Molecular and Agro-Morphological Diversity of Undercharacterized Local Bread Wheat Genetic Resources from Serbia and Bulgaria
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Phenotypic Evaluation
2.3. Molecular Characterization
2.4. Data Analysis
3. Results
3.1. Phenotypic Variation of Plant Height and Flowering Time
3.2. Genetic Diversity Within Cultivars and Landraces Based on SSR Markers
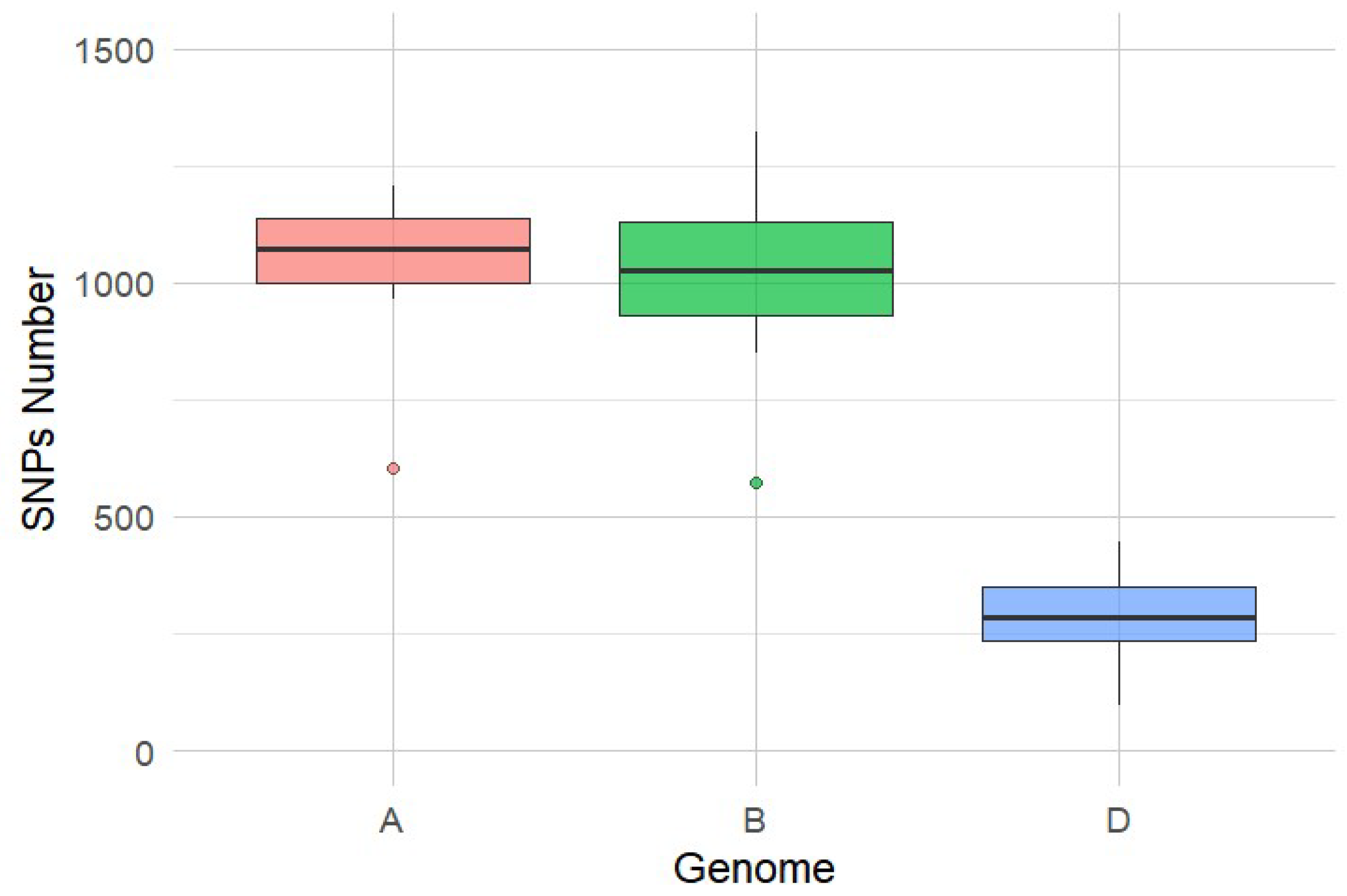
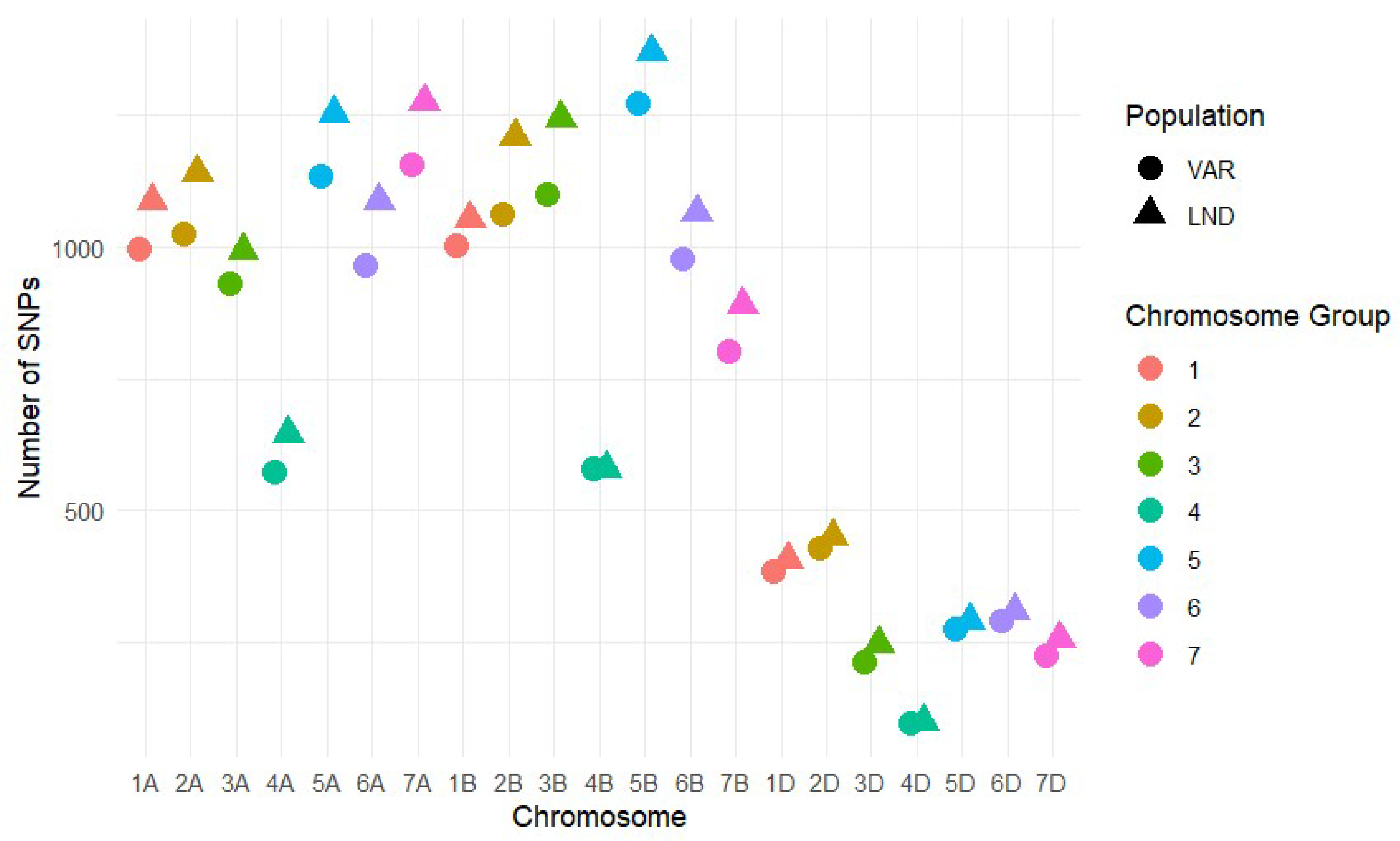
3.3. Genetic Diversity Within Cultivars and Landraces Based on SNPs
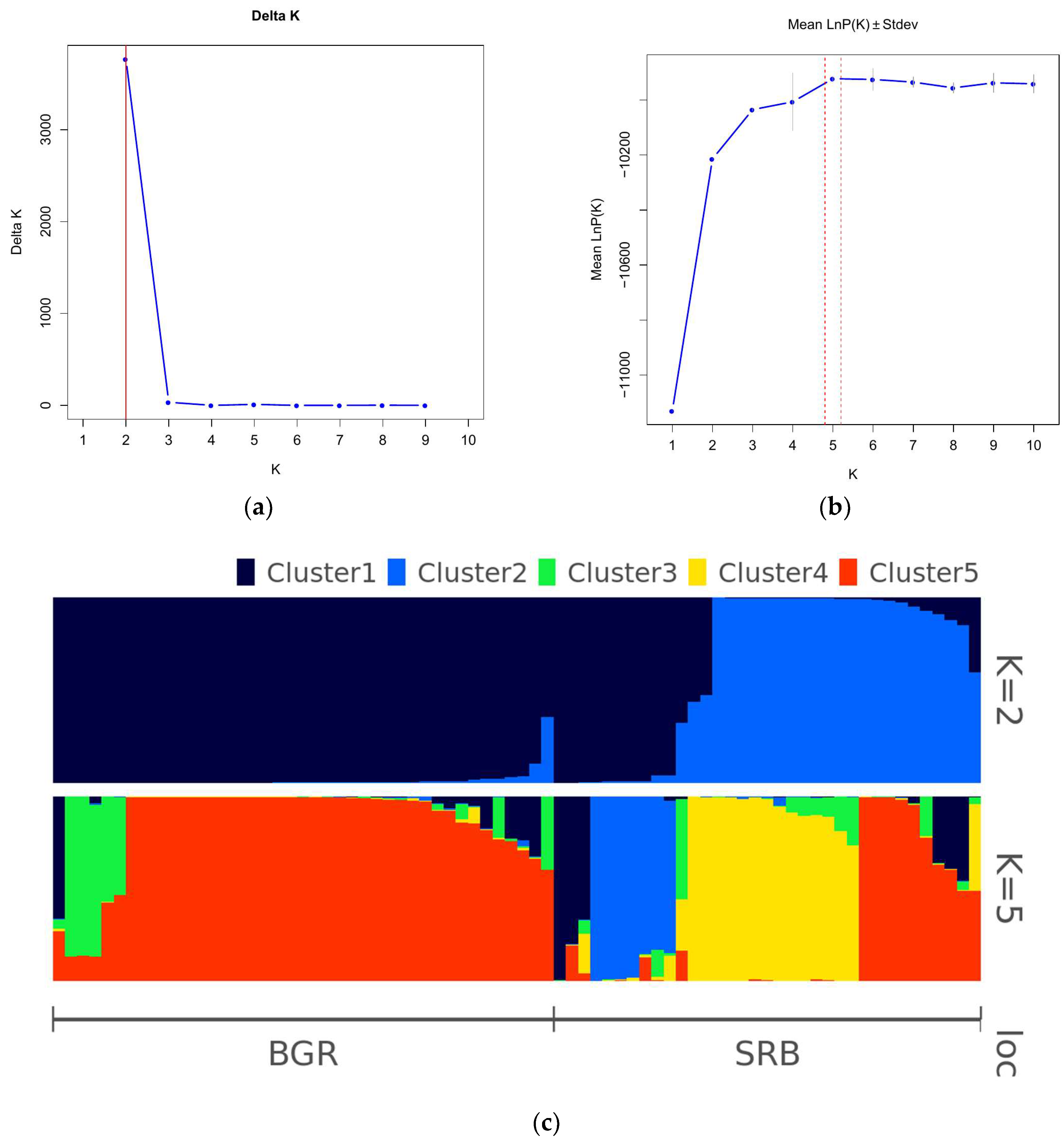
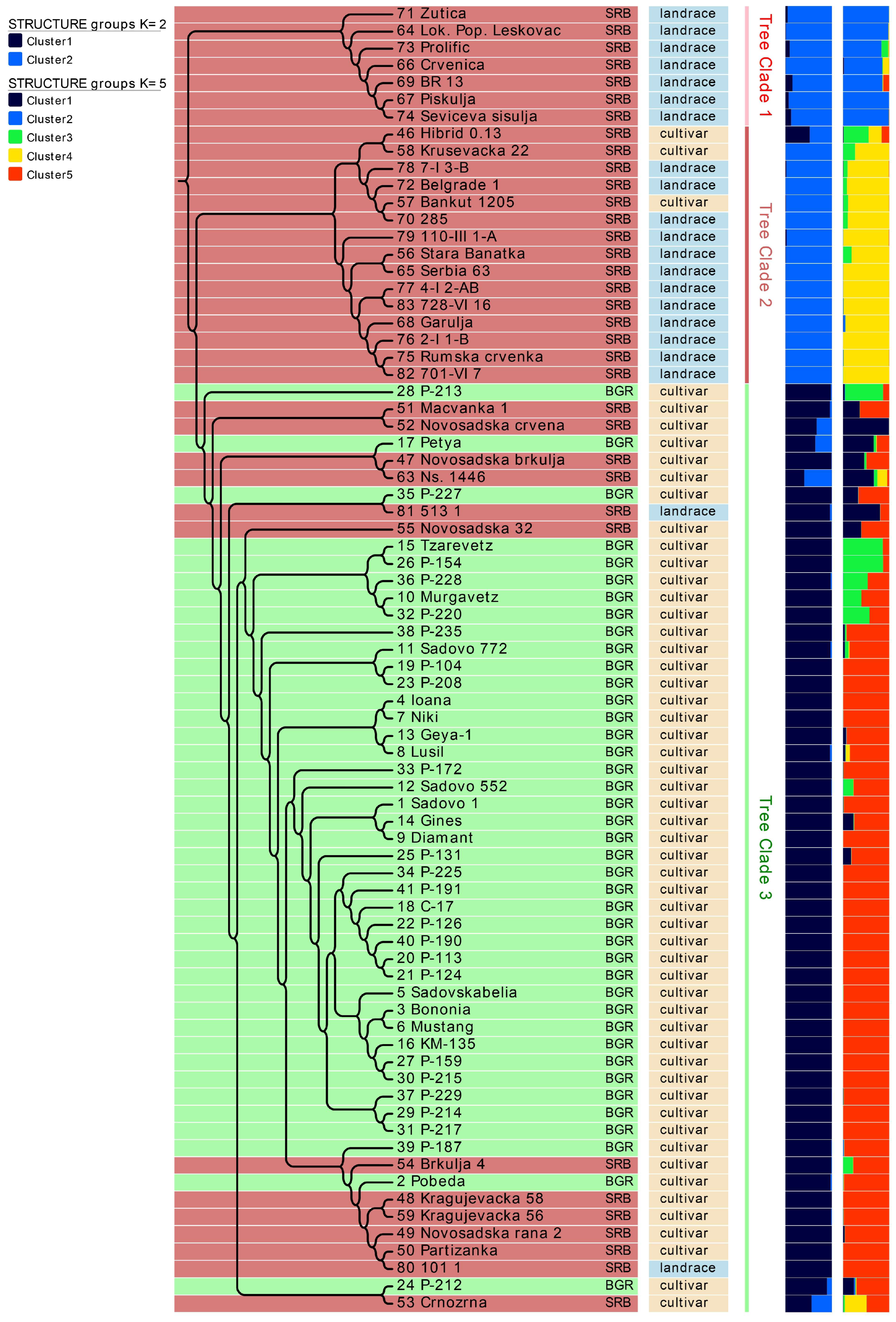
3.4. Population Structure and UPGMA Analysis
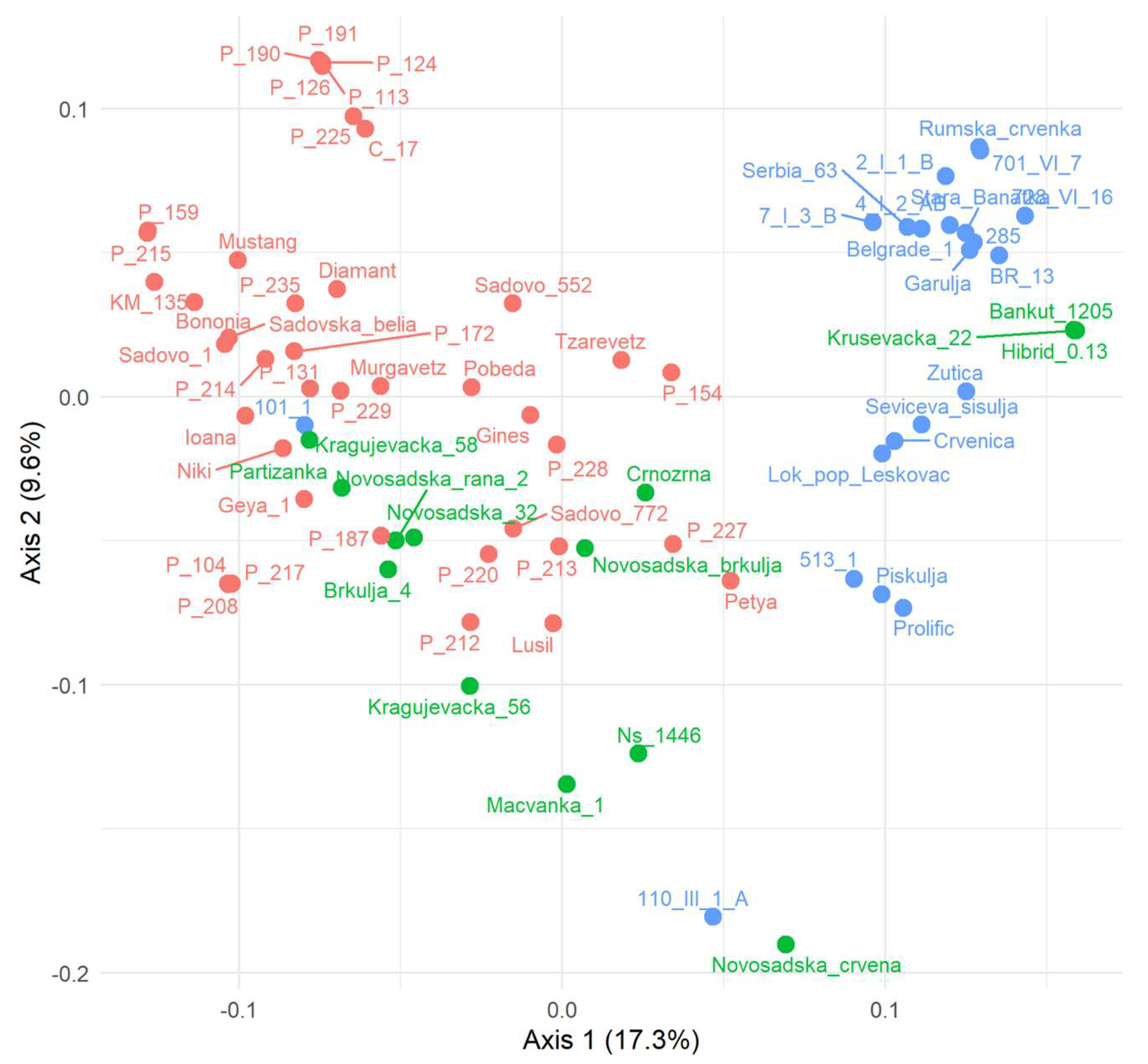
3.5. PCoA Based on SNPs and Clustering Pattern Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PIC | Polymorphism Information Content |
| UPGMA | Unweighted Pair Group Method with Arithmetic mean |
| SSR | Simple Sequence Repeat |
| SNP | Single-Nucleotide Polymorphism |
| FT | Flowering Time |
| PH | Plant Height |
| DNA | DeoxyriboNucleic Acid |
| PCR | Polymerase Chain Reaction |
| CTAB | CetylTrimethylAmmonium Bromide |
| dNTP | DeoxyNucleotideTriPhosphates |
| MAF | Major Allele Frequency |
| ANOVA | Analysis of Variance |
| GD | Gene Diversity |
| He | Expected Heterozigosity |
