Regulation of 17α-Hydroxyprogesterone Production during Induced Oocyte Maturation and Ovulation in Amur Sturgeon (Acipenser schrenckii)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cloning of Amur Sturgeon hsd3b, cyp17a1 and cyp17a2 cDNA
2.3. Phylogenetic Analysis
2.4. Enzyme Activity Determined via Liquid Chromatography/Mass Spectrometry (LC/MS)
2.5. LC/MS
2.6. Tissue-Specific hsd3b and cyp17a1/2 mRNA Expression
2.7. DHP Measurement
2.8. Serum E2, T, and 11KT Measurement
2.9. In Vivo Experiment
2.10. Incubation of Ovarian Follicles
2.11. cDNAs Synthesis and Quantitative PCR
2.12. Statistical Analyses
3. Results
3.1. Isolation of hsd3b and cyp17a1/2 cDNA from Amur Sturgeon
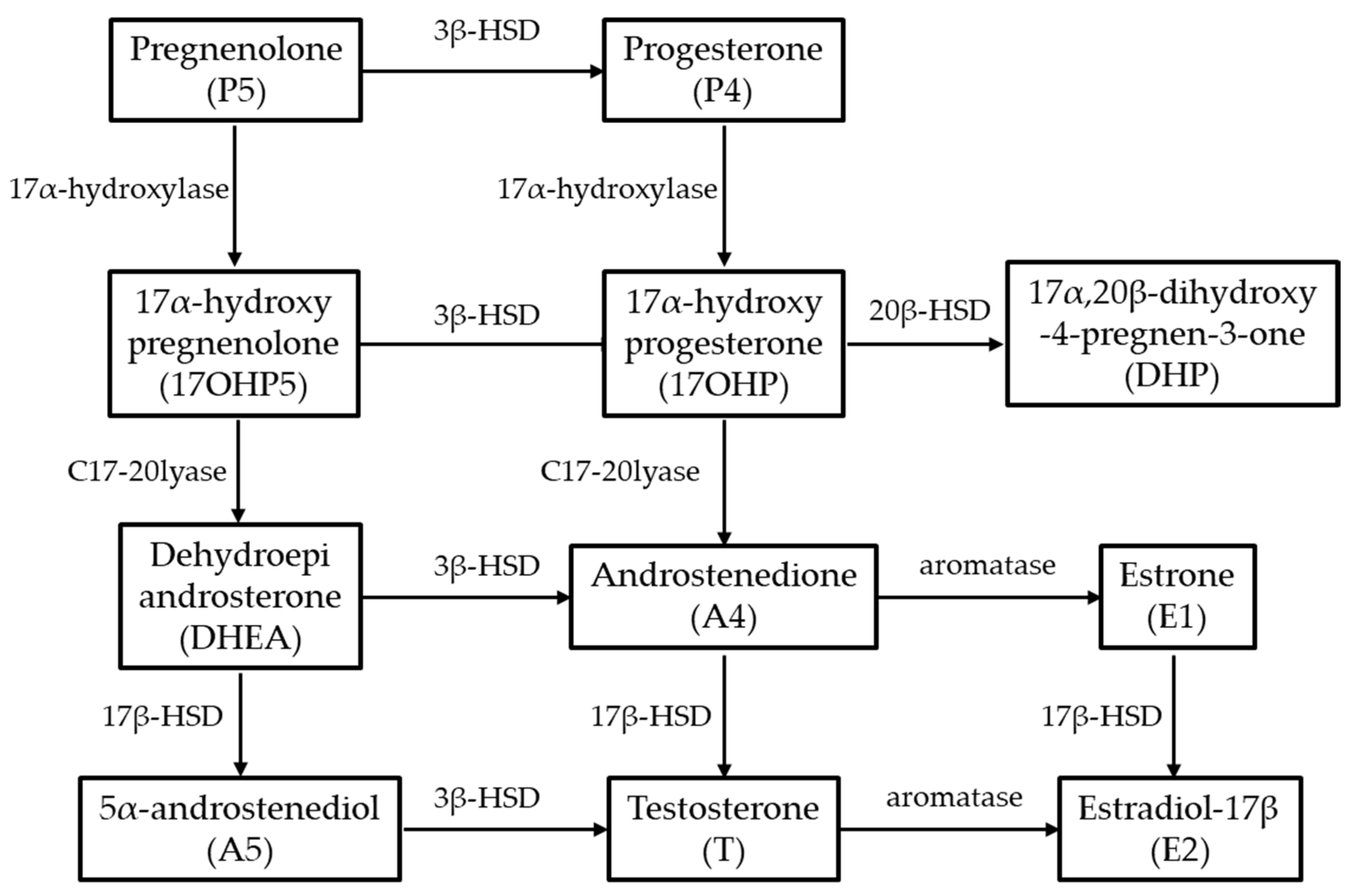
3.2. Enzyme Activity Determined via LC/MS Analysis
3.3. Tissue-Specific hsd3b and cyp17a1/2 mRNA Expression
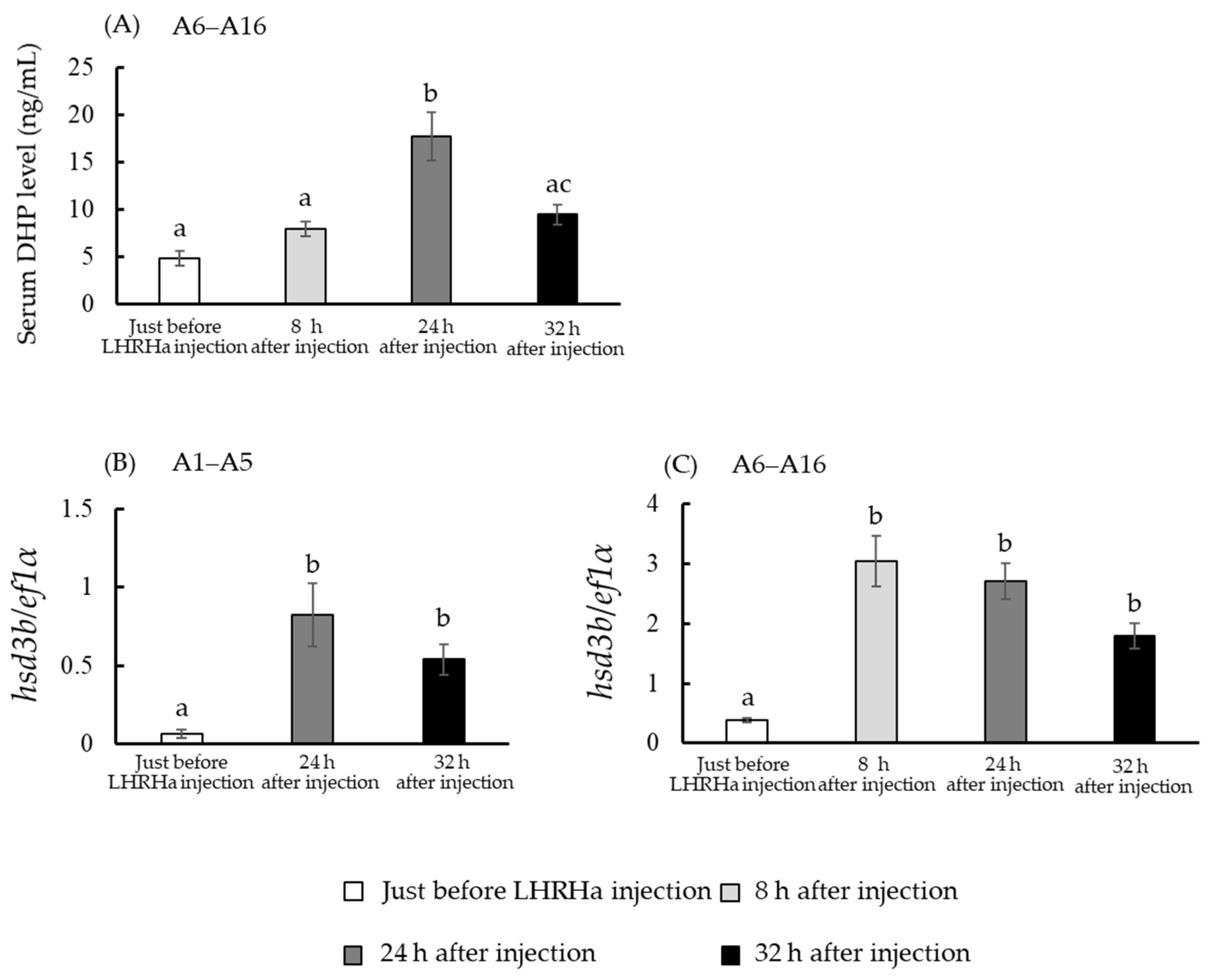
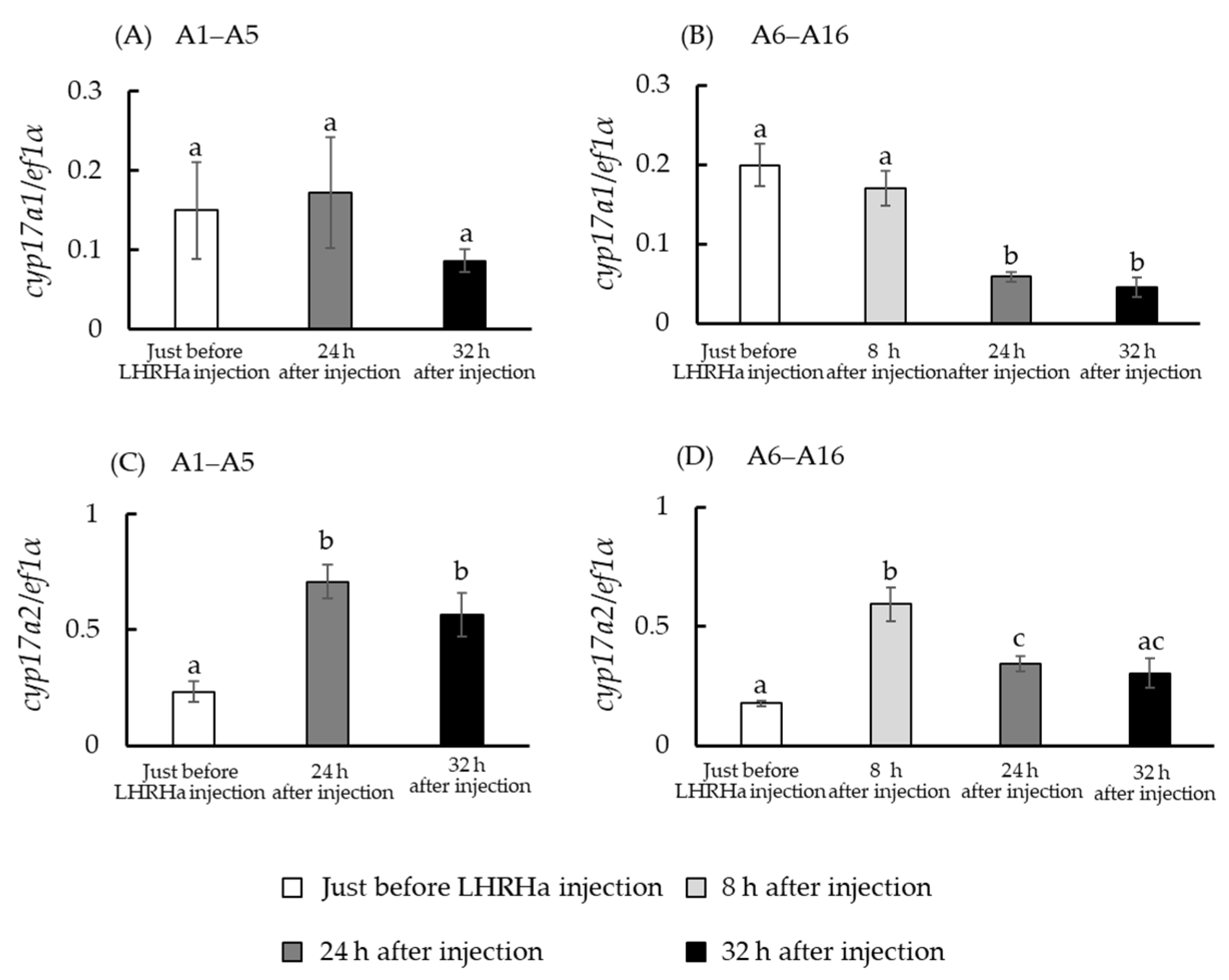
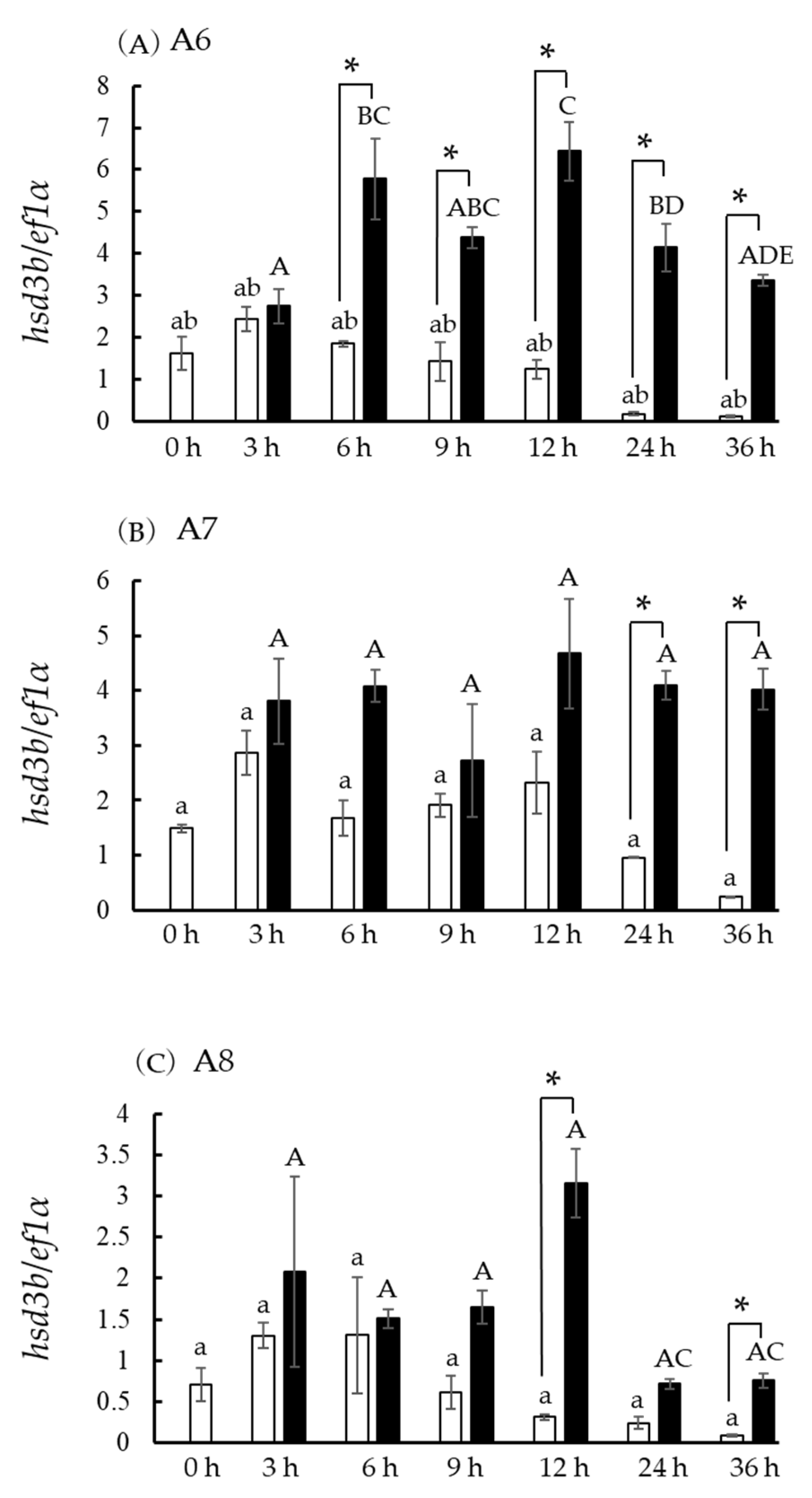
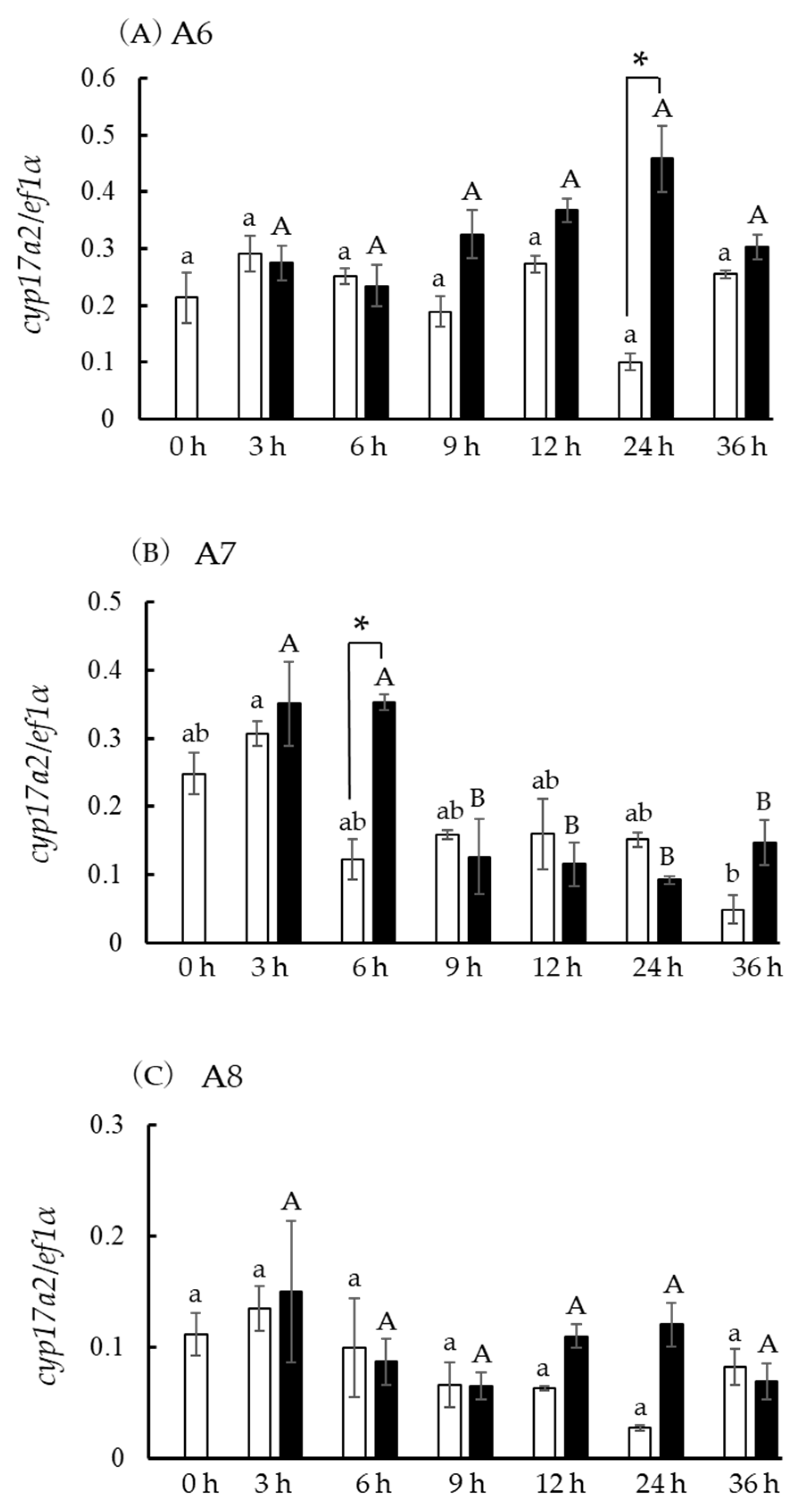
3.4. In Vivo hsd3b and cyp17a1/2 mRNA Levels
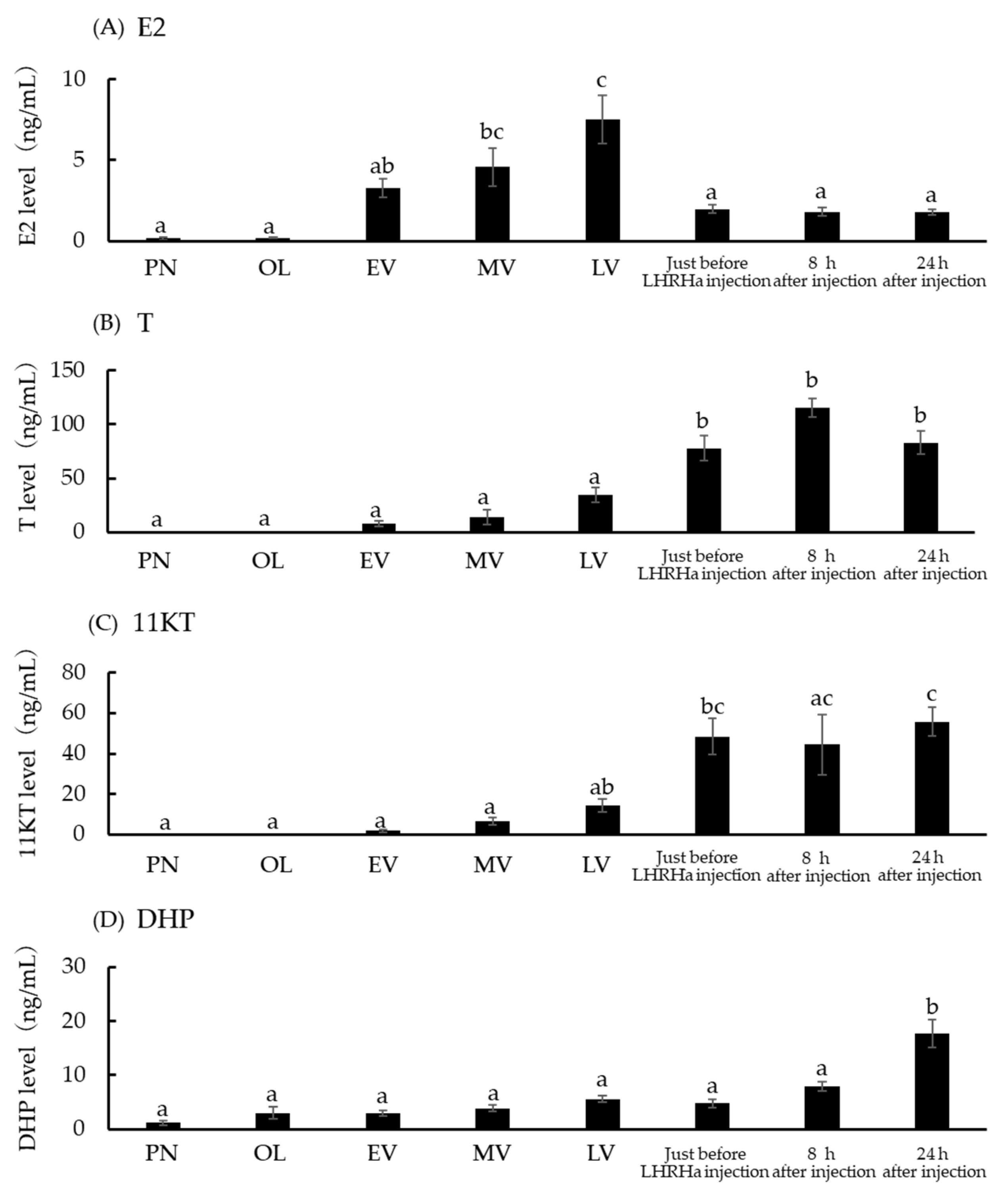
3.5. Serum E2, T, 11KT, and DHP Concentration at Five Ovarian Developmental Stages
3.6. Hsd3b and Cyp17a1/2 mRNA Levels in Ovaries at the Five Developmental Stages
3.7. DHP Concentration in Incubation Media
3.8. Hsd3b and cyp17a1/2 Expression in Incubated Ovarian Follicles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pikitch, E.K.; Doukakis, P.; Laucl, L.; Chakrabatory, P.; Erickson, D.L. Status, trends and management of sturgeon and paddlefish fisheries. Fish Fish. 2005, 6, 233–265. [Google Scholar] [CrossRef]
