Genome-Wide Association Study for the Capacity to Skip the Dry Period in Dairy Goats
Abstract
1. Introduction
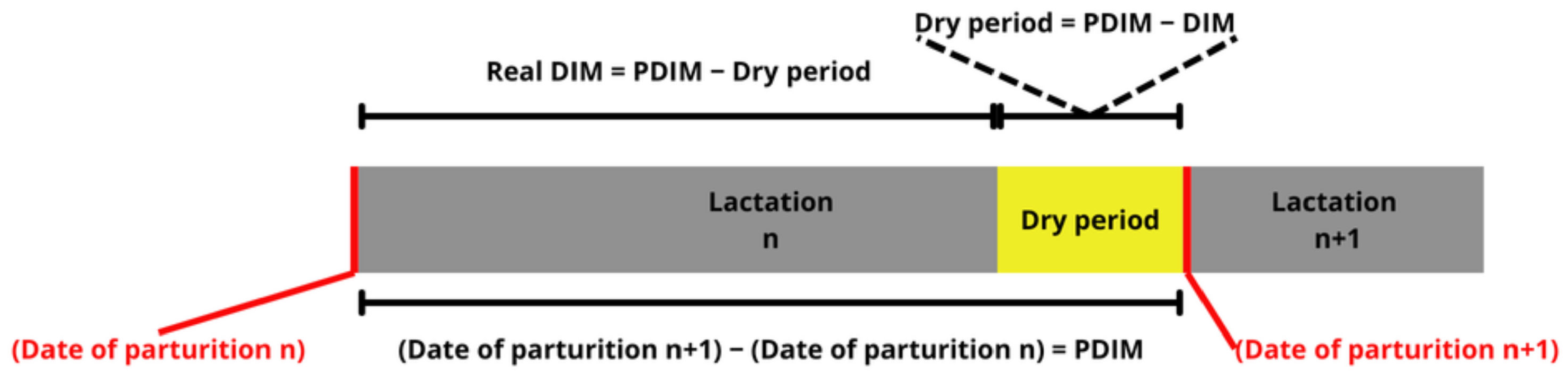
2. Materials and Methods
3. Results
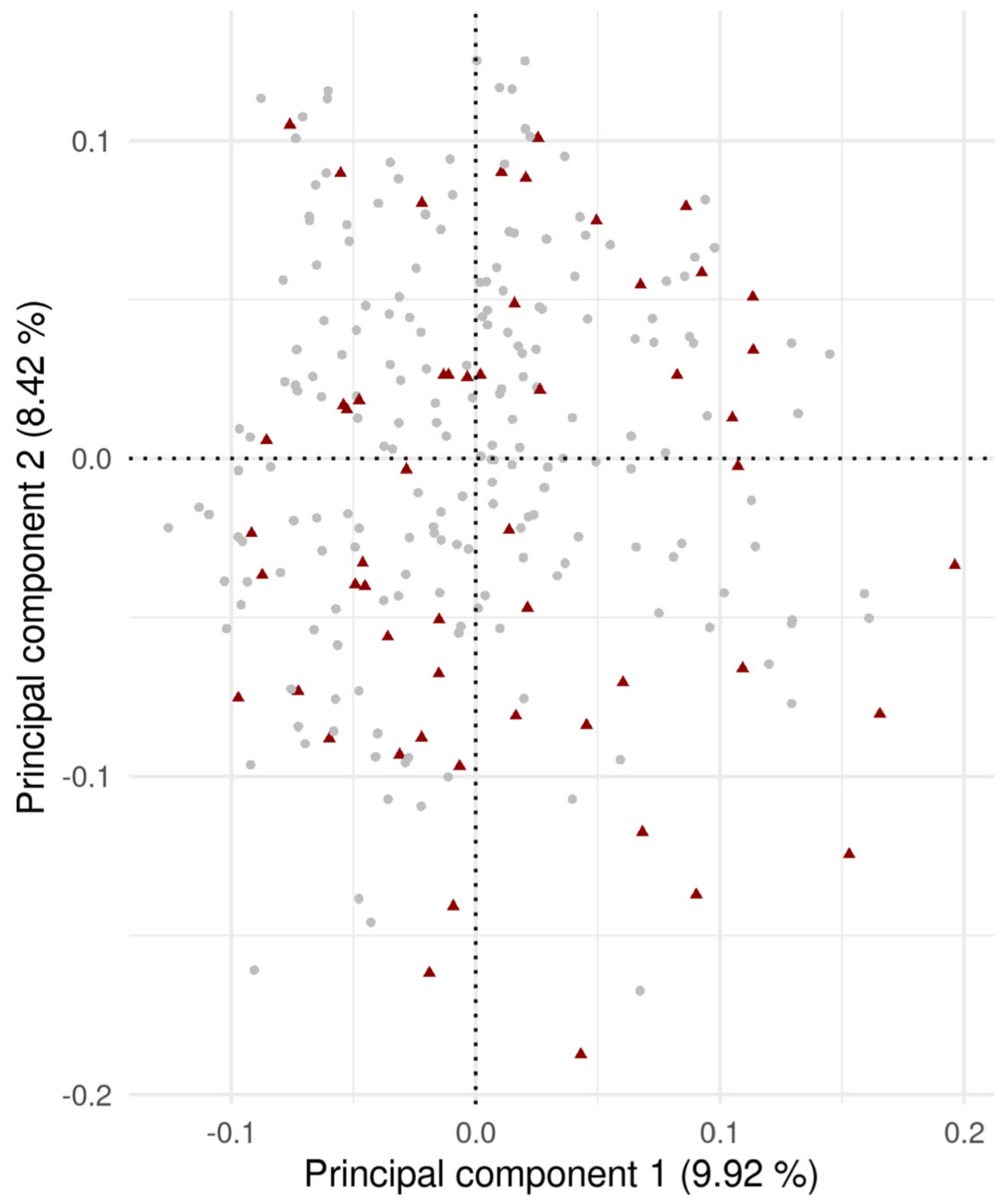
3.1. Variance Components and Heritability Estimation
3.2. GWAS
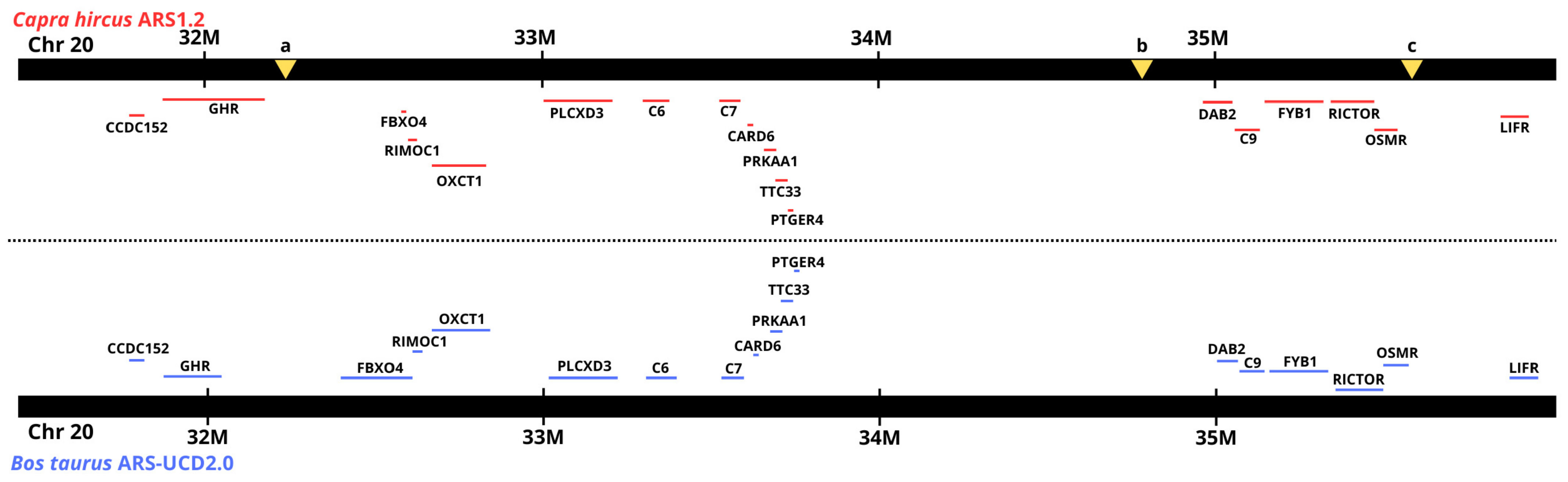
3.3. Gene and QTL Search
3.4. Functional Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Akers, R.M. Overview of Mammary Development. In Lactation and the Mammary Gland; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2016; pp. 3–44. ISBN 978-1-119-26488-0. [Google Scholar]
- Capuco, A.V.; Ellis, S.E.; Hale, S.A.; Long, E.; Erdman, R.A.; Zhao, X.; Paape, M.J. Lactation Persistency: Insights from Mammary Cell Proliferation Studies. J. Anim. Sci. 2003, 81, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Knight, C.H.; Peaker, M. Mammary Development and Regression During Lactation in Goats in Relation to Milk Secretion. Q. J. Exp. Physiol. 1984, 69, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Zobel, G.; Leslie, K.; Weary, D.M.; von Keyserlingk, M.A.G. Ketonemia in Dairy Goats: Effect of Dry Period Length and Effect on Lying Behavior. J. Dairy Sci. 2015, 98, 6128–6138. [Google Scholar] [CrossRef] [PubMed]
