Estimation of Genetic Parameters and Weighted Single-Step Genome-Wide Association Study for Indicators of Colostrum Quality in Chinese Holstein Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Committee Approval
2.2. Data
2.2.1. Phenotypes
2.2.2. Pedigree
2.2.3. Genotypes
2.3. Statistical Model Development
2.4. Estimation of Genetic Parameters
2.5. Weighted Single-Step Genome-Wide Association Study (WssGWAS)
3. Results
3.1. Descriptive Statistics and Factors Influencing Colostrum Quality
3.2. Genetic Parameters of Colostrum Quality
3.2.1. Heritability Estimates
3.2.2. Genetic Correlations and Repeatability Estimates
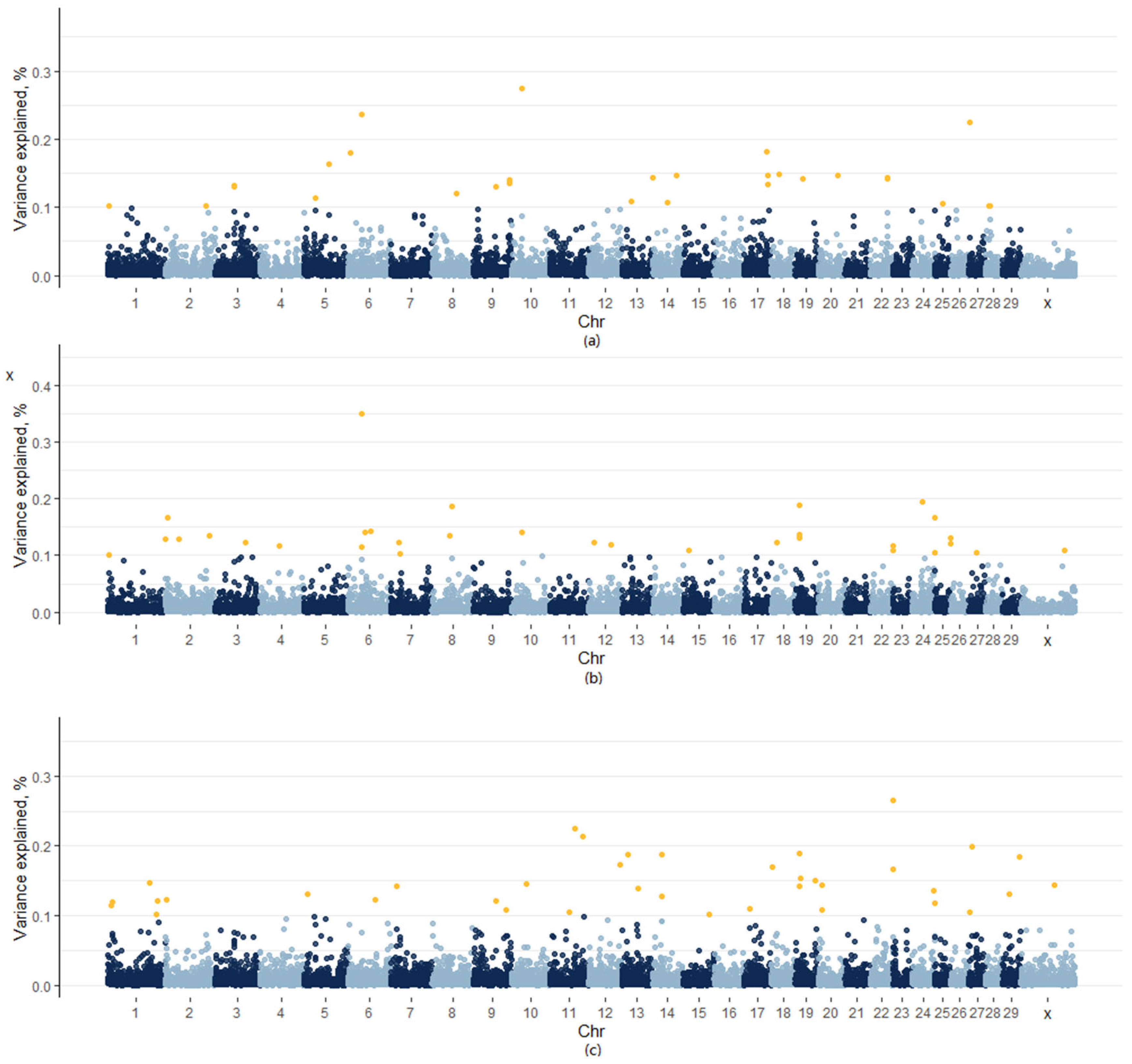
3.3. Weighted Single-Step Genome-Wide Association Studies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Brix1 | Brix measured in first-parity cows |
| Brix2 | Brix measured in second-parity cows |
| Brix3 | Brix measured in third-parity cows |
| BTA | Bos taurus autosome |
| GO | Gene Ontology |
| Ig | Immunoglobulin |
| IgG | Immunoglobulin G |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| WssGWAS | Weighted single-step genome-wide association studies |
References
- Playford, R.J.; Weiser, M.J. Bovine Colostrum: Its Constituents and Uses. Nutrients 2021, 13, 265. [Google Scholar] [CrossRef]
- Cabral, R.G.; Chapman, C.E.; Aragona, K.M.; Clark, E.; Lunak, M.; Erickson, P.S. Predicting colostrum quality from performance in the previous lactation and environmental changes. J. Dairy Sci. 2016, 99, 4048–4055. [Google Scholar] [CrossRef] [PubMed]
- Gulliksen, S.M.; Lie, K.I.; Sølverød, L.; østerås, O. Risk factors associated with colostrum quality in Norwegian dairy cows. J. Dairy Sci. 2008, 91, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Le Cozler, Y.; Guatteo, R.; Le Dréan, E.; Turban, H.; Leboeuf, F.; Pecceu, K.; Guinard-Flament, J. IgG1 variations in the colostrum of Holstein dairy cows. Animal 2016, 10, 230–237. [Google Scholar] [CrossRef]
- Bielmann, V.; Gillan, J.; Perkins, N.R.; Skidmore, A.L.; Godden, S.; Leslie, K.E. An evaluation of Brix refractometry instruments for measurement of colostrum quality in dairy cattle. J. Dairy Sci. 2010, 93, 3713–3721. [Google Scholar] [CrossRef] [PubMed]
- Godden, S.M.; Lombard, J.E.; Woolums, A.R. Colostrum Management for Dairy Calves. Vet. Clin. N. Am. Food. Anim. Pract. 2019, 35, 535–556. [Google Scholar] [CrossRef]
