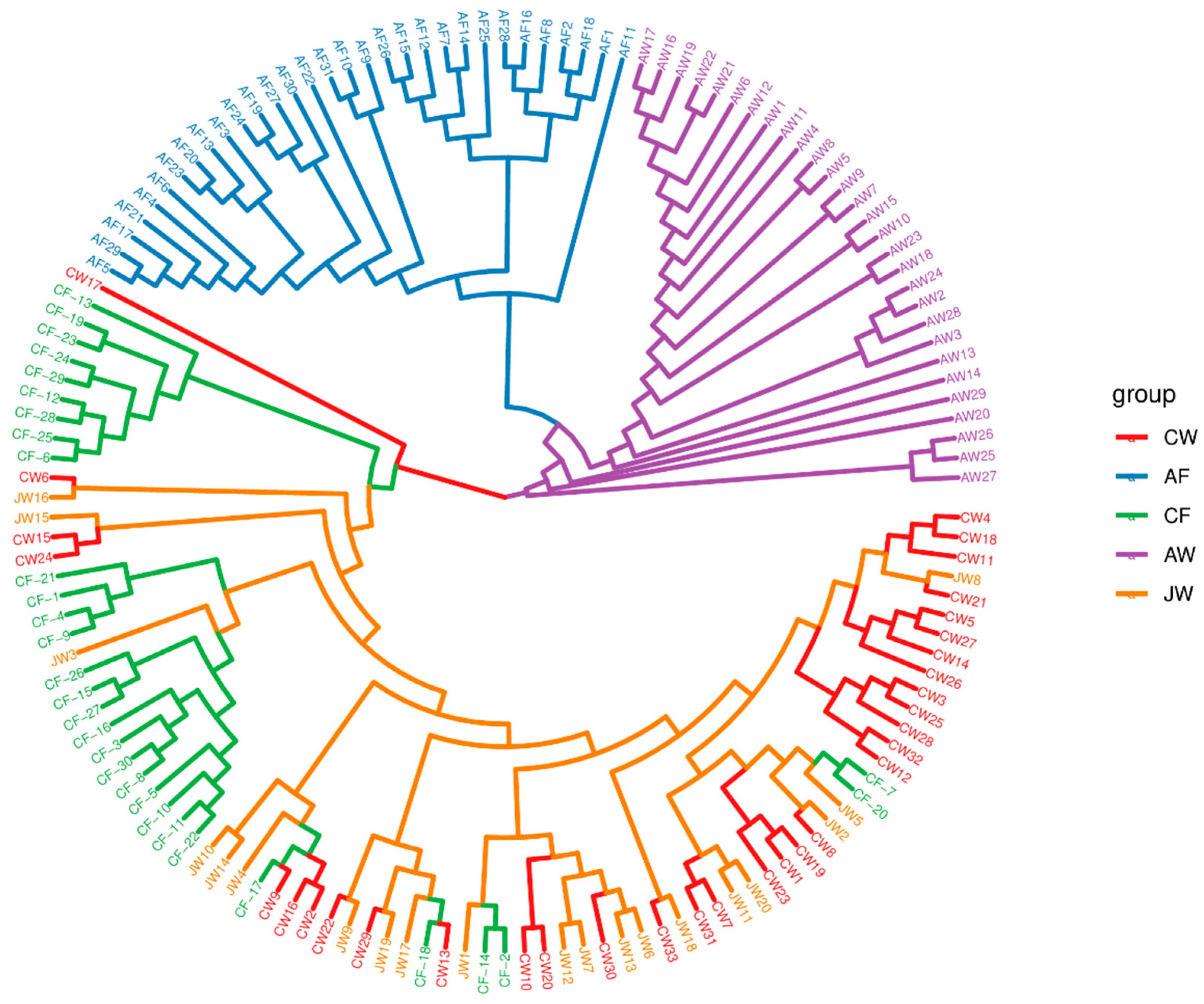
3.2. Yellowtail Kingfish Population Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sicuro, B.; Luzzana, U. The state of Seriola spp. other than yellowtail (S. quinqueradiata) farming in the world. Rev. Fish Sci. Aquac. 2016, 24, 314–325. [Google Scholar] [CrossRef]
- Liu, X.; Xu, Y.; Li, R.; Lu, Y.; Shi, B.; Ning, J.; Wang, B. Analysis and evaluation of nutritional composition of the muscle of yellowtail kingfish (Seriola aureovittata). Prog. Fish. Sci. 2017, 1, 128–135. [Google Scholar]
- Whatmore, P.; Nguyen, N.H.; Miller, A.; Lamont, R.; Powell, D.; D’Antignana, T.; Bubner, E.; Elizur, A.; Knibb, W. Genetic parameters for economically important traits in yellowtail kingfish seriola lalandi. Aquaculture 2013, 400–401, 77–84. [Google Scholar] [CrossRef]
- Zhang, C. Investigation Report on Fishes in the Yellow Sea and Bohai Sea; Science Press: Beijing, China, 1995; pp. 116–119. [Google Scholar]
- Liu, J.; Chen, Y.; Ma, L. Fish Atlas of the Yellow Sea and Bohai Sea; Science Press: Beijing, China, 2015; pp. 171–173. [Google Scholar]
- Liu, X. Great breakthrough has been made in breeding technology of yellowtail kingfish. Ocean. Fish. 2017, 8, 19. [Google Scholar]
- Premachandra, H.K.A.; La Cruz, L.D.; Takeuchi, Y.; Miller, A.; Fielder, S.; O’Connor, W. Genomic DNA variation confirmed seriola lalandi comprises three different populations in the pacific, but with recent divergence. Sci. Rep. 2017, 7, 9386. [Google Scholar] [CrossRef]
- Purcell, C.M.; Chabot, C.L.; Craig, M.T.; Martinez-Takeshita, N.; Allen, L.G. Developing a genetic baseline for the yellowtail amberjack species complex, Seriola lalandi sensu lato, to assess and preserve variation in wild populations of these globally important aquaculture species. Conserv. Genet. 2015, 16, 1475–1488. [Google Scholar] [CrossRef]
- Nugroho, E.; Ferrell, D.J.; Smith, P.; Taniguchi, N. Genetic divergence of kingfish from Japan, Australia and New Zealand inferred by microsatellite DNA and mitochondrial DNA control region markers. Fish Sci. 2001, 67, 843–850. [Google Scholar] [CrossRef]
- Martinez-Takeshita, N.; Purcell, C.M.; Chabot, C.L.; Craig, M.T.; Paterson, C.N.; Hyde, J.R.; Allen, L.G. A tale of three tails: Cryptic speciation in a globally distributed marine fish of the genus seriola. Copeia 2015, 103, 357–368. [Google Scholar] [CrossRef]
- Ai, Q.; Song, L.; Tan, H.; Huang, X.; Bao, B.; Li, C. Genetic and morphological differences between yellowtail kingfish (Seriola lalandi) from the Bohai Sea, China and the Southern Ocean, Australia. Aquac. Fish. 2021, 6, 260–266. [Google Scholar] [CrossRef]
- Shi, B.; Liu, Y.; Liu, X.; Xu, Y.; Li, R.; Song, X.; Zhou, L. Study on the karyotype of yellowtail kingfish (Seriola aureovittata). Prog. Fish. Sci. 2017, 1, 136–141. [Google Scholar]
- Liu, Y.; Liu, X.; Shi, B.; Xu, Y.; Li, R.; Lv, Y.; Song, X.; Wang, B.; Jiang, Y. Analysis of the banding patterns of Seriola aureovittata. J. Fish. China 2018, 9, 1338–1347. [Google Scholar]