- Doukakis, P.; Pikitch, E.K.; Rothschil, A.; Desalle, R.; Amato, G.; Kolokotronis, S.O. Testing the effectivenedd of an international conservation agreement: Marketplace foren-sics and Cites caviar trade regulation. PLoS ONE 2012, 7, e40907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Y.; Valbo-Jørgensen, J. Effects of IUU fishing and stock enhancement on and restoration strategies for the stellate stur-geon fishery in the Caspian Sea. Fish. Res. 2012, 131, 21–29. [Google Scholar] [CrossRef]
- Fujii, K.; Hirose, K.; Hara, A.; Shiraishi, M.; Maruyama, T. Use of vitellogenine level as a maturational indicator for artificial spawning of cultured hybrid sturgeon, Huso huso × Acipenser ruthenus. In Acipenser; CEMAGREF Publication: Bordeaux, France, 1991; pp. 381–388. [Google Scholar]
- Omoto, N.; Maebayashi, M.; Adachi, S.; Arai, K.; Yamauchi, K. The influence of oocyte maturational stage on hatching and triploidy rates in hybrid (bester) sturgeon, Huso huso × Acipenser ruthenus. Aquaculture 2005, 245, 287–294. [Google Scholar] [CrossRef]
- Wallace, R.A. Vitellogenesis and oocyte growth in non-mammalian vertebrates. In Developmental Biology; Browder, L.W., Ed.; Plenum Publishing Corporation: New York, NY, USA, 1985; Chapter 3; Volume 1, pp. 127–177. [Google Scholar]
- Endo, T.; Todo, T.; Lokman, P.M.; Kudo, H.; Ijiri, S.; Adachi, S.; Yamauchi, K. Androgens and very low density lipoprotein are essential for the growth of previtellogenic oocytes from Japanese eel, Anguilla japonica, in vitro. Biol. Reprod. 2011, 84, 816–825. [Google Scholar] [CrossRef]