| Ho | Observed Heterozigosity |
| PCoA | Principal Coordinate Analysis |
References
- Korpetis, E.; Ninou, E.; Mylonas, I.; Ouzounidou, G.; Xynias, I.N.; Mavromatis, A.G. Bread wheat landraces adaptability to low-input agriculture. Plants 2023, 12, 2561. [Google Scholar] [CrossRef]
- Villa, T.C.C.; Maxted, N.; Scholten, M.; Ford-Lloyd, B. Defining and identifying crop landraces. Plant Genet. Resour. 2005, 3, 373–384. [Google Scholar] [CrossRef]
- Berg, T. Landraces and folk varieties: A conceptual reappraisal of terminology. Euphytica 2009, 166, 423–430. [Google Scholar] [CrossRef]
- Landjeva, S.; Ganeva, G.; Korzun, V.; Palejev, D.; Chebotar, S.; Kudrjavtsev, A. Genetic diversity of old bread wheat germplasm from the Black Sea region evaluated by microsatellites and agronomic traits. Plant Genet. Resour. 2015, 13, 119–130. [Google Scholar] [CrossRef]
- Khoury, C.K.; Brush, S.; Costich, D.E.; Curry, H.A.; De Haan, S.; Engels, J.M.; Guarino, L.; Hoban, S.; Mercer, K.L.; Miller, A.J.; et al. Crop genetic erosion: Understanding and responding to loss of crop diversity. New Phytol. 2022, 233, 84–118. [Google Scholar] [CrossRef]
- Cseh, A.; Poczai, P.; Kiss, T.; Balla, K.; Berki, Z.; Horváth, Á.; Kuti, C.; Karsai, I. Exploring the legacy of Central European historical winter wheat landraces. Sci. Rep. 2021, 11, 23915. [Google Scholar] [CrossRef]