- Costa, A.; Visentin, G.; Goi, A.; De Marchi, M.; Penasa, M. Genetic characteristics of colostrum refractive index and its use as a proxy for the concentration of immunoglobulins in Holstein cattle. Genet. Sel. Evol. 2022, 54, 79. [Google Scholar] [CrossRef]
- Buranakarl, C.; Thammacharoen, S.; Nuntapaitoon, M.; Semsirmboon, S.; Katoh, K. Validation of Brix refractometer to estimate immunoglobulin G concentration in goat colostrum. Vet. World 2021, 14, 3194–3199. [Google Scholar] [CrossRef]
- van Keulen, P.; Mccoard, S.A.; Dijkstra, J.; Swansson, H.; Khan, M.A. Effect of postpartum collection time and colostrum quality on passive transfer of immunity, performance, and small intestinal development in preweaning calves. J. Dairy Sci. 2021, 104, 11931–11944. [Google Scholar] [CrossRef]
- Sutter, F.; Borchardt, S.; Schuenemann, G.M.; Rauch, E.; Erhard, M.; Heuwieser, W. Evaluation of 2 different treatment procedures after calving to improve harvesting of high-quantity and high-quality colostrum. J. Dairy Sci. 2019, 102, 9370–9381. [Google Scholar] [CrossRef] [PubMed]
- Borchardt, S.; Sutter, F.; Heuwieser, W.; Venjakob, P. Management-related factors in dry cows and their associations with colostrum quantity and quality on a large commercial dairy farm. J. Dairy Sci. 2022, 105, 1589–1602. [Google Scholar] [CrossRef] [PubMed]
- Kessler, E.C.; Bruckmaier, R.M.; Gross, J.J. Short communication: Comparative estimation of colostrum quality by Brix refractometry in bovine, caprine, and ovine colostrum. J. Dairy Sci. 2021, 104, 2438–2444. [Google Scholar] [CrossRef]
- Röder, M.; Borchardt, S.; Heuwieser, W.; Rauch, E.; Sargent, R.; Sutter, F. Evaluation of laboratory and on-farm tests to estimate colostrum quality for dairy cows. J. Dairy Sci. 2023, 106, 9164–9173. [Google Scholar] [CrossRef]
- Arslan, A.; Kaplan, M.; Duman, H.; Bayraktar, A.; Ertürk, M.; Henrick, B.M.; Frese, S.A.; Karav, S. Bovine Colostrum and Its Potential for Human Health and Nutrition. Front. Nutr. 2021, 8, 651721. [Google Scholar] [CrossRef]
- Moore, M.; Tyler, J.W.; Chigerwe, M.; Dawes, M.E.; Middleton, J.R. Effect of delayed colostrum collection on colostral IgG concentration in dairy cows. J. Am. Vet. Med. Assoc. 2005, 226, 1375–1377. [Google Scholar] [CrossRef]
- Cordero-Solorzano, J.; de Koning, D.; Tråvén, M.; de Haan, T.; Jouffroy, M.; Larsson, A.; Myrthe, A.; Arts, J.A.J.; Parmentier, H.K.; Bovenhuis, H.; et al. Genetic parameters of colostrum and calf serum antibodies in Swedish dairy cattle. Genet. Sel. Evol. 2022, 54, 68. [Google Scholar] [CrossRef]