- Shi, B.; Liu, X.; Liu, Y.; Zhang, Y.; Gao, Q.; Xu, Y.; Wang, B.; Jiang, Y.; Song, X. Complete sequence and gene organization of the mitochondrial genome of Seriola aureovittata. J. Fish. Sci. China 2019, 26, 405–415. [Google Scholar] [CrossRef]
- Cui, A.; Wang, B.; Jiang, Y.; Liu, X.; Xu, Y. Development of SNP markers for yellowtail kingfish (Seriola lalandi) by 2b-RAD simplified genome sequencing. Conserv. Genet. Resour. 2020, 12, 403–407. [Google Scholar] [CrossRef]
- Wang, S.; Meyer, E.; Mckay, J.K.; Matz, M.V. 2b-rad: A simple and flexible method for genome-wide genotyping. Nat. Methods 2012, 9, 808–810. [Google Scholar] [CrossRef]
- Pecoraro, C.; Babbucci, M.; Villamor, A.; Franch, R.; Papetti, C.; Leroy, B.; Cariani, A. Methodological assessment of 2b-RAD genotyping technique for population structure inferences in yellowfin tuna (Thunnus albacares). Mar. Genom. 2016, 25, 43–48. [Google Scholar] [CrossRef]
- Dou, J.; Li, X.; Fu, Q.; Jiao, W.; Li, Y.; Li, T. Evaluation of the 2b-rad method for genomic selection in scallop breeding. Sci. Rep. 2016, 6, 19244. [Google Scholar] [CrossRef]
- Yi, S.; Li, Y.; Shi, L.; Zhang, L.; Li, Q.; Chen, J. Characterization of population genetic structure of red swamp crayfish, Procambarus clarkii, in China. Sci. Rep. 2018, 8, 5586. [Google Scholar] [CrossRef]
- Manuzzi, A.; Zane, L.; Muñoz-Merida, A.; Griffiths, A.M.; Veríssimo, A. Population genomics and phylogeography of a benthic coastal shark (Scyliorhinus canicula) using 2b-RAD single nucleotide polymorphisms. Biol. J. Linn. Soc. 2018, 126, 289–303. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2013, 30, 614–620. [Google Scholar] [CrossRef]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, Y.; Kristiansen, K. SOAP: Short oligonucleotide alignment program. Bioinformatic 2008, 24, 713–714. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Dou, J.; Mao, J.; Su, H.; Jiao, W.; Zhang, L.; Hu, X.; Huang, X.; Wang, S.; Bao, Z. RADtyping: An integrated package for accurate de novo codominant and dominant RAD genotyping in mapping populations. PLoS ONE 2013, 8, e79960. [Google Scholar] [CrossRef] [PubMed]
- Rousset, F. GENEPOP‘ 007: A complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Toddbrown, K. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Chakraborty, R.; Weiss, K.M. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc. Natl. Acad. Sci. USA 1988, 85, 9119–9123. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Cam, T.L.; Shashaank, V.; Purcell, S.M.; Lee, J.J. Second-generation plink: Rising to the challenge of larger and richer datasets. Giga Sci. 2015, 4, 7. [Google Scholar] [CrossRef]
- Vilella, A.; Severin, J.; Ureta-Vidal, A.; Heng, L.; Durbin, R.; Birney, E. Ensembl Compara Gene Trees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009, 19, 327–335. [Google Scholar] [CrossRef]