- Mezzetti, M.; Minuti, A.; Cattaneo, L.; Trevisi, E.; Bencetti, F.; Piccioli-Cappelli, F. Effects of Dry-off or Continuous Lactation in Alpine and Saanen Dairy Goats Carrying Single or Double Kids on Peripartum Metabolic Profile, Performances, and Milk Composition. Ital. J. Anim. Sci. 2024, 23, 227–240. [Google Scholar] [CrossRef]
- Caja, G.; Salama, A.A.K.; Such, X. Omitting the Dry-Off Period Negatively Affects Colostrum and Milk Yield in Dairy Goats. J. Dairy Sci. 2006, 89, 4220–4228. [Google Scholar] [CrossRef]
- NASA Prediction of Worldwide Energy Resources (POWER) POWER|DAV. Available online: https://power.larc.nasa.gov/data-access-viewer/ (accessed on 25 February 2025).
- Purcell, S.; Chang, C. PLINK 1.9. Available online: www.cog-genomics.org/plink/1.9/ (accessed on 25 February 2025).
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. GigaSci 2015, 4, 7. [Google Scholar] [CrossRef]
- Purcell, S.; Chang, C. PLINK 2.00 Alpha. Available online: www.cog-genomics.org/plink/2.0/ (accessed on 25 February 2025).
- Relationship Inference in KING. Available online: https://www.kingrelatedness.com/manual.shtml (accessed on 19 December 2024).
- Manichaikul, A.; Mychaleckyj, J.C.; Rich, S.S.; Daly, K.; Sale, M.; Chen, W.-M. Robust Relationship Inference in Genome-Wide Association Studies. Bioinformatics 2010, 26, 2867–2873. [Google Scholar] [CrossRef]
- Bickhart, D.M.; Rosen, B.D.; Koren, S.; Sayre, B.L.; Hastie, A.R.; Chan, S.; Lee, J.; Lam, E.T.; Liachko, I.; Sullivan, S.T.; et al. Single-Molecule Sequencing and Chromatin Conformation Capture Enable de Novo Reference Assembly of the Domestic Goat Genome. Nat. Genet. 2017, 49, 643–650. [Google Scholar] [CrossRef]
