Synteny Patterns of Class 1 Integrons Reflect Microbial Adaptation and Soil Health in Agroecosystems
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Collection
2.2. DNA Extraction, Library Preparation, and Sequencing
2.3. Bioinformatic and Statistical Analyses
3. Results
3.1. Gene-Cassette Signatures
3.2. Gene-Cassette Synteny
3.3. Machine Learning Clustering
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gayan, A.; Borah, P.; Nath, D.; Kataki, R. Soil Microbial Diversity, Soil Health and Agricultural Sustainability. In Sustainable Agriculture and the Environment; Elsevier: Amsterdam, The Netherlands, 2023; pp. 107–126. ISBN 978-0-323-90500-8. [Google Scholar]
- Banerjee, S.; Van Der Heijden, M.G.A. Soil Microbiomes and One Health. Nat. Rev. Microbiol. 2023, 21, 6–20. [Google Scholar] [CrossRef]
- Visca, A.; Di Gregorio, L.; Clagnan, E.; Bevivino, A. Sustainable Strategies: Nature-Based Solutions to Tackle Antibiotic Resistance Gene Proliferation and Improve Agricultural Productivity and Soil Quality. Environ. Res. 2024, 248, 118395. [Google Scholar] [CrossRef]
- Ali, N.; Ali, I.; Din, A.U.; Akhtar, K.; He, B.; Wen, R. Integrons in the Age of Antibiotic Resistance: Evolution, Mechanisms, and Environmental Implications: A Review. Microorganisms 2024, 12, 2579. [Google Scholar] [CrossRef]
- Singh, H.; Pandya, S.; Jasani, S.; Patel, M.; Kaur, T.; Rustagi, S.; Shreaz, S.; Yadav, A.N. Integrons: The Hidden Architects of Bacterial Adaptation, Evolution, and the Challenges of Antimicrobial Resistance. Antonie Leeuwenhoek 2025, 118, 90. [Google Scholar] [CrossRef]
- Fonseca, É.L.; Vicente, A.C. Integron Functionality and Genome Innovation: An Update on the Subtle and Smart Strategy of Integrase and Gene Cassette Expression Regulation. Microorganisms 2022, 10, 224. [Google Scholar] [CrossRef]
- Ding, Y.; Wang, J.; Chen, Y.; Yang, Y.; Liu, X. Natural Transformation of Antibiotic Resistance Genes and the Enhanced Adaptability in Bacterial Biofilm under Antibiotic and Heavy Metal Stresses. J. Hazard. Mater. 2025, 490, 137740. [Google Scholar] [CrossRef]
- Shintani, M.; Vestergaard, G.; Milaković, M.; Kublik, S.; Smalla, K.; Schloter, M.; Udiković-Kolić, N. Integrons, Transposons and IS Elements Promote Diversification of Multidrug Resistance Plasmids and Adaptation of Their Hosts to Antibiotic Pollutants from Pharmaceutical Companies. Environ. Microbiol. 2023, 25, 3035–3051. [Google Scholar] [CrossRef] [PubMed]