- Nagahama, Y. Gonadotropin action on gametogenesis and steroidogenesis in teleost gonad. Zool. Sci. 1987, 4, 209–222. [Google Scholar]
- Nagahama, Y.; Adachi, S. Identification of maturation-inducing steroid in a teleost, the amago salmon (Oncorhynchus rhodurus). Dev. Biol. 1985, 109, 428–435. [Google Scholar] [CrossRef]
- Nagahama, Y. Cytodifferentiation of ovarian follicle cells during oocyte growth and maturation. Cell. Differ. Dev. 1988, 25, 9–14. [Google Scholar] [CrossRef]
- Adachi, S.; Ijiri, S.; Kazeto, Y.; Yamauchi, Y. Oogenesis in the Japanese eel, Anguilla japonica. In Eel Biology; Aida, K., Tsukamoto, K., Yamauchi, K., Eds.; Springer: Tokyo, Japan, 2003; pp. 301–317. [Google Scholar]
- Simard, J.; Ricketts, M.L.; Gingras, S.; Soucy, P.; Feltus, F.A.; Melner, M.H. Molecular biology of the 3beta-hydroxysteroid dehydrogenase/delta5-delta4 isomerase gene family. Endocr. Rev. 2005, 26, 525–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.C.; Hu, S.; Ho, P.H.; Hsu, H.J.; Ostlethwait, J.H.; Chung, B. Two zebrafish hsd3b genes are distinct in function, expression, and evolution. Endocrinology 2015, 156, 2854–2862. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, I.; Evans, J.C.; Kusakabe, M.; Nagahama, Y.; Young, G. Changes in steroidogenic enzyme and steroidogenic acute regulatory protein messenger RNAs in ovarian follicles during ovarian development of rainbow trout (Oncorhynchus mykiss). Gen. Comp. Endocrinol. 2005, 144, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Guzmán, J.M.; Luckenbach, J.A.; Yamamoto, Y.; Swanson, P. Expression profiles of Fsh-regulated ovarian genes during oogenesis in coho salmon. PLoS ONE 2014, 9, e114176. [Google Scholar] [CrossRef] [Green Version]