- Butler, D.G.; Cullis, B.R.; Gilmour, A.R.; Gogel, B.J.; Thompson, R. ASReml-R Reference Manual Version 4.2; VSN International Ltd.: Hemel Hempstead, UK, 2023. [Google Scholar]
- Genetic Data Analysis Software—ASReml-SA. Available online: https://vsni.co.uk/software/asreml-sa/ (accessed on 2 January 2025).
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A Tool for Genome-Wide Complex Trait Analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Krishna Kumar, S.; Feldman, M.W.; Rehkopf, D.H.; Tuljapurkar, S. Limitations of GCTA as a Solution to the Missing Heritability Problem. Proc. Natl. Acad. Sci. USA 2016, 113, E61–E70. [Google Scholar] [CrossRef]
- Goddard, M.E.; Hayes, B.J.; Meuwissen, T.H.E. Using the Genomic Relationship Matrix to Predict the Accuracy of Genomic Selection. J. Anim. Breed. Genet. 2011, 128, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Brito, L.F.; Jafarikia, M.; Grossi, D.A.; Kijas, J.W.; Porto-Neto, L.R.; Ventura, R.V.; Salgorzaei, M.; Schenkel, F.S. Characterization of Linkage Disequilibrium, Consistency of Gametic Phase and Admixture in Australian and Canadian Goats. BMC Genet. 2015, 16, 67. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. Dplyr: A Grammar of Data Manipulation. Available online: https://dplyr.tidyverse.org/ (accessed on 19 December 2024).