- Martin, P.; Vinet, A.; Denis, C.; Grohs, C.; Chanteloup, L.; Dozias, D.; Maupetit, D.; Sapa, J.; Renand, G.; Blanc, F. Determination of immunoglobulin concentrations and genetic parameters for colostrum and calf serum in Charolais animals. J. Dairy Sci. 2021, 104, 3240–3249. [Google Scholar] [CrossRef] [PubMed]
- Soufleri, A.; Banos, G.; Panousis, N.; Fletouris, D.; Arsenos, G.; Valergakis, G.E. Genetic parameters of colostrum traits in Holstein dairy cows. J. Dairy Sci. 2019, 102, 11225–11232. [Google Scholar] [CrossRef]
- Kiser, J.N.; Cornmesser, M.A.; Gavin, K.; Hoffman, A.; Moore, D.A.; Neibergs, H.L. Rapid Communication: Genome-wide association analyses identify loci associated with colostrum production in Jersey cattle1. J. Anim. Sci. 2019, 97, 1117–1123. [Google Scholar] [CrossRef]
- Lin, S.; Ke, C.; Liu, L.; Gao, Y.; Xu, L.; Han, B.; Zhao, Y.; Zhang, S.; Sun, D. Genome-wide association studies for immunoglobulin concentrations in colostrum and serum in Chinese Holstein. BMC Genom. 2022, 23, 41. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Misztal, I.; Tsuruta, S.; Lourenco, D.; Masuda, Y.; Aguilar, I.; Legarra, A.; Vitezica, Z. Manual for BLUPF90 Family of Programs; University of Georgia: Athens, USA, 2014. [Google Scholar]
- Aguilar, I.; Misztal, I.; Johnson, D.L.; Legarra, A.; Tsuruta, S.; Lawlor, T.J. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 2010, 93, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Vanraden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Fernando, R.L.; Vitezica, Z.; Okimoto, R.; Wing, T.; Hawken, R.; Muir, W.M. Genome-wide association mapping including phenotypes from relatives without genotypes in a single-step (ssGWAS) for 6-week body weight in broiler chickens. Front. Genet. 2014, 5, 134. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Rosen, B.D.; Bickhart, D.M.; Schnabel, R.D.; Koren, S.; Elsik, C.G.; Tseng, E.; Rowan, T.N.; Low, W.Y.; Zimin, A.; Couldrey, C.; et al. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience 2020, 9, giaa021. [Google Scholar] [CrossRef]
- Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; Hill, D.P.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein. Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Matsuura, Y.; Ishiguro-Watanabe, M. KEGG: Biological systems database as a model of the real world. Nucleic. Acids. Res. 2025, 53, D672–D677. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic. Acids. Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation-Amsterdam 2021, 2, 100141. [Google Scholar] [CrossRef]
