Exploring the Root-Associated Bacterial Community of Tomato Plants in Response to Salt Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Trial and Treatments
2.2. Rhizophere and Leaf Sample Collection
2.3. EC Measurement of Commercial Substrate
2.4. Total DNA Extraction and Illumina Sequencing from Rhizosphere Samples
2.5. Oxylipins, Phytohormones and Fatty Acids Extraction and Analyses
2.6. Metabolomic Statistical Analysis
2.7. 16S rDNA Statistical Analysis
3. Results
3.1. Biodiversity of Rhizosphere Microbiome
3.2. Differential Abundance of Taxa Among Time Points and Treatment
3.3. Changes in Oxylipins, Hormones and Fatty Acids Between Treatments
3.4. Functional Genes That Potentially Alleviate Salt Stress for Plants
3.5. Non-Linear Correlation Analysis Bacterial Genera, Functional Genes and Plant Metabolites
4. Discussion
4.1. Short-Term Rhizosphere Bacterial Community Changes Under Salt Stress
4.2. How Bacterial Communties Reacted over Time Under Salt Stress
4.3. Effect of Salt-Induced Stress on Plant Metabolism
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| JA | Jasmonic acid |
| SA | Salicylic acid |
| MRM | Multiple reaction monitoring |
| SIM | Single ion monitoring |
| FFA | Free fatty acid |
| PUFA | Polyunsaturated fatty acid |
| SFA | Saturated fatty acid |
| 9-HODE | 9-hydroxyoctadecenoic acid |
| 9-HpODE | 9-hydroperoxyoctadienoic acid |
| 9-HpOTre | 9-hydroperoxyoctatrienoic acid |
| 9-HOTre | 9-oxo-octadecatrienoic acid |
| 9-OxoDE | 9-oxo-octadecenoic acid |
| 9-OxoTrE | 9-oxo-octadecatrienoic acid |
| 13-HODE | 13-hydroxyoctadecenoic acid |
| 13-HOTre | 13-OH-9Z,11E,15Z-octadecatrienoic acid |
| 13- HpOTre | 13-hydroperoxy-9,11E,15Z-octadecatrienoic acid |
| 13-OxoDE | 13-oxo-octadecenoic acid |
| 13-OxoTrE | 13-oxo-octadecatrienoic acid |
| C16:0 | Palmitic acid |
| C16:1 | Palmitoleic acid |
| C18:0 | Stearic acid |
| C18:1 | Oleic acid |
| C18:2 | Linoleic acid |
| C18:3 | α-Linolenic acid |
| C20:4 | Arachidonic acid |
| C22:0 | Decosanoic acid |
| C24:0 | Lignoceric acid |
| 10-HODE | 10-hydroxyoctadecenoic acid |
| 12,13-DiHOME | 12,13-dihydroxyoctamonoenoic acid |
| 12,13-EpOME | 12,13-dihydroxyoctamonoenoic acid |
| 8,13 diHODE | 8,13-dihydroxy-9,11-octadecadienoic acid |
| 8-HODE | 8-hydroxyoctadecenoic acid |
| 8-HpODE | 8-hydroperoxyoctadienoic acid |
| 9,10-EpOME | 9,10-epoxyoctamonoenoic acid |
References
- Manuel, R.; Machado, A.; Serralheiro, R.P.; Alvino, A.; Freire, M.I.; Ferreira, R. Soil Salinity: Effect on Vegetable Crop Growth. Management Practices to Prevent and Mitigate Soil Salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- Shrivastava, P.; Kumar, R. Soil Salinity: A Serious Environmental Issue and Plant Growth Promoting Bacteria as One of the Tools for Its Alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef]
- Sliti, A.; Singh, V.; Pande, A.; Shin, J.-H. Soil Microbial Holobiont Interplay and Its Role in Protecting Plants against Salinity Stress. Pedosphere 2024, 35, 97–115. [Google Scholar] [CrossRef]
- Canfora, L.; Bacci, G.; Pinzari, F.; Lo Papa, G.; Dazzi, C.; Benedetti, A. Salinity and Bacterial Diversity: To What Extent Does the Concentration of Salt Affect the Bacterial Community in a Saline Soil? PLoS ONE 2014, 9, e106662. [Google Scholar] [CrossRef] [PubMed]
- Richards, L.A. Diagnosis and Improvement of Saline and Alkali Soils. Soil Sci. 1954, 78, 154. [Google Scholar] [CrossRef]
- Flowers, T.J.; Munns, R.; Colmer, T.D. Sodium Chloride Toxicity and the Cellular Basis of Salt Tolerance in Halophytes. Ann. Bot. 2015, 115, 419–431. [Google Scholar] [CrossRef]
- Yadav, S.; Irfan, M.D.; Ahmad, A.; Hayat, S. Causes of Salinity and Plant Manifestations to Salt Stress: A Review. J. Environ. Biol. 2011, 32, 667–685. [Google Scholar]