- Sabbagh, P.; Rajabnia, M.; Maali, A.; Ferdosi-Shahandashti, E. Integron and Its Role in Antimicrobial Resistance: A Literature Review on Some Bacterial Pathogens. Iran. J. Basic Med. Sci. 2021, 24, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Néron, B.; Littner, E.; Haudiquet, M.; Perrin, A.; Cury, J.; Rocha, E. IntegronFinder 2.0: Identification and Analysis of Integrons across Bacteria, with a Focus on Antibiotic Resistance in Klebsiella. Microorganisms 2022, 10, 700. [Google Scholar] [CrossRef] [PubMed]
- Ghaly, T.M.; Tetu, S.G.; Penesyan, A.; Qi, Q.; Rajabal, V.; Gillings, M.R. Discovery of Integrons in Archaea: Platforms for Cross-Domain Gene Transfer. Sci. Adv. 2022, 8, eabq6376. [Google Scholar] [CrossRef]
- Bhat, B.A.; Mir, R.A.; Qadri, H.; Dhiman, R.; Almilaibary, A.; Alkhanani, M.; Mir, M.A. Integrons in the Development of Antimicrobial Resistance: Critical Review and Perspectives. Front. Microbiol. 2023, 14, 1231938. [Google Scholar] [CrossRef]
- Corno, G.; Ghaly, T.; Sabatino, R.; Eckert, E.M.; Galafassi, S.; Gillings, M.R.; Di Cesare, A. Class 1 Integron and Related Antimicrobial Resistance Gene Dynamics along a Complex Freshwater System Affected by Different Anthropogenic Pressures. Environ. Pollut. 2023, 316, 120601. [Google Scholar] [CrossRef] [PubMed]
- Quintela-Baluja, M.; Frigon, D.; Abouelnaga, M.; Jobling, K.; Romalde, J.L.; Gomez Lopez, M.; Graham, D.W. Dynamics of Integron Structures across a Wastewater Network–Implications to Resistance Gene Transfer. Water Res. 2021, 206, 117720. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.R.; Gaze, W.H.; Pruden, A.; Smalla, K.; Tiedje, J.M.; Zhu, Y.-G. Using the Class 1 Integron-Integrase Gene as a Proxy for Anthropogenic Pollution. ISME J. 2015, 9, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Tavares, R.D.S.; Fidalgo, C.; Rodrigues, E.T.; Tacão, M.; Henriques, I. Integron-Associated Genes Are Reliable Indicators of Antibiotic Resistance in Wastewater despite Treatment- and Seasonality-Driven Fluctuations. Water Res. 2024, 258, 121784. [Google Scholar] [CrossRef]
- Djordjevic, S.P.; Jarocki, V.M.; Seemann, T.; Cummins, M.L.; Watt, A.E.; Drigo, B.; Wyrsch, E.R.; Reid, C.J.; Donner, E.; Howden, B.P. Genomic Surveillance for Antimicrobial Resistance—A One Health Perspective. Nat. Rev. Genet. 2024, 25, 142–157. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, C.-F. Antibiotic Resistance in the Environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Castañeda-Barba, S.; Top, E.M.; Stalder, T. Plasmids, a Molecular Cornerstone of Antimicrobial Resistance in the One Health Era. Nat. Rev. Microbiol. 2024, 22, 18–32. [Google Scholar] [CrossRef]