- Gillanders, B.M.; Ferrell, D.J.; Andrew, N.L. Estimates of movement and life-history parameters of yellowtail kingfish (seriola lalandi): How useful are data from a cooperative tagging programme? Mar. Freshw. Res. 2001, 52, 179–192. [Google Scholar] [CrossRef]
- Miller, P.A.; Fitch, A.J.; Gardner, M.; Hutson, K.S.; Mair, G. Genetic population structure of yellowtail kingfish (Seriola lalandi) in temperate Australasian waters inferred from microsatellite markers and mitochondrial DNA. Aquaculture 2011, 319, 328–336. [Google Scholar] [CrossRef]
- Wolvaardt, C. Initiation of Company Coverage—IPO, Western Kingfish Ltd.; State One Stockbroking Ltd.: Sydney, Australia, 2007; pp. 1–9. [Google Scholar]
- Poulin, E.; Cardenas, L.; Hernandez, C.E.; Kornfield, I.; Ojeda, F.P. Resolution of the taxonomic status of Chilean and Californian jack mackerels using mitochondrial DNA sequence. J. Fish Biol. 2004, 65, 1160–1164. [Google Scholar] [CrossRef]
- Díaz-Jaimes, P.; Uribe-Alcocer, M.; Rocha-Olivares, A.; García-de-León, F.J.; Nortmoon, P.; Durand, J.D. Global phylogeography of the dolphinfish (coryphaena hippurus): The influence of large effective population size and recent dispersal on the divergence of a marine pelagic cosmopolitan species. Mol. Phylogenet Evol. 2010, 57, 1209–1218. [Google Scholar] [CrossRef]
- Sepúlveda, F.A. and González, M.T. Spatio-temporal patterns of genetic variations in populations of yellowtail kingfish Seriola lalandi from the south-eastern Pacific Ocean and potential implications for its fishery management. J. Fish Biol. 2017, 90, 249–264. [Google Scholar] [CrossRef]
- Sala, E.; Aburtooropeza, O.; Paredes, G.; Thompson, G. Spawning aggregations and reproductive behavior of reef fishes in the gulf of California. B Mar. Sci. 2003, 72, 103–121. [Google Scholar]
- Erisman, B.; Mascarenas, I.; Paredes, G.; Mitcheson, Y.S.D.; Aburto-Oropeza, O.; Hastings, P. Seasonal, annual, and long-term trends in commercial fisheries for aggregating reef fishes in the gulf of California, Mexico. Fish Res. 2010, 106, 279–288. [Google Scholar] [CrossRef]
- Poortenaar, C.; Hooker, S.; Sharp, N. Assessment of yellowtail kingfish (Seriola lalandi) reproductive physiology, as a basis for aquaculture development. Aquaculture 2001, 201, 271–286. [Google Scholar] [CrossRef]
- Iguchi, J.; Takashima, Y.; Namikoshi, A.; Yamashita, M. Species identification method for marine products of seriola and related species. Fish Sci. 2012, 78, 197–206. [Google Scholar] [CrossRef]
- Moran, D.; Smith, C.K.; Gara, B.; Poortenaar, C.W. Reproductive behaviour and early development in yellowtail kingfish (Seriola lalandi Valenciennes 1833). Aquaculture 2007, 262, 95–104. [Google Scholar] [CrossRef]
- Moran, D.; Gara, B.; Wells, R.M.G. Energetics and metabolism of yellowtail kingfish (Seriola lalandi Valenciennes 1833) during embryogenesis. Aquaculture 2007, 265, 359–369. [Google Scholar] [CrossRef]
- National Oceanic and Atmospheric Administration. NOAA Optimum Interpolation Sea Surface Temperature Analysis; NOAA/OAR/ESRL PSD (United States Department of Commerce): Boulder, CO, USA, 2011.