- Otlewska, A.; Migliore, M.; Dybka-Stępień, K.; Manfredini, A.; Struszczyk-Świta, K.; Napoli, R.; Białkowska, A.; Canfora, L.; Pinzari, F. When Salt Meddles Between Plant, Soil, and Microorganisms. Front. Plant Sci. 2020, 11, 553087. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Verma, J.P. Does Plant—Microbe Interaction Confer Stress Tolerance in Plants: A Review? Microbiol. Res. 2018, 207, 41–52. [Google Scholar] [CrossRef]
- Parida, A.K.; Das, A.B. Salt Tolerance and Salinity Effects on Plants: A Review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef]
- Rath, K.M.; Murphy, D.N.; Rousk, J. The Microbial Community Size, Structure, and Process Rates along Natural Gradients of Soil Salinity. Soil Biol. Biochem. 2019, 138, 107607. [Google Scholar] [CrossRef]
- Haj-Amor, Z.; Araya, T.; Kim, D.G.; Bouri, S.; Lee, J.; Ghiloufi, W.; Yang, Y.; Kang, H.; Jhariya, M.K.; Banerjee, A.; et al. Soil Salinity and Its Associated Effects on Soil Microorganisms, Greenhouse Gas Emissions, Crop Yield, Biodiversity and Desertification: A Review. Sci. Total Environ. 2022, 843, 156946. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Bai, J.; Zhai, Y.; Jia, J.; Zhao, Q.; Wang, W.; Hu, X. Microbial Diversity and Functions in Saline Soils: A Review from a Biogeochemical Perspective. J. Adv. Res. 2024, 59, 129–140. [Google Scholar] [CrossRef]
- Rath, K.M.; Fierer, N.; Murphy, D.V.; Rousk, J. Linking Bacterial Community Composition to Soil Salinity along Environmental Gradients. ISME J. 2019, 13, 836–846. [Google Scholar] [CrossRef]
- Katerji, N.; Van Hoorn, J.W.; Hamdy, A.; Mastrorilli, M. Salinity Effect on Crop Development and Yield, Analysis of Salt Tolerance According to Several Classification Methods. Agric. Water Manag. 2003, 62, 37–66. [Google Scholar] [CrossRef]
- Maas, E.V.; Hoffman, G.J. Crop salt tolerance—Current assessment. J. Irrig. Drain. Div. 1977, 103, 115–134. [Google Scholar] [CrossRef]
- Panno, S.; Davino, S.; Caruso, A.G.; Bertacca, S.; Crnogorac, A.; Mandić, A.; Noris, E.; Matić, S. A Review of the Most Common and Economically Important Diseases That Undermine the Cultivation of Tomato Crop in the Mediterranean Basin. Agronomy 2021, 11, 2188. [Google Scholar] [CrossRef]
- Roșca, M.; Mihalache, G.; Stoleru, V. Tomato Responses to Salinity Stress: From Morphological Traits to Genetic Changes. Front. Plant Sci. 2023, 14, 1118383. [Google Scholar] [CrossRef]
- Cuartero, J.; Fernández-Muñoz, R. Tomato and Salinity. Sci. Hortic. 1998, 78, 83–125. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant Salt-Tolerance Mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef]
- Meng, X.; Zhou, J.; Sui, N. Mechanisms of Salt Tolerance in Halophytes: Current Understanding and Recent Advances. Open Life Sci. 2018, 13, 149. [Google Scholar] [CrossRef]
- Liu, S.; Tian, Y.; Jia, M.; Lu, X.; Yue, L.; Zhao, X.; Jin, W.; Wang, Y.; Zhang, Y.; Xie, Z.; et al. Induction of Salt Tolerance in Arabidopsis Thaliana by Volatiles from Bacillus Amyloliquefaciens FZB42 via the Jasmonic Acid Signaling Pathway. Front. Microbiol. 2020, 11, 562934. [Google Scholar] [CrossRef]
- Pedranzani, H.; Racagni, G.; Alemano, S.; Miersch, O.; Ramírez, I.; Peña-Cortés, H.; Taleisnik, E.; Machado-Domenech, E.; Abdala, G. Salt Tolerant Tomato Plants Show Increased Levels of Jasmonic Acid. Plant Growth Regul. 2003, 41, 149–158. [Google Scholar] [CrossRef]
- Ali, M.S.; Baek, K.H. Jasmonic Acid Signaling Pathway in Response to Abiotic Stresses in Plants. Int. J. Mol. Sci. 2020, 21, 621. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Shah, L.R.; Mir, N.; Gulzar, G.; Mushtaq, B.; Rashid, R.; Afroza, B. The Roles of Oxylipins in Plant Systemic Resistance. In Phyto-Oxylipins; CRC Press: Boca Raton, FL, USA, 2023; pp. 151–173. [Google Scholar] [CrossRef]