- Aleksandrov, V.; Kartseva, T.; Alqudah, A.M.; Kocheva, K.; Tasheva, K.; Börner, A.; Misheva, S. Genetic diversity, linkage disequilibrium and population structure of Bulgarian bread wheat assessed by genome-wide distributed SNP markers: From old germplasm to semi-dwarf cultivars. Plants 2021, 10, 1116. [Google Scholar] [CrossRef]
- Dimitrijević, M.; Petrović, S.; Cîmpeanu, C.; Bucur, D.; Belić, M. Cereals and Aegilops genus biodiversity survey in the west Balkans: Erosion and preservation. J. Food Agric. Environ. 2011, 9, 219–225. [Google Scholar]
- Petrović, S.; Dimitrijević, M. Genetic erosion of diversity in cereals. Genetika 2012, 44, 217–226. [Google Scholar] [CrossRef]
- Louafi, S.; Manzella, D. The benefit-sharing mechanisms under the International Treaty: Heterogeneity and equity in global resources management. In The Commons, Plant Breeding and Agricultural Research; Routledge: Abingdon, UK, 2018; pp. 255–271. [Google Scholar]
- Adhikari, S.; Kumari, J.; Jacob, S.R.; Prasad, P.; Gangwar, O.P.; Lata, C.; Thakur, R.; Singh, A.K.; Bansal, R.; Kumar, S.; et al. Landraces-potential treasure for sustainable wheat improvement. Genet. Resour. Crop Evol. 2022, 69, 499–523. [Google Scholar]