- Nakajin, S.; Shinoda, M.; Haniu, M.; Shively, J.E.; Hall, P.F. C21 steroid side chain cleavage enzyme from porcineadrenal microsomes. J. Biol. Chem. 1984, 259, 3971–3976. [Google Scholar] [CrossRef]
- Zuber, M.X.; Simpson, E.R.; Waterman, M.R. Expression of bovine 17a-hydroxylase cytochrome P450 cDNA in non-steroidogenic (COS-1) cells. Science 1986, 234, 1258–1261. [Google Scholar] [CrossRef]
- Zhou, L.-Y.; Wang, D.-S.; Kobayashi, T.; Yano, A.; Paul-Prasanth, B.; Suzuki, A.; Sakai, F.; Nagahama, Y. A novel type of P450c17 lacking the lyase activity is responsible for C21-steroid biosynthesis in the fish ovary and head kidney. Endocrinology 2007, 148, 4282–4291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.-Y.; Wang, D.-S.; Shibata, Y.; Paul-Prasanth, B.; Suzuki, A.; Nagahama, Y. Characterization, expression and transcriptional regulation of P450c17-Iand-II in the medaka, Oryzias latipes. Biochem. Biophys. Res. Commun. 2007, 362, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Kazeto, Y.; Ijiri, S.; Todo, T.; Adachi, S.; Yamauchi, K. Molecular cloning and characterization of Japanese eel ovarian P450c17 (CYP17) cDNA. Gen. Comp. Endocrinol. 2000, 18, 123–133. [Google Scholar] [CrossRef]
- Su, T.; Ijiri, S.; Kanbara, H.; Hagihara, S.; Wang, D.S.; Adachi, S. Characterization and expression of cDNAs encoding P450c17-II (cyp17a2) in Japanese eel during induced ovarian development. Gen. Comp. Endocrinol. 2015, 221, 134–143. [Google Scholar] [CrossRef]