- Savchenko, T.V.; Zastrijnaja, O.M.; Klimov, V.V. Oxylipins and Plant Abiotic Stress Resistance. Biochemistry 2014, 79, 362–375. [Google Scholar] [CrossRef]
- Christensen, S.A.; Kolomiets, M.V. The Lipid Language of Plant-Fungal Interactions. Fungal Genet. Biol. 2011, 48, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Martínez, E.; Cosnahan, R.K.; Wu, M.; Gadila, S.K.; Quick, E.B.; Mobley, J.A.; Campos-Gómez, J. Oxylipins Mediate Cell-to-Cell Communication in Pseudomonas Aeruginosa. Commun. Biol. 2019, 2, 66. [Google Scholar] [CrossRef]
- Beccaccioli, M.; Pucci, N.; Salustri, M.; Scortichini, M.; Zaccaria, M.; Momeni, B.; Loreti, S.; Reverberi, M.; Scala, V. Fungal and Bacterial Oxylipins Are Signals for Intra- and Inter-Cellular Communication within Plant Disease. Front. Plant Sci. 2022, 13, 823233. [Google Scholar] [CrossRef]
- Liang, Y.; Huang, Y.; Liu, C.; Chen, K.; Li, M. Functions and Interaction of Plant Lipid Signalling under Abiotic Stresses. Plant Biol. 2023, 25, 361–378. [Google Scholar] [CrossRef]
- Blée, E. Impact of Phyto-Oxylipins in Plant Defense. Trends Plant Sci. 2002, 7, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Goswami, D.; Thakker, J.N.; Dhandhukia, P.C. Portraying Mechanics of Plant Growth Promoting Rhizobacteria (PGPR): A Review. Cogent Food Agric. 2016, 2, 1127500. [Google Scholar] [CrossRef]
- Vurukonda, S.S.K.P.; Vardharajula, S.; Shrivastava, M.; SkZ, A. Enhancement of Drought Stress Tolerance in Crops by Plant Growth Promoting Rhizobacteria. Microbiol. Res. 2016, 184, 13–24. [Google Scholar] [CrossRef]
- Kumar, A.; Patel, J.S.; Meena, V.S.; Ramteke, P.W. Plant Growth-Promoting Rhizobacteria: Strategies to Improve Abiotic Stresses under Sustainable Agriculture. J. Plant Nutr. 2019, 42, 1402–1415. [Google Scholar] [CrossRef]
- Hoque, M.N.; Hannan, A.; Imran, S.; Paul, N.C.; Mondal, M.F.; Sadhin, M.M.R.; Bristi, J.M.; Dola, F.S.; Hanif, M.A.; Ye, W.; et al. Plant Growth-Promoting Rhizobacteria-Mediated Adaptive Responses of Plants Under Salinity Stress. J. Plant Growth Regul. 2022, 42, 1307–1326. [Google Scholar] [CrossRef]
- Trinchera, A.; Leita, L.; Sequi, P. Ministero delle Politiche Agricole e Forestali. Osservatorio Nazionale Pedologico e per la Qualità del Suolo Agricolo e Forestale; Consiglio per la Ricerca e la Sperimentazione in Agricoltura; Istituto Sperimentale per la Nutrizione delle Piante. In Metodi Di Analisi per i Fertilizzanti; Consiglio per la Ricerca e la Sperimentazione in Agricoltura: Rome, Italy, 2006. [Google Scholar]
- Turner, S.; Pryer, K.M.; Miao, V.P.W.; Palmer, J.D. Investigating Deep Phylogenetic Relationships among Cyanobacteria and Plastids by Small Subunit RRNA Sequence Analysis. J. Eukaryot. Microbiol. 1999, 46, 327–338. [Google Scholar] [CrossRef]
- Kisand, V.; Cuadros, R.; Wikner, J. Phylogeny of Culturable Estuarine Bacteria Catabolizing Riverine Organic Matter in the Northern Baltic Sea. Appl. Environ. Microbiol. 2002, 68, 379. [Google Scholar] [CrossRef] [PubMed]
- Scala, V.; Reverberi, M.; Salustri, M.; Pucci, N.; Modesti, V.; Lucchesi, S.; Loreti, S. Lipid Profile of Xylella Fastidiosa Subsp. pauca Associated With the Olive Quick Decline Syndrome. Front. Microbiol. 2018, 9, 1839. [Google Scholar] [CrossRef]
- Del Duca, S.; Mocali, S.; Vitali, F.; Fabiani, A.; Cucu, M.A.; Valboa, G.; d’Errico, G.; Binazzi, F.; Storchi, P.; Perria, R.; et al. Impacts of Soil Management and Sustainable Plant Protection Strategies on Soil Biodiversity in a Sangiovese Vineyard. Land 2024, 13, 599. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2′s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Lahti, L.; Shetty, S. Tools for Microbiome Analysis in R. Microbiome Package. 2018. Available online: https://microbiome.github.io/tutorials (accessed on 30 December 2024).