- Nardone, A.; Lacetera, N.; Bernabucci, U.; Ronchi, B. Composition of colostrum from dairy heifers exposed to high air temperatures during late pregnancy and the early postpartum period. J. Dairy Sci. 1997, 80, 838–844. [Google Scholar] [CrossRef]
- Bernabucci, U.; Basiricò, L.; Morera, P. Impact of hot environment on colostrum and milk composition. Cell Mol. Biol. 2013, 59, 67–83. [Google Scholar]
- Conneely, M.; Berry, D.P.; Sayers, R.; Murphy, J.P.; Lorenz, I.; Doherty, M.L.; Kennedy, E. Factors associated with the concentration of immunoglobulin G in the colostrum of dairy cows. Animal 2013, 7, 1824–1832. [Google Scholar] [CrossRef]
- Tyler, J.W.; Steevens, B.J.; Hostetler, D.E.; Holle, J.M.; Denbigh, J.J. Colostral immunoglobulin concentrations in Holstein and Guernsey cows. Am. J. Vet. Res. 1999, 60, 1136–1139. [Google Scholar] [CrossRef] [PubMed]
- Soufleri, A.; Banos, G.; Panousis, N.; Fletouris, D.; Arsenos, G.; Kougioumtzis, A.; Valergakis, G.E. Evaluation of Factors Affecting Colostrum Quality and Quantity in Holstein Dairy Cattle. Animals 2021, 11, 2005. [Google Scholar] [CrossRef] [PubMed]
- Lokke, M.M.; Engelbrecht, R.; Wiking, L. Covariance structures of fat and protein influence the estimation of IgG in bovine colostrum. J. Dairy. Res. 2016, 83, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Lund, M.S.; Wang, Y.; Guo, G.; Dong, G.; Madsen, P.; Su, G. Variance components and correlations of female fertility traits in Chinese Holstein population. J. Anim. Sci. Biotechnol. 2017, 8, 56. [Google Scholar] [CrossRef]
- Wiltbank, M.; Lopez, H.; Sartori, R.; Sangsritavong, S.; Gümen, A. Changes in reproductive physiology of lactating dairy cows due to elevated steroid metabolism. Theriogenology 2006, 65, 17–29. [Google Scholar] [CrossRef]
- Luo, H.; Hu, L.; Brito, L.F.; Dou, J.; Sammad, A.; Chang, Y.; Ma, L.; Guo, G.; Liu, L.; Zhai, L.; et al. Weighted single-step GWAS and RNA sequencing reveals key candidate genes associated with physiological indicators of heat stress in Holstein cattle. J. Anim. Sci. Biotechnol. 2022, 13, 108. [Google Scholar] [CrossRef]