- Hasegawa, Y.; Ijiri, S.; Surugaya, R.; Sakai, R.; Adachi, S. 17β-Hydroxysteroid dehydrogenase type 12 is associated with maturation-inducing steroid synthesis during induced oocyte maturation and ovulation in sturgeons. Aquaculture 2022, 546, 15. [Google Scholar] [CrossRef]
- Amiri, B.; Maebayashi, M.; Hara, A.; Adachi, S.; Yamauchi, K. Ovarian development and serum sex steroid and vitellogenin profiles in the female cultured sturgeon hybrid, the bester. J. Fish Biol. 1996, 48, 1164–1178. [Google Scholar] [CrossRef]
- Kagawa, H.; Takano, K.; Nagahama, Y. Correlation of plasma estradiol-17β and progesterone levels with ultrastructure and histochemistry of ovarian follicles in the white-spotted char, Salvelinus leucomaenis. Cell. Tissue Res. 1981, 218, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Yamada, H.; Satoh, R.; Ogoh, M.; Takaji, K.; Fujimoto, Y.; Hakuba, T.; Chiba, H.; Kambegawa, A.; Iwata, M. Circadian changes in serum concentrations of steroids in Japanese char Salvelinus leucomaenis at the stage of final maturation. Zool. Sci. 2002, 19, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Ono, H.; Iwasaki, M.; Sakamoto, N.; Mizuno, S. cDNA cloning and sequence analysis of a chicken gene expressed during the gonadal development and homologous to mammalian cytochrome P-450c17. Gene 1988, 66, 77–85. [Google Scholar] [PubMed]
- Wang, Y.; GE, W. Cloning of zebrafish ovarian P450c17 (CYP17, 17α-hydroxylase/17, 20-lyase) and characterization of its expression in gonadal and extra-gonadal tissues. Gen. Comp. Endocrinol. 2004, 135, 241–249. [Google Scholar] [CrossRef]