- Soriano, J.M.; Villegas, D.; Aranzana, M.J.; García del Moral, L.F.; Royo, C. Genetic structure of modern durum wheat cultivars and Mediterranean landraces matches with their agronomic performance. PLoS ONE 2016, 11, e0160983. [Google Scholar] [CrossRef]
- Rufo, R.; Alvaro, F.; Royo, C.; Soriano, J.M. From landraces to improved cultivars: Assessment of genetic diversity and population structure of Mediterranean wheat using SNP markers. PLoS ONE 2019, 14, e0219867. [Google Scholar] [CrossRef]
- Velimirović, A.; Jovović, Z.; Perović, D.; Lehnert, H.; Mikić, S.; Mandić, D.; Pržulj, N.; Mangini, G.; Finetti-Sialer, M.M. SNP diversity and genetic structure of “Rogosija”, an old Western Balkan durum wheat collection. Plants 2023, 12, 1157. [Google Scholar] [CrossRef] [PubMed]
- Kobiljski, B.; Denčić, S.; Pilipović, J. Molecular screening of domestic germplasm for allelic variants at the dwarfing gene Rht8 locus in wheat. Genetika 2006, 38, 67–74. [Google Scholar] [CrossRef]
- Kondić-Špika, A.; Denčić, S.; Mladenov, N.; Trkulja, D.; Mikić, S.; Hristov, N.; Marjanović-Jeromela, A. Polymorphism of microsatellite loci in bread wheat (Triticum aestivum L.): And related species. Zb. Matice Srp. Prir. Nauk. 2016, 81–89. [Google Scholar] [CrossRef]
- Mladenov, V.; Dimitrijević, M.; Petrović, S.; Boćanski, J.; Banjac, B.; Kondić-Špika, A.; Trkulja, D. Genetic analysis of spike length in wheat. Genetika 2019, 51, 167–178. [Google Scholar] [CrossRef]
