Detection and Segmentation of Chip Budding Graft Sites in Apple Nursery Using YOLO Models
Abstract
1. Introduction
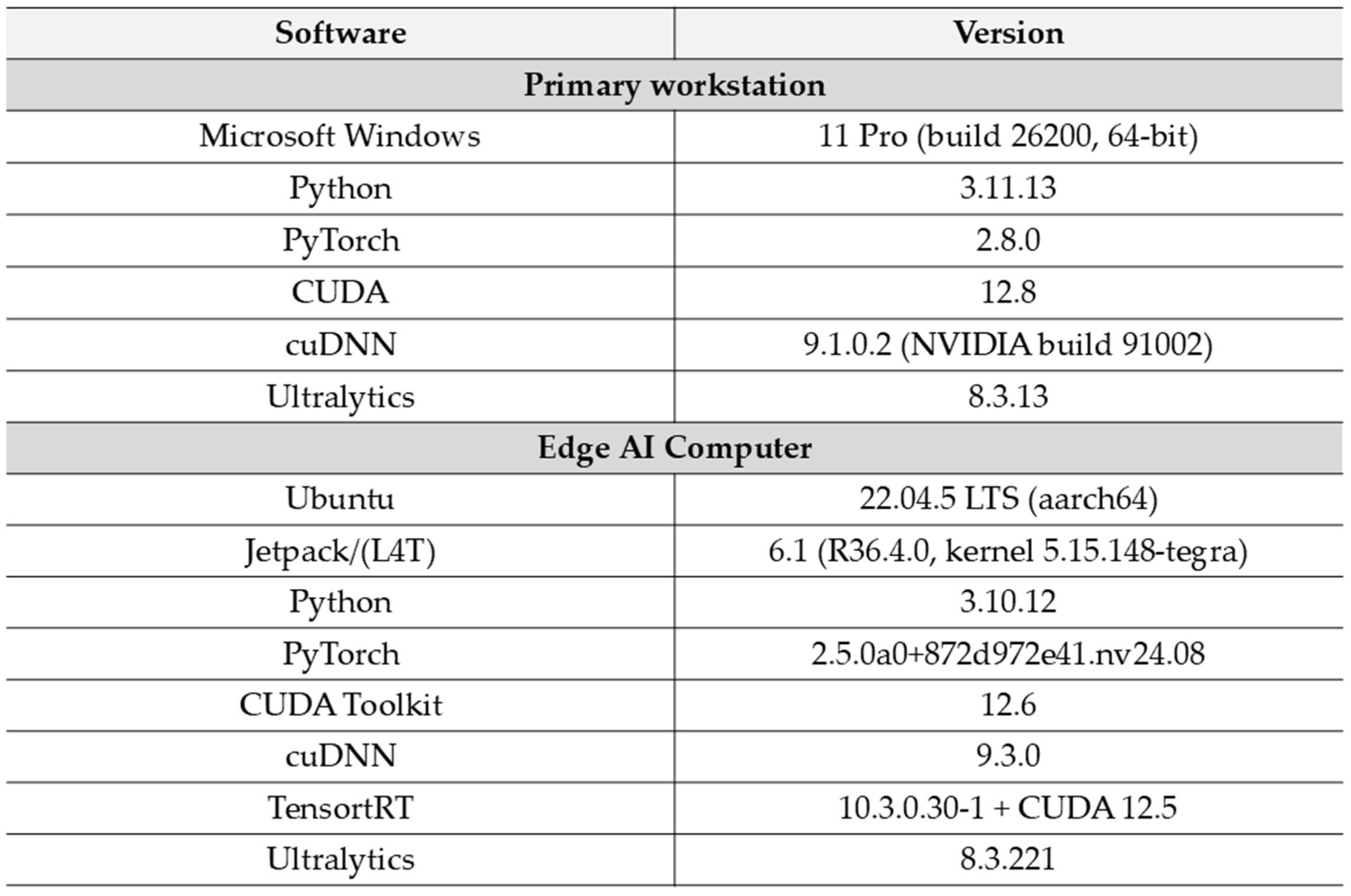
2. Materials and Methods
2.1. Data Collection
2.2. Data Preprocessing
2.3. YOLO Models Training
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jin, S.; Li, W.; Cao, Y.; Jones, G.; Chen, J.; Li, Z.; Chang, Q.; Yang, G.; Frewer, L.J. Identifying Barriers to Sustainable Apple Production: A Stakeholder Perspective. J. Environ. Manag. 2022, 302, 114082. [Google Scholar] [CrossRef]
- Cheng, J.; Yu, J.; Tan, D.; Wang, Q.; Zhao, Z. Life Cycle Assessment of Apple Production and Consumption under Different Sales Models in China. Sustain. Prod. Consum. 2025, 55, 100–116. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations (FAO). Available online: https://www.fao.org/faostat/en/#data/qcl (accessed on 19 October 2025).