- Ozols, J.; Heinemann, F.S.; Johnson, E.F. Amino acid sequence of an analogous peptide from two forms of cytochrome P-450. J. Biol. Chem. 1981, 256, 11405–11408. [Google Scholar] [CrossRef]
- Gotoh, O.; Tagashira, Y.; Iizuka, T.; Fujii-Kuriyama, Y. Structural characteristics of cytochrome P-450. Possible location of the heme-binding cysteine in determined amino-acid sequences. J. Biochem. 1983, 93, 807–817. [Google Scholar] [CrossRef]
- Miller, W.L. Minireview: Regulation of steroidogenesis by electron transfer. Endocrinology 2005, 146, 2544–2550. [Google Scholar] [CrossRef] [Green Version]
- Ijiri, S.; Shibata, Y.; Takezawa, N.; Kazeto, Y.; Takatsuka, N.; Kato, E.; Hagihara, S.; Ozaki, Y.; Adachi, S.; Yamauchi, K.; et al. 17β-HSD type 12-Like is responsible for maturation-inducing hormone synthesis during oocyte maturation in masu salmon. Endocrinology 2017, 158, 627–639. [Google Scholar] [PubMed] [Green Version]
- Aranyakanont, C.; Ijiri, S.; Hasegawa, Y.; Adachi, S. 17β-Hydroxysteroid dehydrogenase type 12 is responsible for maturation-inducing steroid synthesis during oocyte maturation in Nile tilapia. Gen. Com. Endocrinol. 2020, 290, 113399. [Google Scholar] [CrossRef]
- Tokarz, J.; Mindnich, R.; Norton, W.; Moeller, G.; de Angelis, M.H.; Adamski, J. Discovery of a novel enzyme mediating glucocorticoid catabolism in fish: 20β-Hydroxysteroid dehydrogenase type 2. Mol. Cell. Endocrinol. 2012, 349, 202–213. [Google Scholar] [CrossRef] [PubMed]





| Purpose | Name | Sequence (5′–3′) | PCR Product Sizes | Annealing Temperature |
|---|---|---|---|---|
| Hsd3b | ||||