- Ganeva, G.; Korzun, V. Microsatellite genetic diversity analysis and allelic variation comparison of obsolete and modern Bulgarian bread wheat (Triticum aestivum L.) varieties. Cereal Res. Commun. 2012, 40, 14–23. [Google Scholar] [CrossRef]
- Tsonev, S.; Christov, N.K.; Mihova, G.; Dimitrova, A.; Todorovska, E.G. Genetic diversity and population structure of bread wheat varieties grown in Bulgaria based on microsatellite and phenotypic analyses. Biotechnol. Biotechnol. Equip. 2021, 35, 1520–1533. [Google Scholar] [CrossRef]
- Chen, Y.; Dong, H.-B.; Peng, C.-J.; Du, X.-J.; Li, C.-X.; Han, X.-L.; Sun, W.-X.; Zhang, Y.-M.; Hu, L. Phenotypic plasticity of flowering time and plant height related traits in wheat. BMC Plant Biol. 2025, 25, 636. [Google Scholar] [CrossRef]
- Khobra, R.; Sareen, S.; Meena, B.K.; Kumar, A.; Tiwari, V.; Singh, G.P. Exploring the traits for lodging tolerance in wheat genotypes: A review. Physiol. Mol. Biol. Plants 2019, 25, 589–600. [Google Scholar] [CrossRef]
- Shavrukov, Y.; Kurishbayev, A.; Jatayev, S.; Shvidchenko, V.; Zotova, L.; Koekemoer, F.; De Groot, S.; Soole, K.; Langridge, P. Early flowering as a drought escape mechanism in plants: How can it aid wheat production? Front. Plant Sci. 2017, 8, 1950. [Google Scholar] [CrossRef]