- Wickham, H. Data Analysis. In ggplot2: Elegant Graphics for Data Analysis; Wickham, H., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 189–201. ISBN 978-3-319-24277-4. [Google Scholar]
- Slowikowski, K. Ggrepel: Automatically Position Non-Overlapping Text Labels with “Ggplot2”. Available online: https://ggrepel.slowkow.com/ (accessed on 19 December 2024).
- Holtz, Y. Manhattan Plot in R: A Review. Available online: https://www.r-graph-gallery.com/101_Manhattan_plot.html (accessed on 19 December 2024).
- Fonseca, P.A.S.; Suárez-Vega, A.; Marras, G.; Cánovas, Á. GALLO: An R Package for Genomic Annotation and Integration of Multiple Data Sources in Livestock for Positional Candidate Loci. GigaScience 2020, 9, giaa149. [Google Scholar] [CrossRef] [PubMed]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. G:Profiler—Interoperable Web Service for Functional Enrichment Analysis and Gene Identifier Mapping (2023 Update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]
- Falconer, D.S.; Mackay, T.F.C. Chapter 8—Variance. In Introduction to Quantitative Genetics; Pearson, Prentice Hall: Harlow, UK, 1996; ISBN 0-582-24302-5. [Google Scholar]
- Rangwala, S.H.; Rudnev, D.V.; Ananiev, V.V.; Oh, D.-H.; Asztalos, A.; Benica, B.; Borodin, E.A.; Bouk, N.; Evgeniev, V.I.; Kodali, V.K.; et al. The NCBI Comparative Genome Viewer (CGV) Is an Interactive Visualization Tool for the Analysis of Whole-Genome Eukaryotic Alignments. PLoS Biol. 2024, 22, e3002405. [Google Scholar] [CrossRef]
- Jena, M.K.; Jaswal, S.; Kumar, S.; Mohanty, A.K. Molecular Mechanism of Mammary Gland Involution: An Update. Dev. Biol. 2019, 445, 145–155. [Google Scholar] [CrossRef]
- Watson, C.J.; Kreuzaler, P.A. Remodeling Mechanisms of the Mammary Gland during Involution. Int. J. Dev. Biol. 2011, 55, 757–762. [Google Scholar] [CrossRef]
- Lund, L.R.; Rømer, J.; Thomasset, N.; Solberg, H.; Pyke, C.; Bissell, M.J.; Danø, K.; Werb, Z. Two Distinct Phases of Apoptosis in Mammary Gland Involution: Proteinase-Independent and -Dependent Pathways. Development 1996, 122, 181–193. [Google Scholar] [CrossRef]
- OSMR Oncostatin M Receptor [Capra Hircus (Goat)]—Gene—NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene?term=(OSMR)%20AND%20Capra%5BOrganism%5D (accessed on 9 August 2024).
- Lantieri, F.; Bachetti, T. OSM/OSMR and Interleukin 6 Family Cytokines in Physiological and Pathological Condition. Int. J. Mol. Sci. 2022, 23, 11096. [Google Scholar] [CrossRef]
- Tiffen, P.G.; Omidvar, N.; Marquez-Almuina, N.; Croston, D.; Watson, C.J.; Clarkson, R.W.E. A Dual Role for Oncostatin M Signaling in the Differentiation and Death of Mammary Epithelial Cells in Vivo. Mol. Endocrinol. 2008, 22, 2677–2688. [Google Scholar] [CrossRef]
- Guan, D.; Landi, V.; Luigi-Sierra, M.G.; Delgado, J.V.; Such, X.; Castelló, A.; Cabrera, B.; Mármol-Sánchez, E.; Fernández-Alvarez, J.; de la Torre Casañas, J.L.R.; et al. Analyzing the Genomic and Transcriptomic Architecture of Milk Traits in Murciano-Granadina Goats. J. Anim. Sci. Biotechnol. 2020, 11, 35. [Google Scholar] [CrossRef]
- STAT3 Signal Transducer and Activator of Transcription 3 [Homo Sapiens (Human)]—Gene—NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/6774 (accessed on 16 August 2024).