- Wieczorek, R.; Zydlik, Z.; Zydlik, P. Biofumigation Treatment Using Tagetes Patula, Sinapis Alba and Raphanus Sativus Changes the Biological Properties of Replanted Soil in a Fruit Tree Nursery. Agriculture 2024, 14, 1023. [Google Scholar] [CrossRef]
- Szot, I.; Lipa, T. Apple trees yielding and fruit quality depending on the crop load, branch type and position in the crown. Acta Sci. Pol. Hortorum Cultus 2019, 18, 205–215. [Google Scholar] [CrossRef]
- Kapłan, M.; Jurkowski, G.; Krawiec, M.; Borowy, A.; Wójcik, I.; Palonka, S. Wpływ Zabiegów Stymulujących Rozgałęzianie Na Jakość Okulantów Jabłoni. Ann. Hortic. 2018, 27, 5–20. [Google Scholar] [CrossRef]
- Kapłan, M.; Klimek, K.E.; Buczyński, K. Analysis of the Impact of Treatments Stimulating Branching on the Quality of Maiden Apple Trees. Agriculture 2024, 14, 1757. [Google Scholar] [CrossRef]
- Kapłan, M.; Klimek, K.; Borkowska, A.; Buczyński, K. Effect of Growth Regulators on the Quality of Apple Tree Whorls. Appl. Sci. 2023, 13, 11472. [Google Scholar] [CrossRef]
- Kumawat, K.L.; Raja, W.H.; Nabi, S.U. Quality of Nursery Trees Is Critical for Optimal Growth and Inducing Precocity in Apple. Appl. Fruit Sci. 2024, 66, 2135–2143. [Google Scholar] [CrossRef]
- Zydlik, Z.; Zydlik, P.; Jarosz, Z.; Wieczorek, R. The Use of Organic Additives for Replanted Soil in Apple Tree Production in a Fruit Tree Nursery. Agriculture 2023, 13, 973. [Google Scholar] [CrossRef]
- Muder, A.; Garming, H.; Dreisiebner-Lanz, S.; Kerngast, K.; Rosner, F.; Kličková, K.; Kurthy, G.; Cimer, K.; Bertazzoli, A.; Altamura, V.; et al. Apple Production and Apple Value Chains in Europe. Eur. J. Hortic. Sci. 2022, 87, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Baima, L.; Nari, L.; Nari, D.; Bossolasco, A.; Blanc, S.; Brun, F. Sustainability Analysis of Apple Orchards: Integrating Environmental and Economic Perspectives. Heliyon 2024, 10, e38397. [Google Scholar] [CrossRef] [PubMed]
- Mika, A. Szczepienie i Inne Metody Rozmnażania Roślin Sadowniczych; Powszechne Wydawnictwo Rolnicze i Leśne SP. z o.o.: Warszawa, Poland, 2014. [Google Scholar]
- Lipecki, J.; Jacyna, T.; Lipa, T.; Szot, I. The Quality of Apple Nursery Trees of Knip-Boom Type as Affected by the Methods of Propagation. Acta Sci. Pol. Hortorum Cultus 2013, 12, 157–165. [Google Scholar]
- Lipecki, J.; Szot, I.; Lipa, T. The Effect of Cultivar on the Growth and Relations between Growth Characters in “Knip-Boom” Apple Trees. Acta Sci. Pol. Hortorum Cultus 2014, 13, 139–148. [Google Scholar]