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical Analysis of Taxonomic and Functional Profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Zheng, Y.; Xu, Z.; Liu, H.; Liu, Y.; Zhou, Y.; Meng, C.; Ma, S.; Xie, Z.; Li, Y.; Zhang, C.-S. Patterns in the Microbial Community of Salt-Tolerant Plants and the Functional Genes Associated with Salt Stress Alleviation. Microbiol. Spectr. 2021, 9, e0076721. [Google Scholar] [CrossRef]
- Thiem, D.; Gołebiewski, M.; Hulisz, P.; Piernik, A.; Hrynkiewicz, K. How Does Salinity Shape Bacterial and Fungal Microbiomes of Alnus Glutinosa Roots? Front. Microbiol. 2018, 9, 318754. [Google Scholar] [CrossRef]
- Schmitz, L.; Yan, Z.; Schneijderberg, M.; de Roij, M.; Pijnenburg, R.; Zheng, Q.; Franken, C.; Dechesne, A.; Trindade, L.M.; van Velzen, R.; et al. Synthetic Bacterial Community Derived from a Desert Rhizosphere Confers Salt Stress Resilience to Tomato in the Presence of a Soil Microbiome. ISME J. 2022, 16, 1907–1920. [Google Scholar] [CrossRef]
- Yaghoubi Khanghahi, M.; Crecchio, C.; Verbruggen, E. Shifts in the Rhizosphere and Endosphere Colonizing Bacterial Communities Under Drought and Salinity Stress as Affected by a Biofertilizer Consortium. Microb. Ecol. 2022, 84, 483–495. [Google Scholar] [CrossRef]
- Zhou, Y.; He, Z.; Lin, Q.; Lin, Y.; Long, K.; Xie, Z.; Hu, W. Salt Stress Affects the Bacterial Communities in Rhizosphere Soil of Rice. Front. Microbiol. 2024, 15, 1505368. [Google Scholar] [CrossRef]
- Philippot, L.; Hallin, S.; Börjesson, G.; Baggs, E.M. Biochemical Cycling in the Rhizosphere Having an Impact on Global Change. Plant Soil 2008, 321, 61–81. [Google Scholar] [CrossRef]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A Communal Catalogue Reveals Earth’s Multiscale Microbial Diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Zhao, X.; Ren, J.; Dong, J.; Zhang, H.; Dong, Q.; Jiang, C.; Zhong, C.; Zhou, Y.; Yu, H. Influence of Peanut, Sorghum, and Soil Salinity on Microbial Community Composition in Interspecific Interaction Zone. Front. Microbiol. 2021, 12, 678250. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Ma, B.; McLaughlin, N.B.; Niu, Y.; Wang, D.; Liu, H.; Li, M.; Sun, Z. The Impact of Salinization on Soil Bacterial Diversity, Yield and Quality of Glycyrrhiza Uralensis Fisch. Front. Microbiol. 2024, 15, 1448301. [Google Scholar] [CrossRef]