- STAT3 Signal Transducer and Activator of Transcription 3 [Capra Hircus (Goat)]—Gene—NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/102186154 (accessed on 16 August 2024).
- Kritikou, E.A.; Sharkey, A.; Abell, K.; Came, P.J.; Anderson, E.; Clarkson, R.W.E.; Watson, C.J. A Dual, Non-Redundant, Role for LIF as a Regulator of Development and STAT3-Mediated Cell Death in Mammary Gland. Development 2003, 130, 3459–3468. [Google Scholar] [CrossRef] [PubMed]
- Baxter, F.O.; Neoh, K.; Tevendale, M.C. The Beginning of the End: Death Signaling in Early Involution. J. Mammary Gland Biol. Neoplasia 2007, 12, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Walker, E.C.; McGregor, N.E.; Poulton, I.J.; Solano, M.; Pompolo, S.; Fernandes, T.J.; Constable, M.J.; Nicholson, G.C.; Zhang, J.-G.; Nicola, N.A.; et al. Oncostatin M Promotes Bone Formation Independently of Resorption When Signaling through Leukemia Inhibitory Factor Receptor in Mice. J. Clin. Investig. 2010, 120, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Schuler, F.; Baumgartner, F.; Klepsch, V.; Chamson, M.; Müller-Holzner, E.; Watson, C.J.; Oh, S.; Hennighausen, L.; Tymoszuk, P.; Doppler, W.; et al. The BH3-Only Protein BIM Contributes to Late-Stage Involution in the Mouse Mammary Gland. Cell Death Differ. 2016, 23, 41–51. [Google Scholar] [CrossRef]
- Martins, K.R.; Haas, C.S.; Ferst, J.G.; Rovani, M.T.; Goetten, A.L.F.; Duggavathi, R.; Bordignon, V.; Portela, V.V.M.; Ferreira, R.; Gonçalves, P.B.D.; et al. Oncostatin M and Its Receptors mRNA Regulation in Bovine Granulosa and Luteal Cells. Theriogenology 2019, 125, 324–330. [Google Scholar] [CrossRef]
- Mao, S.; Dong, S.; Hou, B.; Li, Y.; Sun, B.; Guo, Y.; Deng, M.; Liu, D.; Liu, G. Transcriptome Analysis Reveals Pituitary lncRNA, circRNA and mRNA Affecting Fertility in High- and Low-Yielding Goats. Front. Genet. 2023, 14, 1303031. [Google Scholar] [CrossRef]
- Banos, G.; Bramis, G.; Bush, S.J.; Clark, E.L.; McCulloch, M.E.B.; Smith, J.; Schulze, G.; Arsenos, G.; Hume, D.A.; Psifidi, A. The Genomic Architecture of Mastitis Resistance in Dairy Sheep. BMC Genom. 2017, 18, 624. [Google Scholar] [CrossRef]
- Tiezzi, F.; Parker-Gaddis, K.L.; Cole, J.B.; Clay, J.S.; Maltecca, C. A Genome-Wide Association Study for Clinical Mastitis in First Parity US Holstein Cows Using Single-Step Approach and Genomic Matrix Re-Weighting Procedure. PLoS ONE 2015, 10, e0114919. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, J.; Sun, D.; Ma, P.; Ding, X.; Yu, Y.; Zhang, Q. Genome Wide Association Studies for Milk Production Traits in Chinese Holstein Population. PLoS ONE 2010, 5, e13661. [Google Scholar] [CrossRef]
- Yang, Z.; Lian, Z.; Liu, G.; Deng, M.; Sun, B.; Guo, Y.; Liu, D.; Li, Y. Identification of Genetic Markers Associated with Milk Production Traits in Chinese Holstein Cattle Based on Post Genome-Wide Association Studies. Anim. Biotechnol. 2021, 32, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Tribout, T.; Croiseau, P.; Lefebvre, R.; Barbat, A.; Boussaha, M.; Fritz, S.; Boichard, D.; Hoze, C.; Sanchez, M.-P. Confirmed Effects of Candidate Variants for Milk Production, Udder Health, and Udder Morphology in Dairy Cattle. Genet. Sel. Evol. 2020, 52, 55. [Google Scholar] [CrossRef] [PubMed]