- Liu, X.; Wang, H.; Li, L.; Deng, C.; Chen, Y.; Ding, H.; Yu, Z. Do Microplastic Biofilms Promote the Evolution and Co-Selection of Antibiotic and Metal Resistance Genes and Their Associations with Bacterial Communities under Antibiotic and Metal Pressures? J. Hazard. Mater. 2022, 424, 127285. [Google Scholar] [CrossRef]
- Han, B.; Ma, L.; Yu, Q.; Yang, J.; Su, W.; Hilal, M.G.; Li, X.; Zhang, S.; Li, H. The Source, Fate and Prospect of Antibiotic Resistance Genes in Soil: A Review. Front. Microbiol. 2022, 13, 976657. [Google Scholar] [CrossRef]
- Shamsizadeh, Z.; Ehrampoush, M.H.; Nikaeen, M.; Farzaneh, M.; Mokhtari, M.; Gwenzi, W.; Khanahmad, H. Antibiotic Resistance and Class 1 Integron Genes Distribution in Irrigation Water-Soil-Crop Continuum as a Function of Irrigation Water Sources. Environ. Pollut. 2021, 289, 117930. [Google Scholar] [CrossRef]
- Tavares, R.D.S.; Tacão, M.; Henriques, I. Integrons Are Key Players in the Spread of Beta-Lactamase-Encoding Genes. Int. J. Antimicrob. Agents 2025, 65, 107421. [Google Scholar] [CrossRef] [PubMed]
- Ali, N.; Lin, Y.; Jiang, L.; Ali, I.; Ahmed, I.; Akhtar, K.; He, B.; Wen, R. Biochar and Manure Applications Differentially Altered the Class 1 Integrons, Antimicrobial Resistance, and Gene Cassettes Diversity in Paddy Soils. Front. Microbiol. 2022, 13, 943880. [Google Scholar] [CrossRef] [PubMed]
- Tu, Z.; Shui, J.; Liu, J.; Tuo, H.; Zhang, H.; Lin, C.; Feng, J.; Feng, Y.; Su, W.; Zhang, A. Exploring the Abundance and Influencing Factors of Antimicrobial Resistance Genes in Manure Plasmidome from Swine Farms. J. Environ. Sci. 2023, 124, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Fernández Rivas, C.; Porphyre, T.; Chase-Topping, M.E.; Knapp, C.W.; Williamson, H.; Barraud, O.; Tongue, S.C.; Silva, N.; Currie, C.; Elsby, D.T.; et al. High Prevalence and Factors Associated with the Distribution of the Integron intI1 and intI2 Genes in Scottish Cattle Herds. Front. Vet. Sci. 2021, 8, 755833. [Google Scholar] [CrossRef]
- Ghaly, T.M.; Gillings, M.R.; Penesyan, A.; Qi, Q.; Rajabal, V.; Tetu, S.G. The Natural History of Integrons. Microorganisms 2021, 9, 2212. [Google Scholar] [CrossRef]
- Ali, N.; Lin, Y.; Qing, Z.; Xiao, D.; Ud Din, A.; Ali, I.; Lian, T.; Chen, B.; Wen, R. The Role of Agriculture in the Dissemination of Class 1 Integrons, Antimicrobial Resistance, and Diversity of Their Gene Cassettes in Southern China. Genes 2020, 11, 1014. [Google Scholar] [CrossRef]