- Gupta, G.; Kumar Pal, S. Applications of AI in Precision Agriculture. Discov. Agric. 2025, 3, 61. [Google Scholar] [CrossRef]
- Raj, M.; Prahadeeswaran, M. Revolutionizing Agriculture: A Review of Smart Farming Technologies for a Sustainable Future. Discov. Appl. Sci. 2025, 7, 937. [Google Scholar] [CrossRef]
- Vijayakumar, S.; Murugaiyan, V.; Ilakkiya, S.; Kumar, V.; Sundaram, R.M.; Kumar, R.M. Opportunities, Challenges, and Interventions for Agriculture 4.0 Adoption. Discov. Food 2025, 5, 265. [Google Scholar] [CrossRef]
- Khan, Z.; Shen, Y.; Liu, H. ObjectDetection in Agriculture: A Comprehensive Review of Methods, Applications, Challenges, and Future Directions. Agriculture 2025, 15, 1351. [Google Scholar] [CrossRef]
- Szeliski, R. Computer Vision: Algorithms and Applications; Texts in Computer Science; Springer International Publishing: Cham, Switzerland, 2022; ISBN 978-3-030-34371-2. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- García-Navarrete, O.L.; Correa-Guimaraes, A.; Navas-Gracia, L.M. Application of Convolutional Neural Networks in Weed Detection and Identification: A Systematic Review. Agriculture 2024, 14, 568. [Google Scholar] [CrossRef]
- Tugrul, B.; Elfatimi, E.; Eryigit, R. Convolutional Neural Networks in Detection of Plant Leaf Diseases: A Review. Agriculture 2022, 12, 1192. [Google Scholar] [CrossRef]
- Gu, J.; Wang, Z.; Kuen, J.; Ma, L.; Shahroudy, A.; Shuai, B.; Liu, T.; Wang, X.; Wang, G.; Cai, J.; et al. Recent Advances in Convolutional Neural Networks. Pattern Recognit. 2018, 77, 354–377. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; IEEE: Las Vegas, NV, USA, 2016; pp. 779–788. [Google Scholar]
- Wang, C.; Song, C.; Xu, T.; Jiang, R. Precision Weeding in Agriculture: A Comprehensive Review of Intelligent Laser Robots Leveraging Deep Learning Techniques. Agriculture 2025, 15, 1213. [Google Scholar] [CrossRef]
- Samsung. Galaxy S25. Available online: https://www.samsung.com/pl/smartphones/galaxy-s25/specs/ (accessed on 3 November 2025).
- Kamilczynski. GitHub Repository 2025. Available online: https://github.com/kamilczynski/Detection-of-Chip-Budding-Graft-Sites-in-Apple-Nursery-Using-YOLO-Models (accessed on 9 November 2025).
- Tzutalin. LabelImg: Image Annotation Tool. Version 1.8.6, GitHub Repository. 2015. Available online: https://github.com/Tzutalin/labelImg (accessed on 1 June 2025).
- Wkentaro. Labelme: Image Annotation Tool. Version 5.9.1, GitHub Repository. 2025. Available online: https://github.com/Wkentaro/Labelme (accessed on 2 June 2025).
- Ultralytics. Models. Available online: https://docs.ultralytics.com/models/ (accessed on 2 November 2025).