- Viale, E.; Tiezzi, F.; Maretto, F.; De Marchi, M.; Penasa, M.; Cassandro, M. Association of Candidate Gene Polymorphisms with Milk Technological Traits, Yield, Composition, and Somatic Cell Score in Italian Holstein-Friesian Sires. J. Dairy Sci. 2017, 100, 7271–7281. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Bissonnette, N.; Lacasse, P.; Miglior, F.; Sargolzaei, M.; Zhao, X.; Ibeagha-Awemu, E.M. Genome-Wide Association Analysis and Pathways Enrichment for Lactation Persistency in Canadian Holstein Cattle. J. Dairy Sci. 2017, 100, 1955–1970. [Google Scholar] [CrossRef]
- Cheng, J.; Wang, W.; Zhang, D.; Zhang, Y.; Li, X.; Zhao, Y.; Xu, D.; Zhao, L.; Li, W.; Wang, J.; et al. Identification of Polymorphic Loci in OSMR and GHR Genes and Analysis of Their Association with Growth Traits in Sheep. Anim. Biotechnol. 2023, 34, 2546–2553. [Google Scholar] [CrossRef]
- Setiawati, R.; Rahardjo, P.; Setiawati, R.; Rahardjo, P. Bone Development and Growth. In Osteogenesis and Bone Regeneration; IntechOpen: London, UK, 2018; ISBN 978-1-78985-768-9. [Google Scholar]


| Chr | SNP rsID | Position | A1 | A2 | MAF | Va | %Va | |
|---|---|---|---|---|---|---|---|---|
| 20 | rs268262366 | 32099030 | G | A | 0.060 | 0.357 | 0.014 | 0.352 |
| 20 | rs268275254 | 34904494 | A | G | 0.183 | 0.232 | 0.016 | 0.391 |
| 20 | rs268283969 | 35561292 | A | G | 0.068 | 0.344 | 0.015 | 0.366 |
| Gene ID | Gene Name | Gene_Biotype |
|---|---|---|
| ENSCHIG00000000538 | RIMOC1 | protein coding |
| ENSCHIG00000000675 | SNORD72 | snoRNA |
| ENSCHIG00000001827 | U2 | snRNA |
| ENSCHIG00000002172 | - | protein coding |
| ENSCHIG00000003882 | - | lincRNA |
| ENSCHIG00000003885 | - | lincRNA |
| ENSCHIG00000009367 | OSMR | protein coding |
| ENSCHIG00000009529 | DAB2 | protein coding |
| ENSCHIG00000012124 | OXCT1 | protein coding |
| ENSCHIG00000012521 | - | protein coding |
| ENSCHIG00000014444 | RPL37 | protein coding |
| ENSCHIG00000014791 | C9 | protein coding |
| ENSCHIG00000016536 | C7 | protein coding |
| ENSCHIG00000016815 | PLCXD3 | protein coding |
| ENSCHIG00000017495 | PRKAA1 | protein coding |
| ENSCHIG00000018413 | FYB1 | protein coding |
| ENSCHIG00000018928 | GHR | protein coding |
| ENSCHIG00000020642 | TTC33 | protein coding |
| ENSCHIG00000020994 | CARD6 | protein coding |
| ENSCHIG00000021210 | C6 | protein coding |
| ENSCHIG00000021750 | FBXO4 | protein coding |
| ENSCHIG00000022858 | RICTOR | protein coding |
| ENSCHIG00000023835 | PTGER4 | protein coding |
| Source | Term Name | Term ID | FDR * |
|---|---|---|---|
| GO:MF | oncostatin-M receptor activity | GO:0004924 | 0.001732702 |
| GO:MF | cytokine receptor activity | GO:0004896 | 0.013572831 |
| GO:MF | growth factor binding | GO:0019838 | 0.013572831 |
| GO:MF | immune receptor activity | GO:0140375 | 0.013572831 |
| GO:BP | oncostatin-M-mediated signaling pathway | GO:0038165 | 0.005157358 |
| GO:CC | oncostatin-M receptor complex | GO:0005900 | 0.001577731 |
| GO:CC | apical plasma membrane | GO:0016324 | 0.04212543 |
| GO:CC | receptor complex | GO:0043235 | 0.04212543 |
| GO:CC | apical part of cell | GO:0045177 | 0.04212543 |