| Full-cloning | 1F | CTAGCCTCGAGAATTGAGGGA ATTTGTATCCCACACG | 1277 bp | 62 °C |
| 1R | TACCACGCGTGAATTAGCCCA CAATACGTCAGGCTGG | |||
| RT-PCR | 2F | CTGGAGCGTTTGGATTCCTGGG | 305 bp | 55 °C |
| 2R | CCAGCAAGAACTGGGTACCTT | |||
| qPCR | 3F | ACCAGCAGCATTGAGGTGGC | ||
| 3R | CTCCCCATTGACCTGTAGAACC | |||
| Cyp17a1 | ||||
| Full-cloning | 4F | CTAGCCTCGAGAATTTGCACACC TAAAGTTTAGGTAAC | 1633 bp | 62 °C |
| 4R | TACCACGCGTGAATTGACTGTATA ATACAAAGCAGTCT | |||
| RT-PCR | 5F | TAATCTGTAGAGAAGCCCGCTCC | 428 bp | 55 °C |
| 5R | TCTGTCAGCCCCACTTCTCTTGC | |||
| qPCR | 6F | GTGAATAACTACCAAGATGCC | ||
| 6R | GAATAGAATGTCTGTAGTCACAG | |||
| Cyp17a2 | ||||
| 5′RACE | ||||
| RT | 7R | GTACAGACCAAAGAGGCCCCCA | ||
| RACE-PCR | ||||
| 1st round | 8R | TTGCACCCCAGCCTGGTGAAGAG | ||
| 2nd round | 9R | ACGATGGGGAAGGAGGGCAGACA | ||
| 3′RACE | ||||
| 1st round | 7F | AACCTTGAGGGGAGATATGGGGT | ||
| 2nd round | 8F | GTAGTCCTGCAGCCAGAAAGCTA | ||
| Full-cloning | 10F | CTAGCCTCGAGAATTGTGTGCCTGA TTTGGGTTGTGCT | 1742 bp | 62 °C |
| 10R | TACCACGCGTGAATTCTGGTACACA AAGACCCTTCCC | |||
| RT-PCR | 11F | ATGGTGACAACAGCTCTGCTG | 353 bp | 55 °C |
| 11R | TACTAGGCTTCCTCTGGCGATG | |||
| qPCR | 12F | CTTGCTTCATCACCCTGAGGTC | ||
| 12R | CCGATACTGGTATTTTCCAAGG | |||
| Ef1α | Ef1α1F | AAACAACCCCCTGCGTCTG | ||
| Ef1α1R | GGGTACAGTTCCAATACCTCCGA |
| Substrate | Product | Conversion Rate | |||
|---|---|---|---|---|---|
| Amur Sturgeon hsd3b Type-I | Amur Sturgeon cyp17a1 Type-I | Amur Sturgeon cyp17a2 Type-I | Non-Transfected HEK293T | ||
| P5 | 17OHP5 | n.d. | 1.6% | 6.2% | n.d. |
| P5 | DHEA | n.d. | 2.3% | n.d. | n.d. |