- Rajabal, V.; Ghaly, T.M.; Egidi, E.; Ke, M.; Penesyan, A.; Qi, Q.; Gillings, M.R.; Tetu, S.G. Exploring the Role of Mobile Genetic Elements in Shaping Plant–Bacterial Interactions for Sustainable Agriculture and Ecosystem Health. Plants People Planet 2024, 6, 408–420. [Google Scholar] [CrossRef]
- Espinosa, E.; Bautista, R.; Larrosa, R.; Plata, O. Advancements in Long-Read Genome Sequencing Technologies and Algorithms. Genomics 2024, 116, 110842. [Google Scholar] [CrossRef]
- Zhang, Y.; Su, Z.; Qiu, X.; Liu, H.; Wen, D.; Chen, L. Distinct ARG Profiles Associated with Class 1 Integrons in Municipal and Industrial Wastewater Treatment Plants. Environ. Sci. Ecotechnology 2025, 26, 100586. [Google Scholar] [CrossRef]
- Li, H. New Strategies to Improve Minimap2 Alignment Accuracy. Bioinformatics 2021, 37, 4572–4574. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Chooi, Y.-H. Clinker & Clustermap. Js: Automatic Generation of Gene Cluster Comparison Figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [CrossRef] [PubMed]
- van der Maaten, L.; Hinton, G. Visualizing High-Dimensional Data Using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Zhang, R.; Kennedy, M.A. Current Understanding of the Structure and Function of Pentapeptide Repeat Proteins. Biomolecules 2021, 11, 638. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xiang, J.; Goh, S.G.; Xie, Y.; Nam, O.C.; Gin, K.Y.-H.; He, Y. Food Waste Compost and Digestate as Novel Fertilizers: Impacts on Antibiotic Resistome and Potential Risks in a Soil-Vegetable System. Sci. Total Environ. 2024, 923, 171346. [Google Scholar] [CrossRef]
- Allegrini, M.; Zabaloy, M.C. Anaerobic Digestates in Agricultural Soils: A Systematic Review of Their Effects on Antibiotic Resistance Genes. Rev. Argent. Microbiol. 2024, 56, 394–401. [Google Scholar] [CrossRef]
- Visca, A.; Rauseo, J.; Spataro, F.; Patrolecco, L.; Grenni, P.; Massini, G.; Mazzurco Miritana, V.; Barra Caracciolo, A. Antibiotics and Antibiotic Resistance Genes in Anaerobic Digesters and Predicted Concentrations in Agroecosystems. J. Environ. Manag. 2022, 301, 113891. [Google Scholar] [CrossRef]
- Feng, G.; Zou, W.; Zhong, Y. Sulfonamides Repress Cell Division in the Root Apical Meristem by Inhibiting Folates Synthesis. J. Hazard. Mater. Adv. 2022, 5, 100045. [Google Scholar] [CrossRef]
- Lin, Z.; Yuan, T.; Zhou, L.; Cheng, S.; Qu, X.; Lu, P.; Feng, Q. Impact Factors of the Accumulation, Migration and Spread of Antibiotic Resistance in the Environment. Environ. Geochem. Health 2021, 43, 1741–1758. [Google Scholar] [CrossRef]