- Xie, Q.; Mayes, S.; Sparkes, D.L. Carpel size, grain filling, and morphology determine individual grain weight in wheat. J. Exp. Bot. 2015, 66, 6715–6730. [Google Scholar] [CrossRef]
- Chapman, E.A.; Orford, S.; Lage, J.; Griffiths, S. Delaying or delivering: Identification of novel NAM-1 alleles that delay senescence to extend wheat grain fill duration. J. Exp. Bot. 2021, 72, 7710–7728. [Google Scholar] [CrossRef]
- Wingler, A.; Soualiou, S. Overcoming physiological trade-offs between flowering time and crop yield: Strategies for a changing climate. J. Exp. Bot. 2025, 76, 2646–2658. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1990, 19, 11–15. [Google Scholar]
- Röder, M.S.; Korzun, V.; Wendehake, K.; Plaschke, J.; Tixier, M.H.; Leroy, P.; Ganal, M.W. A microsatellite map of wheat. Genetics 1998, 149, 2007–2023. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.J.; Shi, J.R.; Singh, S.; Fickus, E.W.; Costa, J.M.; Lewis, J.; Gill, B.S.; Ward, R.; Cregan, P.B. Development and mapping of microsatellite (SSR) markers in wheat. Theor. Appl. Genet. 2005, 110, 550–556. [Google Scholar] [CrossRef] [PubMed]
- GrainGenes, a Database for Triticeae and Avena. Available online: https://graingenes.org/cgi-bin/GG3/browse.cgi (accessed on 9 May 2025).