| P4 | 17OHP | n.d. | 4.2% | 28.1% | n.d. |
| P4 | A4 | n.d. | 5.6% | n.d. | n.d. |
| P5 | P4 | 12.6% | n.d. | n.d. | n.d. |
| P4 | P5 | n.d. | n.d. | n.d. | n.d. |
| 17OHP5 | 17OHP | 11.4% | - | - | n.d. |
| 17OHP | 17OHP5 | n.d. | - | - | n.d. |
| DHEA | A4 | 18% | - | - | n.d. |
| A4 | DHEA | n.d. | - | - | n.d. |
| A5 | T | 17.4% | - | - | n.d. |
| T | A5 | n.d. | - | - | n.d. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasegawa, Y.; Surugaya, R.; Adachi, S.; Ijiri, S. Regulation of 17α-Hydroxyprogesterone Production during Induced Oocyte Maturation and Ovulation in Amur Sturgeon (Acipenser schrenckii). J. Mar. Sci. Eng. 2022, 10, 86. https://doi.org/10.3390/jmse10010086
Hasegawa Y, Surugaya R, Adachi S, Ijiri S. Regulation of 17α-Hydroxyprogesterone Production during Induced Oocyte Maturation and Ovulation in Amur Sturgeon (Acipenser schrenckii). Journal of Marine Science and Engineering. 2022; 10(1):86. https://doi.org/10.3390/jmse10010086
Chicago/Turabian StyleHasegawa, Yuya, Ryohei Surugaya, Shinji Adachi, and Shigeho Ijiri. 2022. "Regulation of 17α-Hydroxyprogesterone Production during Induced Oocyte Maturation and Ovulation in Amur Sturgeon (Acipenser schrenckii)" Journal of Marine Science and Engineering 10, no. 1: 86. https://doi.org/10.3390/jmse10010086
APA StyleHasegawa, Y., Surugaya, R., Adachi, S., & Ijiri, S. (2022). Regulation of 17α-Hydroxyprogesterone Production during Induced Oocyte Maturation and Ovulation in Amur Sturgeon (Acipenser schrenckii). Journal of Marine Science and Engineering, 10(1), 86. https://doi.org/10.3390/jmse10010086