- Li, H.; La, S.; Zhang, X.; Gao, L.; Tian, Y. Salt-Induced Recruitment of Specific Root-Associated Bacterial Consortium Capable of Enhancing Plant Adaptability to Salt Stress. ISME J. 2021, 15, 2865–2882. [Google Scholar] [CrossRef]
- Ma, B.; Gong, J. A Meta-Analysis of the Publicly Available Bacterial and Archaeal Sequence Diversity in Saline Soils. World J. Microbiol. Biotechnol. 2013, 29, 2325–2334. [Google Scholar] [CrossRef] [PubMed]
- Jog, R.; Pandya, M.; Nareshkumar, G.; Rajkumar, S. Mechanism of Phosphate Solubilization and Antifungal Activity of Streptomyces Spp. Isolated from Wheat Roots and Rhizosphere and Their Application in Improving Plant Growth. Microbiology 2014, 160, 778–788. [Google Scholar] [CrossRef]
- Thilagam, R.; Hemalatha, N. Plant Growth Promotion and Chilli Anthracnose Disease Suppression Ability of Rhizosphere Soil Actinobacteria. J. Appl. Microbiol. 2019, 126, 1835–1849. [Google Scholar] [CrossRef]
- Yadav, A.N.; Verma, P.; Kumar, S.; Kumar, V.; Kumar, M.; Kumari Sugitha, T.C.; Singh, B.P.; Saxena, A.K.; Dhaliwal, H.S. Actinobacteria from Rhizosphere: Molecular Diversity, Distributions, and Potential Biotechnological Applications. In New and Future Developments in Microbial Biotechnology and Bioengineering: Actinobacteria: Diversity and Biotechnological Applications; Elsevier: Amsterdam, The Netherlands, 2018; pp. 13–41. [Google Scholar] [CrossRef]
- Narsing Rao, M.P.; Lohmaneeratana, K.; Bunyoo, C.; Thamchaipenet, A. Actinobacteria–Plant Interactions in Alleviating Abiotic Stress. Plants 2022, 11, 2976. [Google Scholar] [CrossRef]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.L.; Touraine, B.; Moënne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Plant Growth-Promoting Rhizobacteria and Root System Functioning. Front. Plant Sci. 2013, 4, 356. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an Ecological Classification of Soil Bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Zhang, G.; Bai, J.; Jia, J.; Wang, W.; Wang, D.; Zhao, Q.; Wang, C.; Chen, G. Soil Microbial Communities Regulate the Threshold Effect of Salinity Stress on SOM Decomposition in Coastal Salt Marshes. Fundam. Res. 2023, 3, 868–879. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Dolfing, J.; Guo, Z.; Chen, R.; Wu, M.; Li, Z.; Lin, X.; Feng, Y. Important Ecophysiological Roles of Non-Dominant Actinobacteria in Plant Residue Decomposition, Especially in Less Fertile Soils. Microbiome 2021, 9, 84. [Google Scholar] [CrossRef]
- Benlloch, S.; López-López, A.; Casamayor, E.O.; Øvreås, L.; Goddard, V.; Daae, F.L.; Smerdon, G.; Massana, R.; Joint, I.; Thingstad, F.; et al. Prokaryotic Genetic Diversity throughout the Salinity Gradient of a Coastal Solar Saltern. Environ. Microbiol. 2002, 4, 349–360. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Knight, R. Global Patterns in Bacterial Diversity. Proc. Natl. Acad. Sci. USA 2007, 104, 11436–11440. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.; Kim, I.; Chhetri, G.; Park, S.; Lee, H.; Yook, S.; Seo, T. Two Novel Bacterial Species, Rhodanobacter lycopersici sp. nov. and Rhodanobacter geophilus sp. nov., Isolated from the Rhizosphere of Solanum lycopersicum with Plant Growth-Promoting Traits. Microorganisms 2024, 12, 2227. [Google Scholar] [CrossRef]
- Shang, X.C.; Zhang, M.; Zhang, Y.; Hou, X.; Yang, L. Waste Seaweed Compost and Rhizosphere Bacteria Pseudomonas Koreensis Promote Tomato Seedlings Growth by Benefiting Properties, Enzyme Activities and Rhizosphere Bacterial Community in Coastal Saline Soil of Yellow River Delta, China. Waste Manag. 2023, 172, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Roychowdhury, R.; Mishra, S.; Anand, G.; Dalal, D.; Gupta, R.; Kumar, A.; Gupta, R. Decoding the Molecular Mechanism Underlying Salicylic Acid (SA)-Mediated Plant Immunity: An Integrated Overview from Its Biosynthesis to the Mode of Action. Physiol. Plant 2024, 176, e14399. [Google Scholar] [CrossRef]