- Wolak, I.; Bajkacz, S.; Harnisz, M.; Stando, K.; Męcik, M.; Korzeniewska, E. Digestate from Agricultural Biogas Plants as a Reservoir of Antimicrobials and Antibiotic Resistance Genes—Implications for the Environment. Int. J. Environ. Res. Public Health 2023, 20, 2672. [Google Scholar] [CrossRef]
- Xie, W.-Y.; Yuan, Y.; Wang, Y.-T.; Liu, D.-Y.; Shen, Q.; Zhao, F.-J. Hazard Reduction and Persistence of Risk of Antibiotic Resistance during Thermophilic Composting of Animal Waste. J. Environ. Manag. 2023, 330, 117249. [Google Scholar] [CrossRef]
- Aguilar-Paredes, A.; Valdés, G.; Araneda, N.; Valdebenito, E.; Hansen, F.; Nuti, M. Microbial Community in the Composting Process and Its Positive Impact on the Soil Biota in Sustainable Agriculture. Agronomy 2023, 13, 542. [Google Scholar] [CrossRef]
- Luo, Y.; Van Veelen, H.P.J.; Chen, S.; Sechi, V.; Ter Heijne, A.; Veeken, A.; Buisman, C.J.N.; Bezemer, T.M. Effects of Sterilization and Maturity of Compost on Soil Bacterial and Fungal Communities and Wheat Growth. Geoderma 2022, 409, 115598. [Google Scholar] [CrossRef]
- Wang, G.; Kong, Y.; Yang, Y.; Ma, R.; Li, L.; Li, G.; Yuan, J. Composting Temperature Directly Affects the Removal of Antibiotic Resistance Genes and Mobile Genetic Elements in Livestock Manure. Environ. Pollut. 2022, 303, 119174. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, X.; Zhang, L.; Zhang, X.; Cao, Y.; Xiao, R.; Bai, Z.; Ma, L. Meta-Analysis Addressing the Potential of Antibiotic Resistance Gene Elimination through Aerobic Composting. Waste Manag. 2024, 182, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Graziano, S.; Caldara, M.; Gullì, M.; Cornali, S.; Vassura, I.; Coralli, I.; Pagano, L.; Marmiroli, M.; Donati, M.; Bevivino, A.; et al. Improving the Sustainability of Tomato Production with Biochar and Biofertilizers in Emilia-Romagna, Italy. Soil Use Manag. 2025, 41, e70091. [Google Scholar] [CrossRef]
- Mayans, B.; Zamora-Martin, S.; Antón-Herrero, R.; García-Delgado, C.; Delgado-Moreno, L.; Guirado, M.; Pérez-Esteban, J.; Segura, M.L.; Escolástico, C.; Eymar, E. Evaluation of the Rhizosphere Resistome of Cultivated Soils Polluted with Antibiotics from Reclaimed Wastewater. Agronomy 2024, 14, 1118. [Google Scholar] [CrossRef]
- Salam, L.B.; Obayori, O.S.; Ilori, M.O.; Amund, O.O. Chromium Contamination Accentuates Changes in the Microbiome and Heavy Metal Resistome of a Tropical Agricultural Soil. World J. Microbiol. Biotechnol. 2023, 39, 228. [Google Scholar] [CrossRef]
- Vasilachi, I.C.; Stoleru, V.; Gavrilescu, M. Analysis of Heavy Metal Impacts on Cereal Crop Growth and Development in Contaminated Soils. Agriculture 2023, 13, 1983. [Google Scholar] [CrossRef]