| GO:CC | plasma membrane signaling receptor complex | GO:0098802 | 0.04212543 |
| HP | cutaneous amyloidosis | HP:0012309 | 0.029205386 |
| HP | lichenification | HP:0100725 | 0.029205386 |
| Source | Term Name | Term ID | FDR * |
|---|---|---|---|
| GO:MF | cytokine receptor activity | GO:0004896 | 0.0374 |
| GO:MF | histone H2B kinase activity | GO:0140998 | 0.0374 |
| GO:MF | histone H2BS36 kinase activity | GO:0140823 | 0.0374 |
| GO:MF | immune receptor activity | GO:0140375 | 0.0374 |
| GO:MF | [acetyl-CoA carboxylase] kinase activity | GO:0050405 | 0.0374 |
| GO:MF | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity | GO:0047322 | 0.0374 |
| GO:MF | succinyl-CoA:3-oxo-acid CoA-transferase activity | GO:0008260 | 0.0374 |
| GO:MF | oncostatin-M receptor activity | GO:0004924 | 0.0374 |
| GO:MF | CoA-transferase activity | GO:0008410 | 0.0493 |
| GO:MF | prostaglandin E receptor activity | GO:0004957 | 0.0493 |
| GO:MF | AMP-activated protein kinase activity | GO:0004679 | 0.0493 |
| GO:CC | membrane attack complex | GO:0005579 | 0.0000 |
| GO:CC | pore complex | GO:0046930 | 0.0001 |
| GO:CC | plasma membrane protein complex | GO:0098797 | 0.0111 |
| GO:CC | growth hormone receptor complex | GO:0070195 | 0.0194 |
| GO:CC | oncostatin-M receptor complex | GO:0005900 | 0.0310 |
| KEGG | systemic lupus erythematosus | KEGG:05322 | 0.0097 |
| KEGG | coronavirus disease—COVID-19 | KEGG:05171 | 0.0097 |
| KEGG | complement and coagulation cascades | KEGG:04610 | 0.0097 |
| HP | decreased circulating terminal complement component concentration | HP:0033057 | 0.0000 |
| HP | abnormality of complement system | HP:0005339 | 0.0022 |
| HP | complement deficiency | HP:0004431 | 0.0022 |
| HP | recurrent meningococcal disease | HP:0005381 | 0.0022 |
| HP | recurrent Neisserial infections | HP:0005430 | 0.0022 |
| HP | recurrent Gram-negative bacterial infections | HP:0005420 | 0.0375 |
| HP | decreased circulating complement C9 concentration | HP:0012308 | 0.0387 |
| HP | decreased circulating complement C6 concentration | HP:0033059 | 0.0387 |
| HP | decreased circulating complement C7 concentration | HP:0033058 | 0.0387 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galindo, B.A.; Massender, E.; Hermisdorff, I.C.; Schenkel, F.S. Genome-Wide Association Study for the Capacity to Skip the Dry Period in Dairy Goats. Agriculture 2025, 15, 622. https://doi.org/10.3390/agriculture15060622
Galindo BA, Massender E, Hermisdorff IC, Schenkel FS. Genome-Wide Association Study for the Capacity to Skip the Dry Period in Dairy Goats. Agriculture. 2025; 15(6):622. https://doi.org/10.3390/agriculture15060622
Chicago/Turabian StyleGalindo, Bruno A., Erin Massender, Isis C. Hermisdorff, and Flavio S. Schenkel. 2025. "Genome-Wide Association Study for the Capacity to Skip the Dry Period in Dairy Goats" Agriculture 15, no. 6: 622. https://doi.org/10.3390/agriculture15060622
APA StyleGalindo, B. A., Massender, E., Hermisdorff, I. C., & Schenkel, F. S. (2025). Genome-Wide Association Study for the Capacity to Skip the Dry Period in Dairy Goats. Agriculture, 15(6), 622. https://doi.org/10.3390/agriculture15060622