- Elsisi, M.; Elshiekh, M.; Sabry, N.; Aziz, M.; Attia, K.; Islam, F.; Chen, J.; Abdelrahman, M. The Genetic Orchestra of Salicylic Acid in Plant Resilience to Climate Change Induced Abiotic Stress: Critical Review. Stress Biol. 2024, 4, 31. [Google Scholar] [CrossRef]
- Borsani, O.; Valpuesta, V.; Botella, M.A. Evidence for a Role of Salicylic Acid in the Oxidative Damage Generated by NaCl and Osmotic Stress in Arabidopsis Seedlings. Plant Physiol. 2001, 126, 1024–1030. [Google Scholar] [CrossRef]
- Yasin Ashraf, M.; Ashraf, M.; Akhtar, M.; Mahmood, K.; Saleem, M. Improvement in Yield, Quality and Reduction in Fruit Drop in Kinnow (Citrus Reticulata Blanco) by Exogenous Application of Plant Growth Regulators, Potassium and Zinc. Pak. J. Bot 2013, 45, 433–440. [Google Scholar]
- Salama, K.H.A.; Mansour, M.M.F.; Ali, F.Z.M.; Abou-Hadid, A.F. NaCl-Induced Changes in Plasma Membrane Lipids and Proteins of Zea Mays L. Cultivars Differing in Their Response to Salinity. Acta Physiol. Plant 2007, 29, 351–359. [Google Scholar] [CrossRef]
- Guo, Q.; Liu, L.; Barkla, B.J. Membrane Lipid Remodeling in Response to Salinity. Int. J. Mol. Sci. 2019, 20, 4264. [Google Scholar] [CrossRef]
- Molina, A.; Bueno, P.; Marín, M.C.; Rodríguez-Rosales, M.P.; Belver, A.; Venema, K.; Donaire, J.P. Involvement of Endogenous Salicylic Acid Content, Lipoxygenase and Antioxidant Enzyme Activities in the Response of Tomato Cell Suspension Cultures to NaCl. New Phytol. 2002, 156, 409–415. [Google Scholar] [CrossRef]
- Ghanem, M.E.; Ghars, M.A.; Frettinger, P.; Pérez-Alfocea, F.; Lutts, S.; Wathelet, J.P.; du Jardin, P.; Fauconnier, M.L. Organ-Dependent Oxylipin Signature in Leaves and Roots of Salinized Tomato Plants (Solanum lycopersicum). J. Plant Physiol. 2012, 169, 1090–1101. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Choi, J.H.; Truong, H.A.; Lee, Y.J.; Lee, H. Enhanced Nitrate Reductase Activity Offers Arabidopsis Ecotype Landsberg Erecta Better Salt Stress Resistance than Col-0. Plant Biol. 2022, 24, 854–862. [Google Scholar] [CrossRef]
- Shabala, S. Salinity and Programmed Cell Death: Unravelling Mechanisms for Ion Specific Signalling. J. Exp. Bot. 2009, 60, 709–712. [Google Scholar] [CrossRef]