- Fernández, G.; Cichero, D.; Patel, A.; Martínez, V. Genetic structure of Chilean populations of Seriola lalandi for the diversification of the national aquaculture in the north of Chile. Lat. Am. J. Aquat. Res. 2015, 43, 374–379. [Google Scholar] [CrossRef]

| Collection | Ho | He | Pi | HW-P |
|---|---|---|---|---|
| All | 0.0824 | 0.2013 | 0.2020 | 0.4112 |
| JW | 0.0833 | 0.0814 | 0.0837 | 0.9239 |
| CW | 0.0718 | 0.0771 | 0.0785 | 0.8872 |
| CF | 0.0828 | 0.0814 | 0.0829 | 0.9093 |
| AW | 0.0902 | 0.0955 | 0.0974 | 0.8591 |
| AF | 0.0940 | 0.0927 | 0.0943 | 0.8996 |
| Collection | JW | CW | CF | AW | AF |
|---|---|---|---|---|---|
| JW | - | 0.0010 | 0.0168 | 1.2932 | 1.3205 |
| CW | 0.00097 | - | 0.0191 | 1.3409 | 1.3653 |
| CF | 0.01662 | 0.01888 | - | 1.3164 | 1.3417 |
| AW | 0.7256 | 0.7384 | 0.7319 | - | 0.0260 |
| AF | 0.7330 | 0.7447 | 0.7386 | 0.02569 | - |
| Collection | JW | CW | CF | AW | AF |
|---|---|---|---|---|---|
| JW | - | ||||
| CW | 256.4499 | - | |||
| CF | 14.7921 | 12.9287 | - | ||
| AW | 0.0953 | 0.0894 | 0.0923 | - | |
| AF | 0.0911 | 0.0864 | 0.0892 | 9.9002 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, A.; Xu, Y.; Kikuchi, K.; Jiang, Y.; Wang, B.; Koyama, T.; Liu, X. Comparative Analysis of Genetic Structure and Diversity in Five Populations of Yellowtail Kingfish (Seriola aureovittata). J. Mar. Sci. Eng. 2023, 11, 1583. https://doi.org/10.3390/jmse11081583
Cui A, Xu Y, Kikuchi K, Jiang Y, Wang B, Koyama T, Liu X. Comparative Analysis of Genetic Structure and Diversity in Five Populations of Yellowtail Kingfish (Seriola aureovittata). Journal of Marine Science and Engineering. 2023; 11(8):1583. https://doi.org/10.3390/jmse11081583
Chicago/Turabian StyleCui, Aijun, Yongjiang Xu, Kiyoshi Kikuchi, Yan Jiang, Bin Wang, Takashi Koyama, and Xuezhou Liu. 2023. "Comparative Analysis of Genetic Structure and Diversity in Five Populations of Yellowtail Kingfish (Seriola aureovittata)" Journal of Marine Science and Engineering 11, no. 8: 1583. https://doi.org/10.3390/jmse11081583
APA StyleCui, A., Xu, Y., Kikuchi, K., Jiang, Y., Wang, B., Koyama, T., & Liu, X. (2023). Comparative Analysis of Genetic Structure and Diversity in Five Populations of Yellowtail Kingfish (Seriola aureovittata). Journal of Marine Science and Engineering, 11(8), 1583. https://doi.org/10.3390/jmse11081583