- Pawikhum, K.; Yang, Y.; He, L.; Heinemann, P. Development of a Machine Vision System for Apple Bud Thinning in Precision Crop Load Management. Comput. Electron. Agric. 2025, 236, 110479. [Google Scholar] [CrossRef]
- Sahu, R.; He, L. Real-Time Bud Detection Using Yolov4 for Automatic Apple Flower Bud Thinning. In Proceedings of the 2023 ASABE Annual International Meeting, Omaha, NE, USA, 9–12 July 2023; American Society of Agricultural and Biological Engineers: St. Joseph, MI, USA, 2023. [Google Scholar]
- Tong, S.; Zhang, J.; Li, W.; Wang, Y.; Kang, F. An Image-Based System for Locating Pruning Points in Apple Trees Using Instance Segmentation and RGB-D Images. Biosyst. Eng. 2023, 236, 277–286. [Google Scholar] [CrossRef]
- Zhang, J.; He, L.; Karkee, M.; Zhang, Q.; Zhang, X.; Gao, Z. Branch Detection for Apple Trees Trained in Fruiting Wall Architecture Using Depth Features and Regions-Convolutional Neural Network (R-CNN). Comput. Electron. Agric. 2018, 155, 386–393. [Google Scholar] [CrossRef]
- Li, G.; Suo, R.; Zhao, G.; Gao, C.; Fu, L.; Shi, F.; Dhupia, J.; Li, R.; Cui, Y. Real-Time Detection of Kiwifruit Flower and Bud Simultaneously in Orchard Using YOLOv4 for Robotic Pollination. Comput. Electron. Agric. 2022, 193, 106641. [Google Scholar] [CrossRef]
- Dang, H.; He, L.; Shi, Y.; Janneh, L.L.; Liu, X.; Chen, C.; Li, R.; Ye, H.; Chen, J.; Majeed, Y.; et al. Growth Characteristics Based Multi-Class Kiwifruit Bud Detection with Overlap-Partitioning Algorithm for Robotic Thinning. Comput. Electron. Agric. 2025, 229, 109715. [Google Scholar] [CrossRef]
- Oliveira, F.; Da Silva, D.Q.; Filipe, V.; Pinho, T.M.; Cunha, M.; Cunha, J.B.; Dos Santos, F.N. Enhancing Grapevine Node Detection to Support Pruning Automation: Leveraging State-of-the-Art YOLO Detection Models for 2D Image Analysis. Sensors 2024, 24, 6774. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, K. Exploring TensorRT to Improve Real-Time Inference for Deep Learning. In Proceedings of the 2022 IEEE 24th Int Conf on High Performance Computing & Communications; 8th Int Conf on Data Science & Systems; 20th Int Conf on Smart City; 8th Int Conf on Dependability in Sensor, Cloud & Big Data Systems & Application (HPCC/DSS/SmartCity/DependSys), Hainan, China, 18–20 December 2022; IEEE: Hainan, China, 2022; pp. 2011–2018. [Google Scholar]
- Ultralytics. Available online: https://docs.ultralytics.com/integrations/tensorrt/ (accessed on 11 November 2025).
- Ultralytics. Available online: https://docs.ultralytics.com/modes/benchmark/ (accessed on 10 November 2025).




| Train | Photos | 2904 |
| Tagged objects | 3887 | |
| Valid | Photos | 363 |
| Tagged objects | 480 | |
| Test | Photos | 363 |
| Tagged objects | 481 |
| Task | Model | Precision | Recall | mAP50 | mAP50:95 | F1-Score |
|---|---|---|---|---|---|---|
| Detection | YOLOv8s | 0.984 | 0.981 | 0.993 | 0.695 | 0.982 |
| YOLOv9s | 0.963 | 0.985 | 0.993 | 0.693 | 0.974 | |
| YOLOv10s | 0.975 | 0.980 | 0.993 | 0.691 | 0.978 | |
| YOLO11s | 0.979 | 0.979 | 0.994 | 0.701 | 0.979 | |
| YOLO12s | 0.992 | 0.975 | 0.994 | 0.706 | 0.983 | |