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Muse, S.P. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Li, Y.-L.; Liu, J.-X. StructureSelector: A web-based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef]
- Francis, R.M. pophelper: An R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Tajima, F.; Tateno, Y. Accuracy of estimated phylogenetic trees from molecular data. J. Mol. Evol. 1983, 19, 153–170. [Google Scholar] [CrossRef]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.-H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, 236–241. [Google Scholar] [CrossRef]
- Tošović-Marić, B.; Kobiljski, B.; Obreht, D.; Vapa, L. Evaluation of wheat Rht genes using molecular markers. Genetika 2008, 40, 31–38. [Google Scholar] [CrossRef]
- Ganeva, G.; Korzun, V.; Landjeva, S.; Tsenov, N.; Atanasova, M. Identification, distribution and effects on agronomic traits of the semi-dwarfing Rht alleles in Bulgarian common wheat cultivars. Euphytica 2005, 145, 305–315. [Google Scholar] [CrossRef]
- Kolev, S.; Vassilev, D.; Kostov, K.; Todorovska, E. Allele variation in loci for adaptive response in Bulgarian wheat cultivars and landraces and its effect on heading date. Plant Genet. Resour. 2011, 9, 251–255. [Google Scholar] [CrossRef]
- Srivastava, N.; Shrivastav, A.K.; Srivastava, M.; Mishra, P.K. Biofuels production using wheat straw. In Recent Developments in Bioenergy Research; Elsevier: Amsterdam, The Netherlands, 2020; pp. 433–441. [Google Scholar]
- Ninkuu, V.; Liu, Z.; Qin, A.; Xie, Y.; Song, X.; Sun, X. Impact of straw returning on soil ecology and crop yield: A review. Heliyon 2025, 11, e41651. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Richards, R.A. Recent Advances in Breeding Wheat for Drought and Salt Stresses. In Advances in Molecular Breeding Toward Drought and Salt Tolerant Crops; Jenks, M.A., Hasegawa, P.M., Jain, S.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 565–585. [Google Scholar]
- Veteläinen, M.; Negri, V.; Maxted, N. European Landraces: On-farm Conservation, Management and Use; Bioversity International: Rome, Italy, 2009. [Google Scholar]
- Takács, G.; Gergely, I.; Ördög, V.; Vörös, L.; Iváncsics, J. Approaches to studying wheat and maize drought stress responses. Plant Soil. 2025, 1–8. [Google Scholar] [CrossRef]
- Lopes, M.S.; El-Basyoni, I.; Baenziger, P.S.; Singh, S.; Royo, C.; Ozbek, K.; Aktas, H.; Ozer, E.; Ozdemir, F.; Manickavelu, A.; et al. Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J. Exp. Bot. 2015, 66, 3477–3486. [Google Scholar] [CrossRef]
- Baresel, J.P.; Bülow, L.; Finckh, M.R.; Frese, L.; Knapp, S.; Schmidhalter, U.; Weedon, O. Performance and evolutionary adaptation of heterogeneous wheat populations. Euphytica 2022, 218, 137. [Google Scholar] [CrossRef]
- Reif, J.C.; Zhang, P.; Dreisigacker, S.; Warburton, M.L.; van Ginkel, M.; Hoisington, D.; Bohn, M.; Melchinger, A.E. Wheat genetic diversity trends during domestication and breeding. Theor. Appl. Genet. 2005, 110, 859–864. [Google Scholar] [CrossRef]
- Fu, Y.-B.; Somers, D.J. Genome-wide reduction of genetic diversity in wheat breeding. Crop Sci. 2009, 49, 161–168. [Google Scholar] [CrossRef]
- Cavanagh, C.R.; Chao, S.; Wang, S.; Huang, B.E.; Stephen, S.; Kiani, S.; Forrest, K.; Saintenac, C.; Brown-Guedira, G.L.; Akhunova, A.; et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. USA 2013, 110, 8057–8062. [Google Scholar] [CrossRef] [PubMed]
- Pont, C.; Leroy, T.; Seidel, M.A.; Tondelli, A.; Duchemin, W.; Armisen, D.; Lang, D.; Bustos-Korts, D.; Goué, N.; Balfourier, F.; et al. Tracing the ancestry of modern bread wheats. Nat. Genet. 2019, 51, 905–911. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Stiller, J.; Wei, Y.; Zheng, Y.L.; Devos, K.M.; Doležel, J.; Liu, C. Extensive pericentric rearrangements in the bread wheat (Triticum aestivum L.) genotype “Chinese Spring” revealed from chromosome shotgun sequence data. Genome Biol. Evol. 2014, 6, 3039–3048. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, J.; Wang, L.; Zhu, T.; Jorgensen, C.M.; Luo, M.C.; Deal, K.R.; Gu, Y.Q.; Gill, B.S.; Distelfeld, A.; Devos, K.M.; et al. Reassessment of the evolution of wheat chromosomes 4A, 5A, and 7B. Theor. Appl. Genet. 2018, 131, 2451–2462. [Google Scholar] [CrossRef]