- Escolà Casas, M.; Matamoros, V. Linking Plant-Root Exudate Changes to Micropollutant Exposure in Aquatic Plants (Lemna Minor and Salvinia Natans). A Prospective Metabolomic Study. Chemosphere 2022, 287, 132056. [Google Scholar] [CrossRef]
- Jutkina, J.; Marathe, N.P.; Flach, C.-F.; Larsson, D.G.J. Antibiotics and Common Antibacterial Biocides Stimulate Horizontal Transfer of Resistance at Low Concentrations. Sci. Total Environ. 2018, 616–617, 172–178. [Google Scholar] [CrossRef]
- Chen, L.; Liu, Y. The Function of Root Exudates in the Root Colonization by Beneficial Soil Rhizobacteria. Biology 2024, 13, 95. [Google Scholar] [CrossRef]
- Zhang, M.; Cai, Z.; Zhang, G.; Zhang, D.; Pan, X. Abiotic Mechanism Changing Tetracycline Resistance in Root Mucus Layer of Floating Plant: The Role of Antibiotic-Exudate Complexation. J. Hazard. Mater. 2021, 416, 125728. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.T.; Nguyen, H.D.; Le, M.H.; Nguyen, T.T.H.; Nguyen, T.D.; Nguyen, D.L.; Nguyen, Q.H.; Nguyen, T.K.O.; Michalet, S.; Dijoux-Franca, M.-G.; et al. Efflux Pump Inhibitors in Controlling Antibiotic Resistance: Outlook under a Heavy Metal Contamination Context. Molecules 2023, 28, 2912. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Xie, Z.; Yang, Z.; Li, D.; Xu, X. A T-SNE Based Classification Approach to Compositional Microbiome Data. Front. Genet. 2020, 11, 620143. [Google Scholar] [CrossRef] [PubMed]
- Aigle, A.; Bourgeois, E.; Marjolet, L.; Houot, S.; Patureau, D.; Doelsch, E.; Cournoyer, B.; Galia, W. Relative Weight of Organic Waste Origin on Compost and Digestate 16S rRNA Gene Bacterial Profilings and Related Functional Inferences. Front. Microbiol. 2021, 12, 667043. [Google Scholar] [CrossRef]
- Wang, N.; Li, H.; Wang, B.; Ding, J.; Liu, Y.; Wei, Y.; Li, J.; Ding, G.-C. Taxonomic and Functional Diversity of Rhizosphere Microbiome Recruited From Compost Synergistically Determined by Plant Species and Compost. Front. Microbiol. 2022, 12, 798476. [Google Scholar] [CrossRef]
- Sanz, C.; Casadoi, M.; Tadic, Đ.; Pastor-López, E.J.; Navarro-Martin, L.; Parera, J.; Tugues, J.; Ortiz, C.A.; Bayona, J.M.; Piña, B. Impact of Organic Soil Amendments in Antibiotic Levels, Antibiotic Resistance Gene Loads, and Microbiome Composition in Corn Fields and Crops. Environ. Res. 2022, 214, 113760. [Google Scholar] [CrossRef]
- Jauregi, L.; González, A.; Garbisu, C.; Epelde, L. Organic Amendment Treatments for Antimicrobial Resistance and Mobile Element Genes Risk Reduction in Soil-Crop Systems. Sci. Rep. 2023, 13, 863. [Google Scholar] [CrossRef]