- Radchenko, M.V.; Waditee, R.; Oshimi, S.; Fukuhara, M.; Takabe, T.; Nakamura, T. Cloning, Functional Expression and Primary Characterization of Vibrio Parahaemolyticus K+/H+ Antiporter Genes in Escherichia coli. Mol. Microbiol. 2006, 59, 651–663. [Google Scholar] [CrossRef]

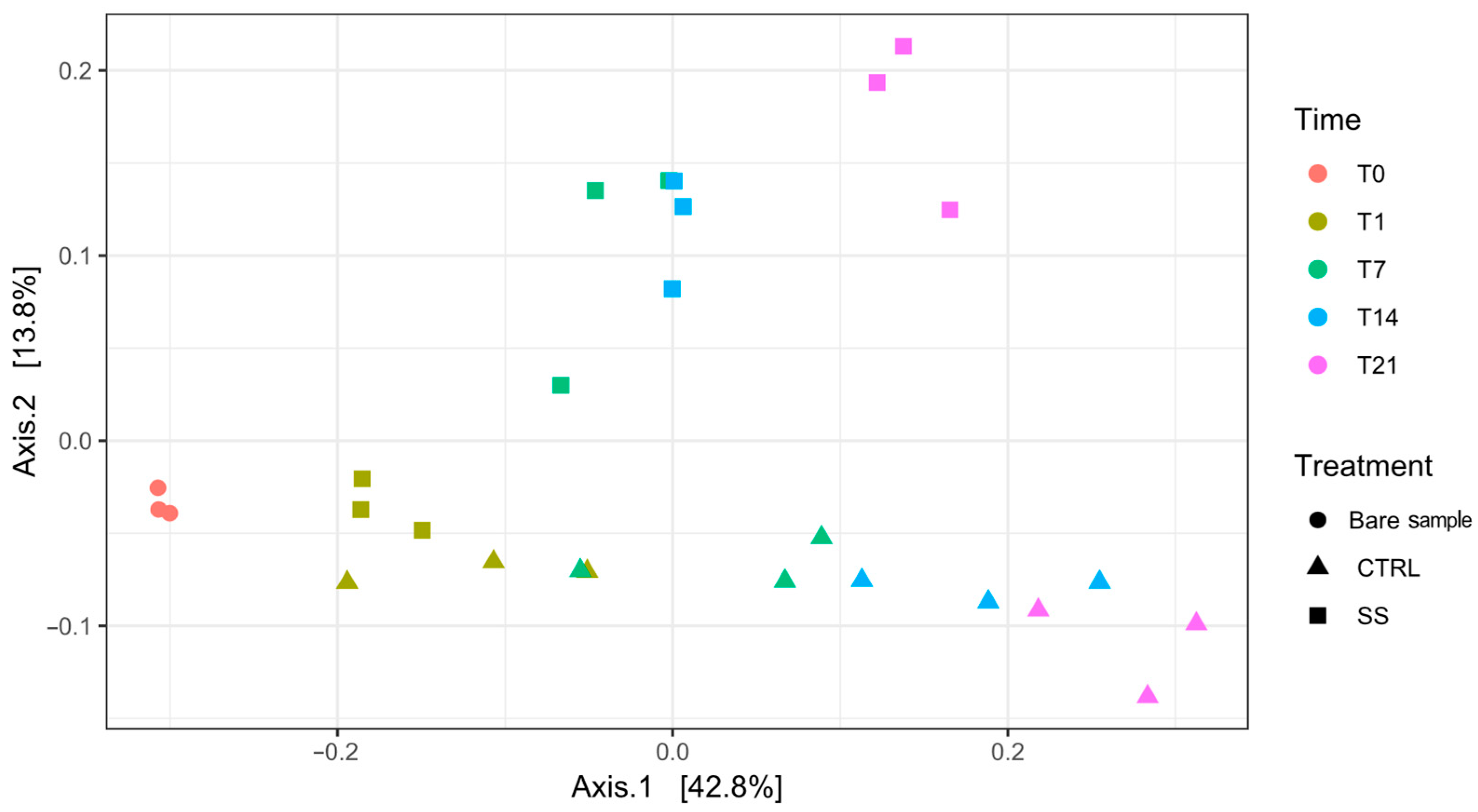
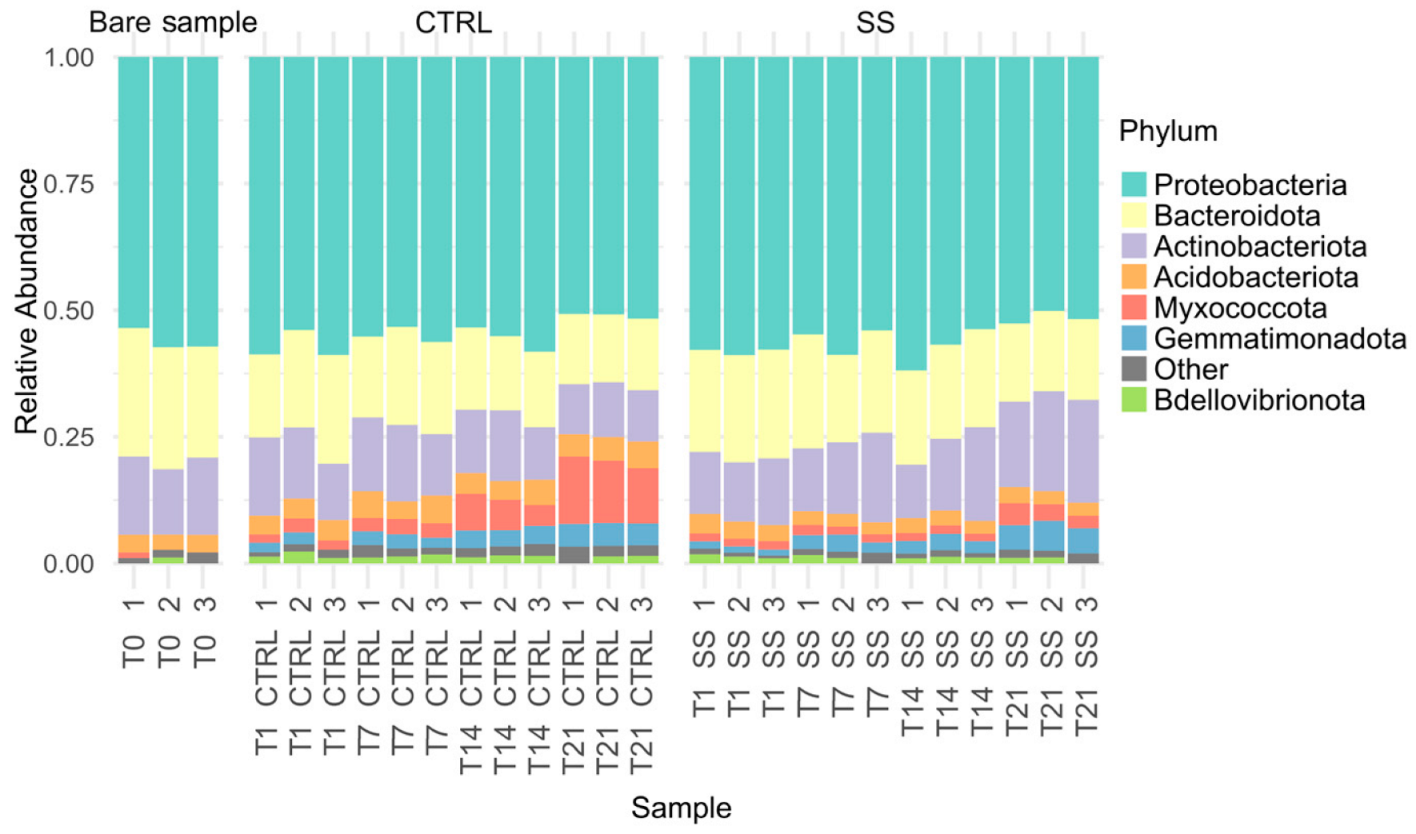
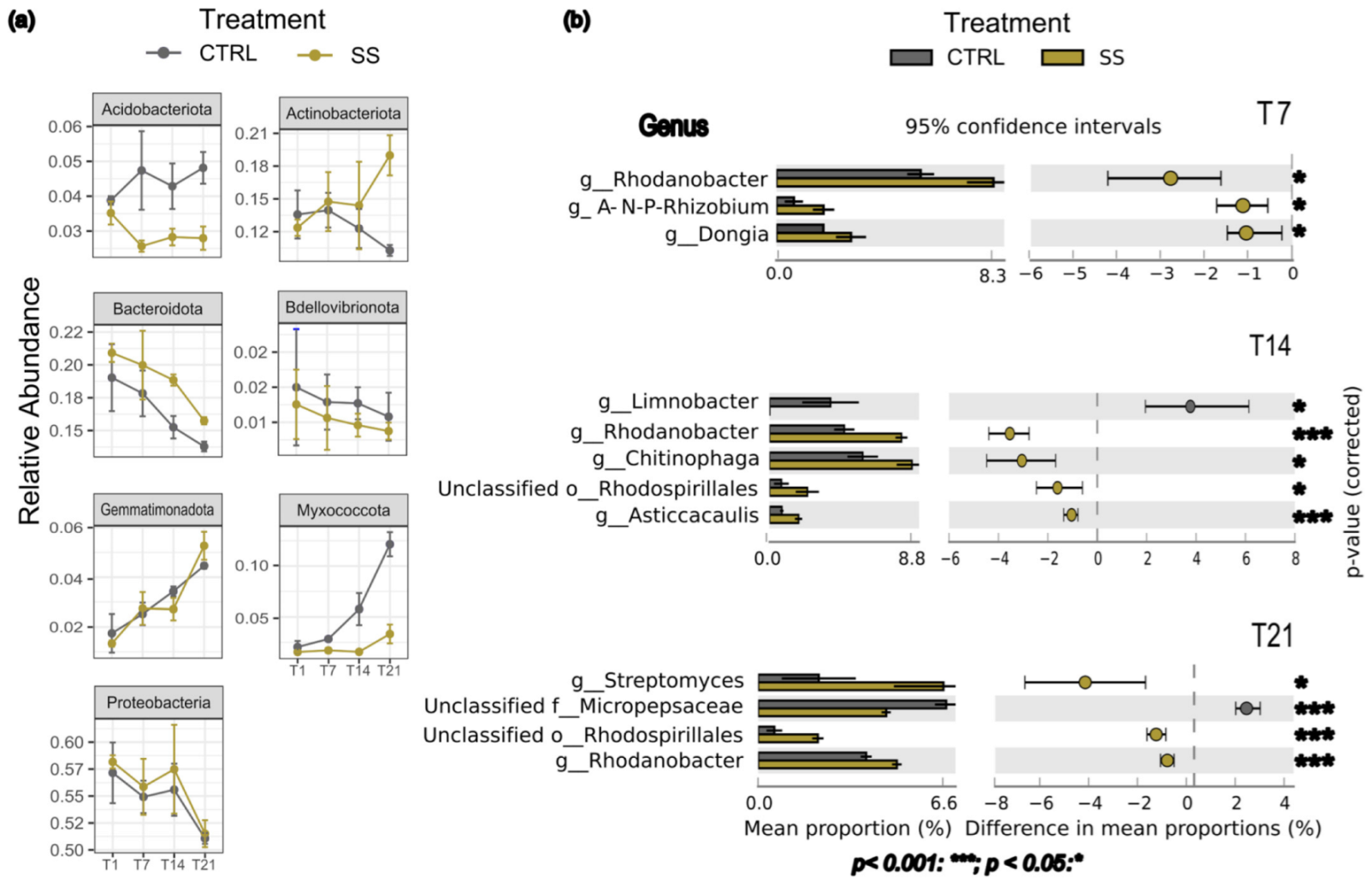
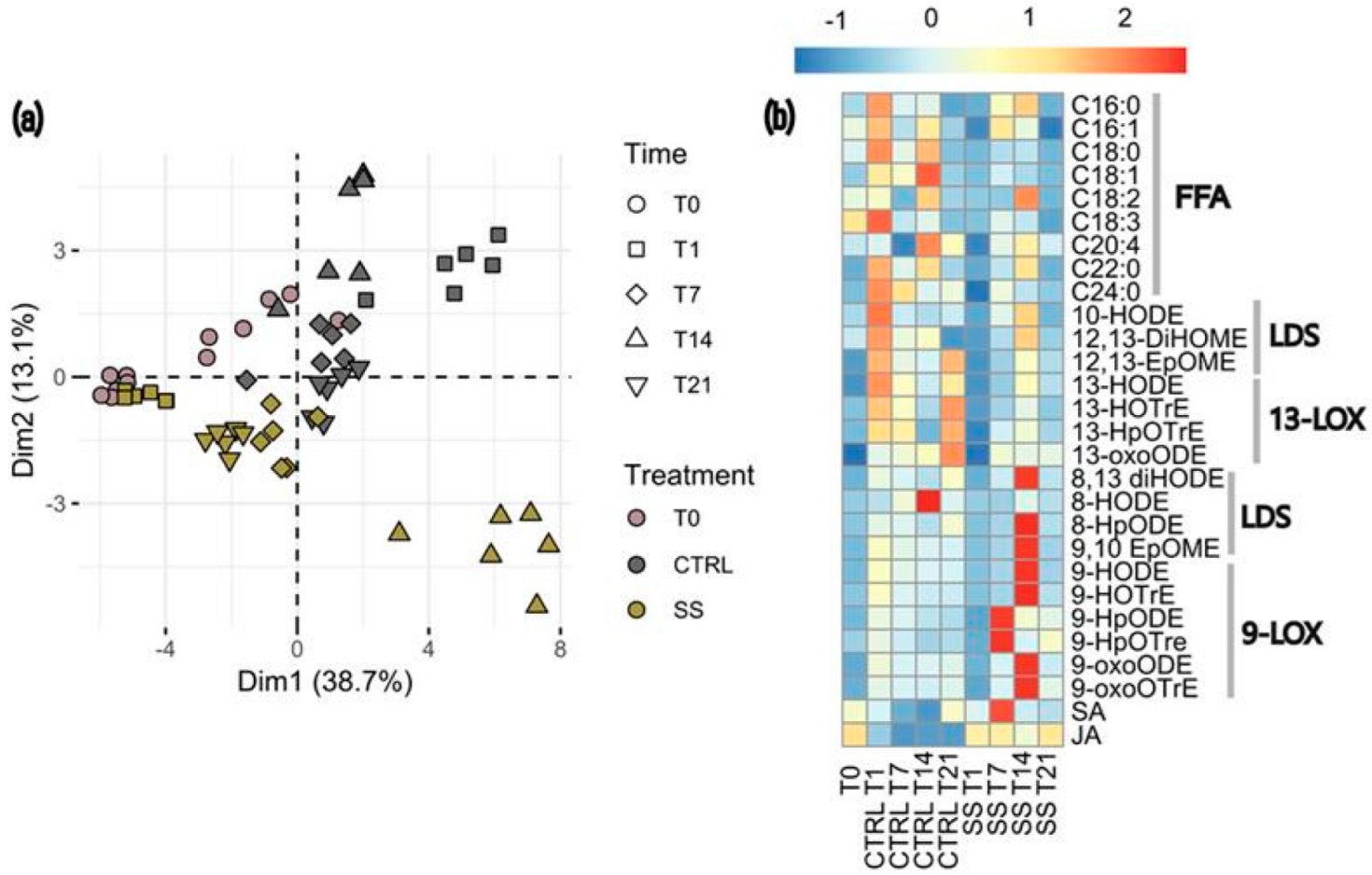

| Comparison | Shannon Index | Pielou’s Evenness | ||
|---|---|---|---|---|
| Diff | p Sig | Diff | p Sig | |
| Time 7–Time 1 | 0.1587463 | * | 0.017834536 | ns |
| Time 14–Time 1 | 0.1960060 | * | 0.024326297 | * |
| Time 21–Time 1 | 0.2796129 | ** | 0.033348564 | ** |
| Time 14–Time 7 | 0.0372597 | ns | 0.006491761 | ns |
| Time 21–Time 7 | 0.1208666 | ns | 0.015514028 | ns |
| Time 21–Time 14 | 0.0836069 | ns | 0.009022267 | ns |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esposito, A.; Scala, V.; Vitali, F.; Beccaccioli, M.; Reverberi, M.; Valboa, G.; Del Duca, S.; Canfora, L.; Mocali, S. Exploring the Root-Associated Bacterial Community of Tomato Plants in Response to Salt Stress. Agriculture 2025, 15, 624. https://doi.org/10.3390/agriculture15060624
Esposito A, Scala V, Vitali F, Beccaccioli M, Reverberi M, Valboa G, Del Duca S, Canfora L, Mocali S. Exploring the Root-Associated Bacterial Community of Tomato Plants in Response to Salt Stress. Agriculture. 2025; 15(6):624. https://doi.org/10.3390/agriculture15060624
Chicago/Turabian StyleEsposito, Antonia, Valeria Scala, Francesco Vitali, Marzia Beccaccioli, Massimo Reverberi, Giuseppe Valboa, Sara Del Duca, Loredana Canfora, and Stefano Mocali. 2025. "Exploring the Root-Associated Bacterial Community of Tomato Plants in Response to Salt Stress" Agriculture 15, no. 6: 624. https://doi.org/10.3390/agriculture15060624
APA StyleEsposito, A., Scala, V., Vitali, F., Beccaccioli, M., Reverberi, M., Valboa, G., Del Duca, S., Canfora, L., & Mocali, S. (2025). Exploring the Root-Associated Bacterial Community of Tomato Plants in Response to Salt Stress. Agriculture, 15(6), 624. https://doi.org/10.3390/agriculture15060624