| Segmentation | YOLOv8s-seg | 0.982 | 0.984 | 0.994 | 0.683 | 0.983 |
| YOLO11s-seg | 0.988 | 0.978 | 0.994 | 0.679 | 0.983 |
| Task | Model | FPS | Latency (ms) | Accuracy Speed Ratio |
|---|---|---|---|---|
| Detection | YOLOv8s | 247.55 ± 1.36 | 4.04 ± 0.02 | 0.0028 ± 0.00002 |
| YOLOv9s | 98.66 ± 0.46 | 10.14 ± 0.05 | 0.0070 ± 0.00003 | |
| YOLOv10s | 209.66 ± 1.04 | 4.77 ± 0.02 | 0.0033 ± 0.00002 | |
| YOLO11s | 199.65 ± 0.50 | 5.01 ± 0.01 | 0.0035 ± 0.00001 | |
| YOLO12s | 148.00 ± 0.19 | 6.76 ± 0.01 | 0.0048 ± 0.00001 | |
| Segmentation | YOLOv8s-seg | 214.90 ± 0.75 | 4.65 ± 0.02 | 0.0032 ± 0.00001 |
| YOLO11s-seg | 177.64 ± 1.27 | 5.63 ± 0.04 | 0.0038 ± 0.00003 |
| Task | Model | FPS | Latency (ms) | Accuracy Speed Ratio |
|---|---|---|---|---|
| Detection | YOLOv8s | 53.61 ± 0.33 | 18.65 ± 0.11 | 0.0130 ± 0.00008 |
| YOLOv9s | 23.93 ± 0.05 | 41.80 ± 0.08 | 0.0290 ± 0.00006 | |
| YOLOv10s | 48.67 ± 0.22 | 20.55 ± 0.09 | 0.0142 ± 0.00006 | |
| YOLO11s | 46.78 ± 0.22 | 21.38 ± 0.10 | 0.0150 ± 0.00007 | |
| YOLO12s | 32.34 ± 0.13 | 30.92 ± 0.12 | 0.0218 ± 0.00009 | |
| Segmentation | YOLOv8s-seg | 44.56 ± 0.13 | 22.44 ± 0.07 | 0.0153 ± 0.00005 |
| YOLO11s-seg | 42.14 ± 0.08 | 23.73 ± 0.05 | 0.0161 ± 0.00003 |
| Task | Model | FPS | +ΔFPS | Latency (ms) | −ΔLatency (ms) | Accuracy Speed Ratio | −ΔAccuracy Speed Ratio |
|---|---|---|---|---|---|---|---|
| Detection | YOLOv8s | 65.33 ± 0.52 | 11.72 (21.86%) | 15.31 ± 0.12 | 3.34 (17.91%) | 0.0106 ± 0.00009 | 0.0024 (18.46%) |
| YOLOv9s | 55.05 ± 0.58 | 31.12 (130.05%) | 18.17 ± 0.19 | 23.63 (56.53%) | 0.0126 ± 0.00013 | 0.0164 (56.55%) | |
| YOLOv10s | 74.52 ± 0.22 | 25.85 (53.11%) | 13.42 ± 0.04 | 7.13 (34.70%) | 0.0093 ± 0.00003 | 0.0049 (34.51%) | |
| YOLO11s | 65.80 ± 0.25 | 19.02 (40.66%) | 15.20 ± 0.06 | 6.18 (28.91%) | 0.0107 ± 0.00004 | 0.0043 (28.67%) | |
| YOLO12s | 48.50 ± 0.41 | 16.16 (49.97%) | 20.62 ± 0.17 | 10.30 (33.31%) | 0.0146 ± 0.00012 | 0.0072 (33.03%) | |
| Segmentation | YOLOv8s-seg | 62.85 ± 1.04 | 18.29 (41.05%) | 15.91 ± 0.26 | 6.53 (29.10%) | 0.0109 ± 0.00018 | 0.0044 (28.76%) |
| YOLO11s-seg | 66.00 ± 0.15 | 23.86 (56.62%) | 15.15 ± 0.03 | 8.58 (36.16%) | 0.0103 ± 0.00002 | 0.0058 (36.02%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kapłan, M.; Wójcik, D.I.; Buczyński, K. Detection and Segmentation of Chip Budding Graft Sites in Apple Nursery Using YOLO Models. Agriculture 2025, 15, 2565. https://doi.org/10.3390/agriculture15242565
Kapłan M, Wójcik DI, Buczyński K. Detection and Segmentation of Chip Budding Graft Sites in Apple Nursery Using YOLO Models. Agriculture. 2025; 15(24):2565. https://doi.org/10.3390/agriculture15242565
Chicago/Turabian StyleKapłan, Magdalena, Damian I. Wójcik, and Kamil Buczyński. 2025. "Detection and Segmentation of Chip Budding Graft Sites in Apple Nursery Using YOLO Models" Agriculture 15, no. 24: 2565. https://doi.org/10.3390/agriculture15242565
APA StyleKapłan, M., Wójcik, D. I., & Buczyński, K. (2025). Detection and Segmentation of Chip Budding Graft Sites in Apple Nursery Using YOLO Models. Agriculture, 15(24), 2565. https://doi.org/10.3390/agriculture15242565