- Liu, Z.-T.; Ma, R.-A.; Zhu, D.; Konstantinidis, K.T.; Zhu, Y.-G.; Zhang, S.-Y. Organic Fertilization Co-Selects Genetically Linked Antibiotic and Metal(Loid) Resistance Genes in Global Soil Microbiome. Nat. Commun. 2024, 15, 5168. [Google Scholar] [CrossRef] [PubMed]
- Ghaly, T.M.; Tetu, S.G.; Gillings, M.R. Predicting the Taxonomic and Environmental Sources of Integron Gene Cassettes Using Structural and Sequence Homology of attC Sites. Commun. Biol. 2021, 4, 946. [Google Scholar] [CrossRef] [PubMed]
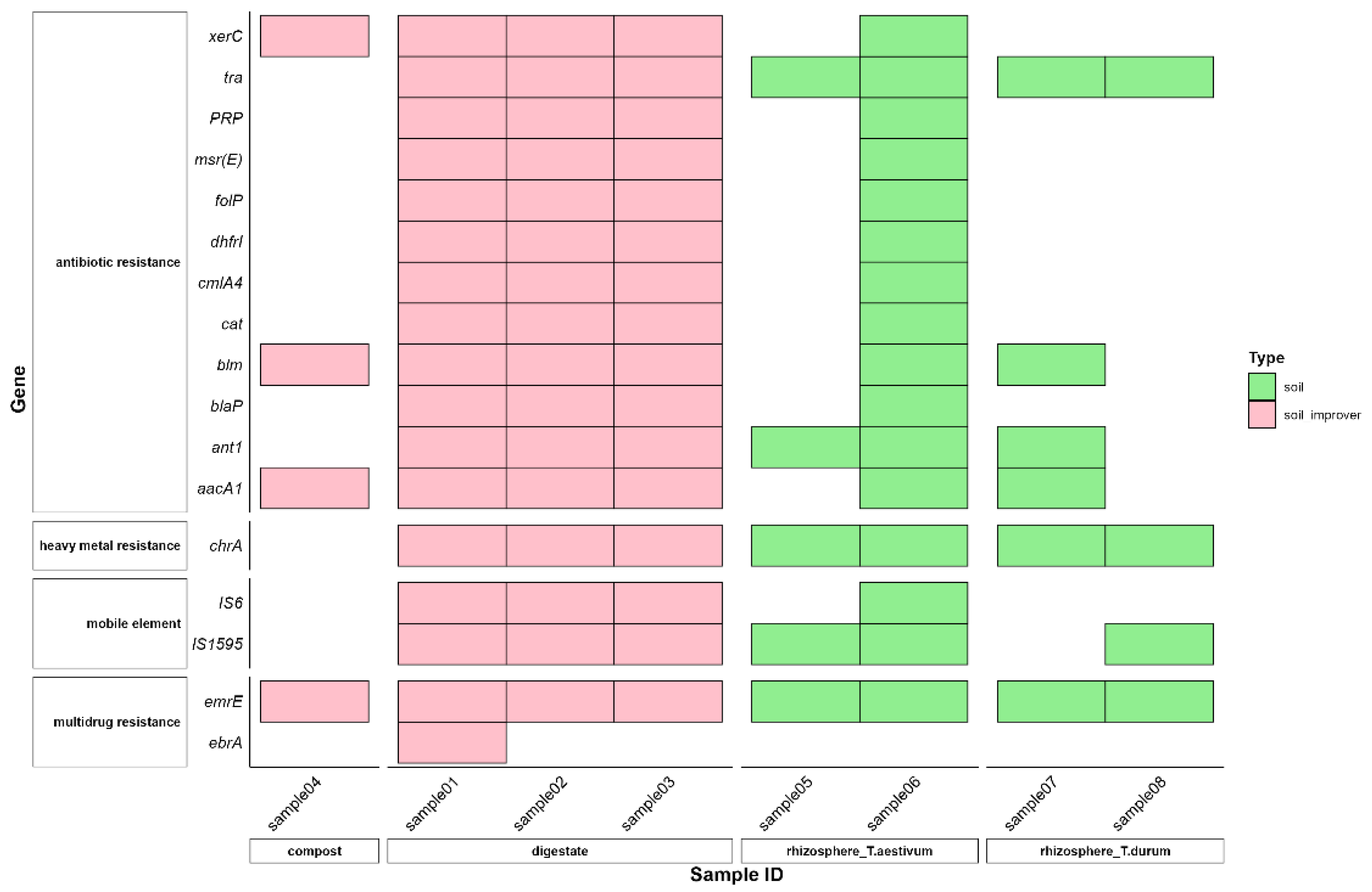
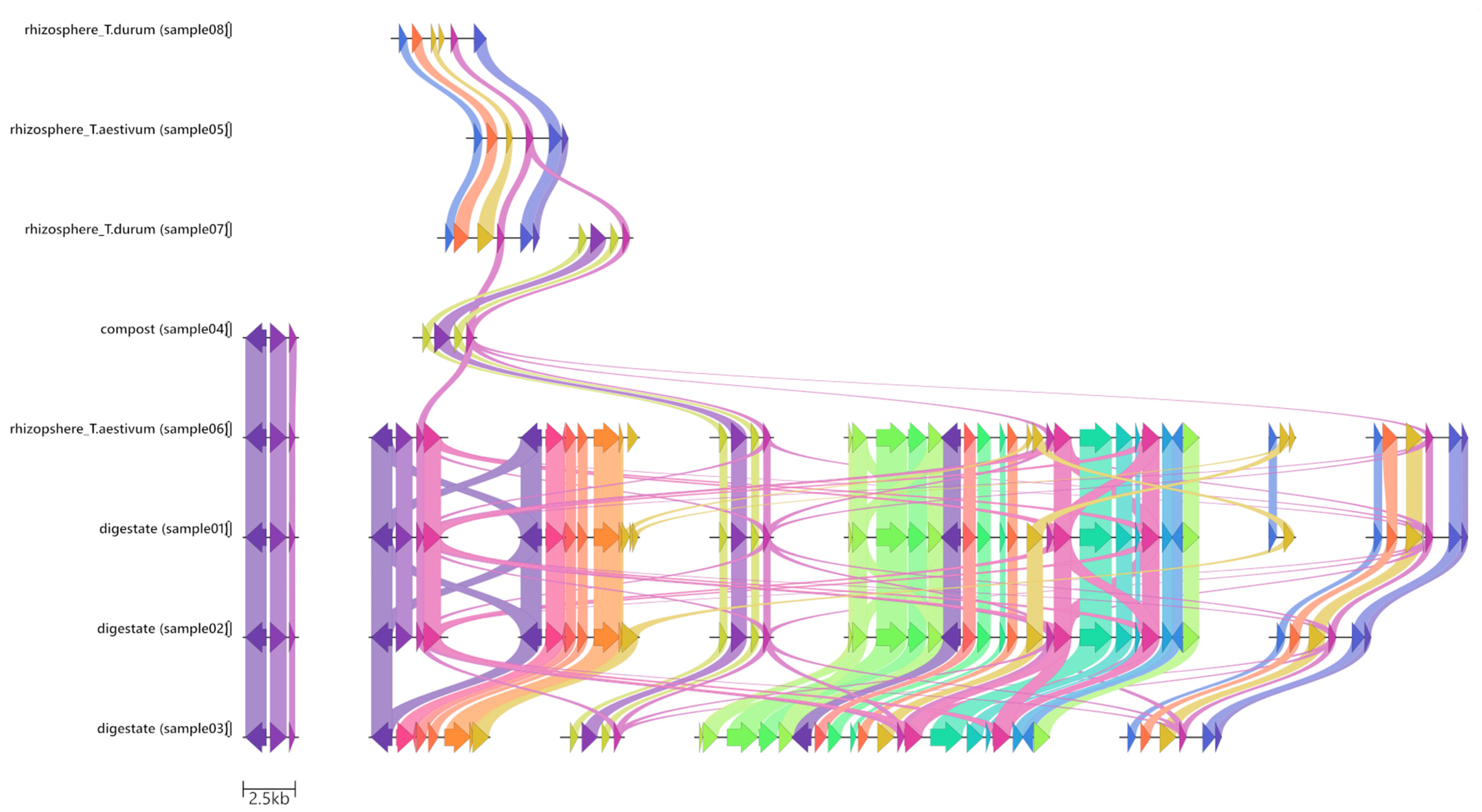

| Sample ID | Type of Sample | Classification | Source |
|---|---|---|---|
| sample 01 | Digestate | Soil improver | Side streams from food industry, municipalities and agriculture |
| sample 02 | Digestate | Soil improver | Side streams from agriculture |
| sample 03 | Digestate | Soil improver | Side streams from food industry and agriculture |
| sample 04 | Compost | Soil improver | Food wastes |
| sample 05 | Soil | Rhizosphere | T. aestivum cv. Taylor rhizosphere (north Italy) |
| sample 06 | Soil | Rhizosphere | T. aestivum cv. Providence rhizosphere (north Italy) |
| sample 07 | Soil | Rhizosphere | T. durum cv. Platone rhizosphere (center Italy) |
| sample 08 | Soil | Rhizosphere | T. durum cv. Shrekan rhizosphere (south Italy) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Visca, A.; Costanzo, M.; Di Gregorio, L.; Nolfi, L.; Bernini, R.; Bevivino, A. Synteny Patterns of Class 1 Integrons Reflect Microbial Adaptation and Soil Health in Agroecosystems. Agriculture 2025, 15, 1833. https://doi.org/10.3390/agriculture15171833
Visca A, Costanzo M, Di Gregorio L, Nolfi L, Bernini R, Bevivino A. Synteny Patterns of Class 1 Integrons Reflect Microbial Adaptation and Soil Health in Agroecosystems. Agriculture. 2025; 15(17):1833. https://doi.org/10.3390/agriculture15171833
Chicago/Turabian StyleVisca, Andrea, Manuela Costanzo, Luciana Di Gregorio, Lorenzo Nolfi, Roberta Bernini, and Annamaria Bevivino. 2025. "Synteny Patterns of Class 1 Integrons Reflect Microbial Adaptation and Soil Health in Agroecosystems" Agriculture 15, no. 17: 1833. https://doi.org/10.3390/agriculture15171833
APA StyleVisca, A., Costanzo, M., Di Gregorio, L., Nolfi, L., Bernini, R., & Bevivino, A. (2025). Synteny Patterns of Class 1 Integrons Reflect Microbial Adaptation and Soil Health in Agroecosystems. Agriculture, 15(17), 1833. https://doi.org/10.3390/agriculture15171833