- Zhao, X.; Fu, X.; Yin, C.; Lu, F. Wheat speciation and adaptation: Perspectives from reticulate evolution. Abiotech 2021, 2, 386–402. [Google Scholar] [CrossRef]
- Liu, Y.; Fu, B.; Zhang, Q.; Cai, J.; Guo, W.; Zhai, W.; Wu, J. Genetic diversity and population structure of wheat landraces in Southern Winter Wheat Region of China. BMC Genom. 2024, 25, 664. [Google Scholar] [CrossRef] [PubMed]
- Fradgley, N.; Gardner, K.A.; Cockram, J.; Elderfield, J.; Hickey, J.M.; Howell, P.; Jackson, R.; Mackay, I.J. A large-scale pedigree resource of wheat reveals evidence for adaptation and selection by breeders. PLoS Biol. 2019, 17, e3000071. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Feng, C.; Wingen, L.U.; Cheng, H.; Riche, A.B.; Jiang, M.; Leverington-Waite, M.; Huang, Z.; Collier, S.; Orford, S.; et al. Harnessing landrace diversity empowers wheat breeding. Nature 2024, 632, 823–831. [Google Scholar] [CrossRef] [PubMed]
| Trait | Group | Min | Max | Q1 | Median | Q3 | Variance | SD | Mean |
|---|---|---|---|---|---|---|---|---|---|
| Plant height (cm) | Bulgarian cultivars | 63 | 129 | 86.0 | 99.0 | 115.7 | 279.9 | 16.7 | 100.9 a |
| Serbian cultivars | 82 | 141 | 96.7 | 112.5 | 121.2 | 257.3 | 16.0 | 111.4 b | |
| Serbian landraces | 97 | 149 | 126.5 | 133.5 | 136.0 | 122.0 | 11.0 | 130.1 c | |
| Flowering time (days) | Bulgarian cultivars | 128 | 139 | 132.0 | 135.0 | 135.0 | 6.4 | 2.5 | 133.8 a |
| Serbian cultivars | 128 | 141 | 134.0 | 135.0 | 137.2 | 8.04 | 2.8 | 135.5 b | |
| Serbian landraces | 132 | 145 | 138.0 | 139.0 | 142.0 | 10.8 | 3.3 | 139.7 c |
| Trait | Source of Variation | df | Sum Square | Mean Square | F Value | p (>F) | Partial η2 |
|---|---|---|---|---|---|---|---|
| Plant height (cm) | Group | 2 | 22,997 | 11,498 | 117.76 | <2 × 10−16 ** | 0.62 |
| Year | 1 | 18,092 | 18,092 | 185.28 | <2 × 10−16 ** | 0.56 | |
| Group:Year | 2 | 3843 | 1921 | 19.68 | 2.71 × 10−8 ** | 0.21 | |
| Residuals | 146 | 14,256 | 97.6 | ||||
| Flowering time (days) | Group | 2 | 129 | 63.5 | 11.95 | 1.561 × 10−5 ** | 0.14 |
| Year | 1 | 260 | 260 | 48.08 | 1.228 × 10−10 ** | 0.25 | |
| Group:Year | 2 | 183 | 183.0 | 33.852 | 8.34 × 10−13 ** | 0.32 | |
| Residuals | 146 | 789 | 5.4 |
| Parameter | All | Bulgarian Cultivars | Serbian Cultivars | Serbian Landraces |
|---|---|---|---|---|
| MAF | 0.469 | 0.578 | 0.496 | 0.429 |
| Sample size | 76 | 41 | 21 | 14 |
| Allele No. | 446 | 277 | 252 | 340 |
| Allele/Locus | 9.49 | 5.89 | 5.36 | 7.23 |
| GD (He) | 0.675 | 0.566 | 0.602 | 0.675 |
| Ho | 0.201 | 0.206 | 0.175 | 0.209 |
| PIC | 0.646 | 0.534 | 0.597 | 0.671 |
| No. of private alleles | - | 42 | 30 | 106 |
| No. of rare alleles (<5%) | - | 45 | 18 | 73 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mikić, S.; Christov, N.K.; Tsonev, S.; Todorovska, E.G.; Trkulja, D.; Kondić-Špika, A.; Zelić, V. Molecular and Agro-Morphological Diversity of Undercharacterized Local Bread Wheat Genetic Resources from Serbia and Bulgaria. Agriculture 2025, 15, 2127. https://doi.org/10.3390/agriculture15202127
Mikić S, Christov NK, Tsonev S, Todorovska EG, Trkulja D, Kondić-Špika A, Zelić V. Molecular and Agro-Morphological Diversity of Undercharacterized Local Bread Wheat Genetic Resources from Serbia and Bulgaria. Agriculture. 2025; 15(20):2127. https://doi.org/10.3390/agriculture15202127
Chicago/Turabian StyleMikić, Sanja, Nikolai Kirilov Christov, Stefan Tsonev, Elena Georgieva Todorovska, Dragana Trkulja, Ankica Kondić-Špika, and Verica Zelić. 2025. "Molecular and Agro-Morphological Diversity of Undercharacterized Local Bread Wheat Genetic Resources from Serbia and Bulgaria" Agriculture 15, no. 20: 2127. https://doi.org/10.3390/agriculture15202127
APA StyleMikić, S., Christov, N. K., Tsonev, S., Todorovska, E. G., Trkulja, D., Kondić-Špika, A., & Zelić, V. (2025). Molecular and Agro-Morphological Diversity of Undercharacterized Local Bread Wheat Genetic Resources from Serbia and Bulgaria. Agriculture, 15(20), 2127. https://doi.org/10.3390/agriculture15202127