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Muir, W.M. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet. Res. 2012, 94, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Simkins, T.; Crawford, R.B.; Goudreau, J.L.; Lookingland, K.J.; Kaplan, B.L. Enhanced humoral immunity in mice lacking CB1 and CB2 receptors (Cnr1-/-/Cnr2-/- mice) is not due to increased splenic noradrenergic neuronal activity. J. Neuroimmune Pharm. 2014, 9, 544–557. [Google Scholar] [CrossRef] [PubMed]
- Karmaus, P.W.; Chen, W.; Crawford, R.B.; Harkema, J.R.; Kaplan, B.L.; Kaminski, N.E. Deletion of cannabinoid receptors 1 and 2 exacerbates APC function to increase inflammation and cellular immunity during influenza infection. J. Leukoc. Biol. 2011, 90, 983–995. [Google Scholar] [CrossRef]
- Kaplan, B.L. The role of CB1 in immune modulation by cannabinoids. Pharmacol. Ther. 2013, 137, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Zachut, M.; Butenko, Y.; Dos, S.S.P. International Symposium on Ruminant Physiology: The involvement of the endocannabinoid system in metabolic and inflammatory responses in dairy cows during negative energy balance. J. Dairy Sci. 2025, 108, 7643–7661. [Google Scholar] [CrossRef]
- Zachut, M.; Kra, G.; Moallem, U.; Livshitz, L.; Levin, Y.; Udi, S.; Nemirovski, A.; Tam, J. Characterization of the endocannabinoid system in subcutaneous adipose tissue in periparturient dairy cows and its association to metabolic profiles. PLoS ONE 2018, 13, e0205996. [Google Scholar] [CrossRef]
- Jambunathan, S. Post-Translational Modification of the Major Histocompatibility Complex Class II Gene Regulator, ZXDC; Cleveland State University: Cleveland, OH, USA, 2006. [Google Scholar]
- Al-Kandari, W.; Koneni, R.; Navalgund, V.; Aleksandrova, A.; Jambunathan, S.; Fontes, J.D. The zinc finger proteins ZXDA and ZXDC form a complex that binds CIITA and regulates MHC II gene transcription. J. Mol. Biol. 2007, 369, 1175–1187. [Google Scholar] [CrossRef]
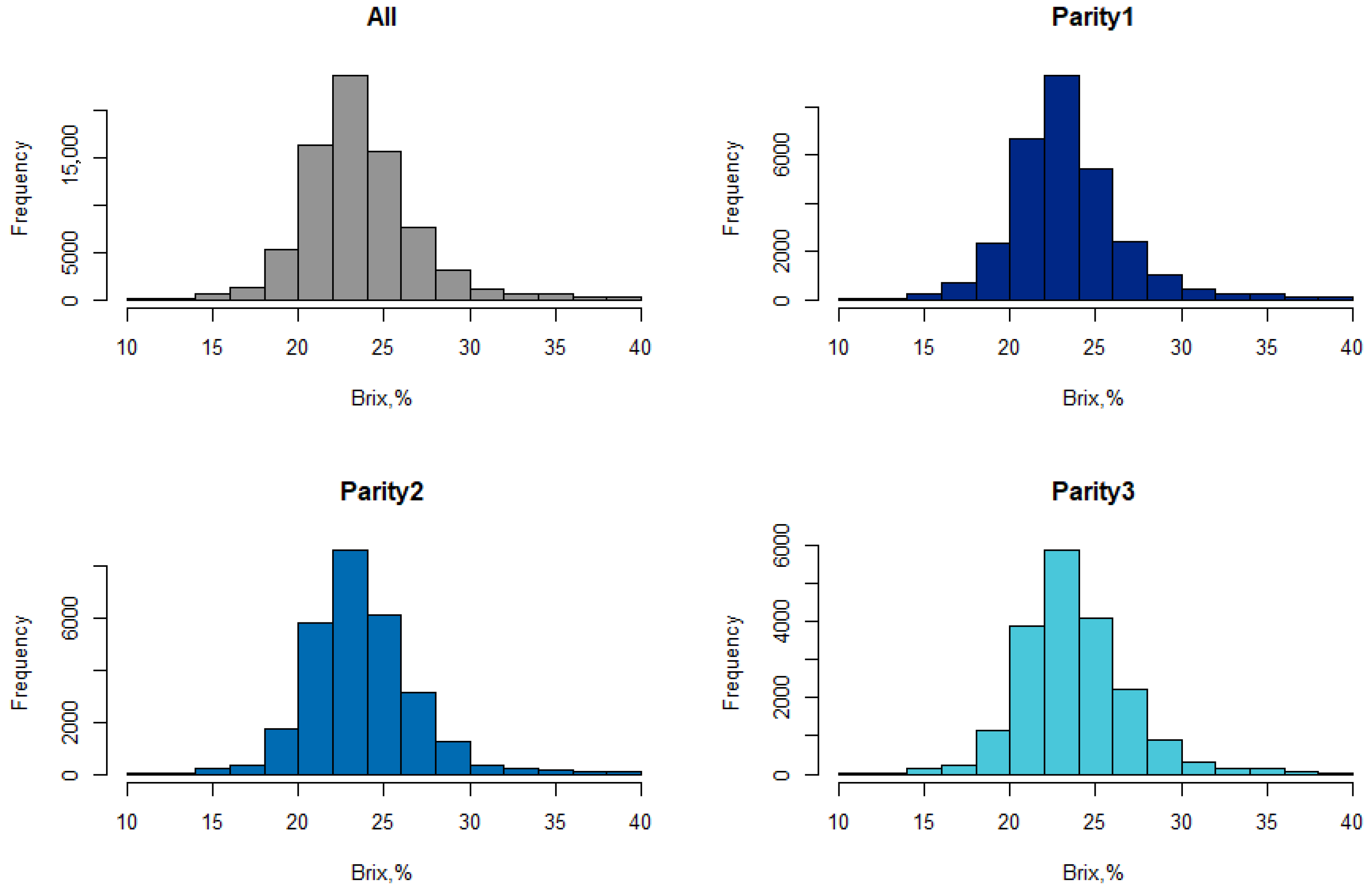

| Trait | Parity | No. of Records | Mean | SD | Min | Max | CV |
|---|---|---|---|---|---|---|---|
| Colostrum quality (Brix, %) | 1 | 28,624 | 23.50 | 3.32 | 10.2 | 40 | 14.13% |
| 2 | 27,688 | 23.89 | 3.19 | 10 | 40 | 13.35% | |
| 3 | 18,921 | 23.98 | 3.21 | 11 | 40 | 13.39% | |
| All | 75,233 | 23.76 | 3.25 | 10 | 40 | 13.68% |
| Effects | Level | N 1 | LSM ± SE 1 |
|---|---|---|---|
| Calving year | 2016 | 4943 | 23.85 ± 0.05 Ee |
| 2017 | 13,644 | 24.03 ± 0.04 Dd | |
| 2018 | 10,970 | 24.12 ± 0.04 Dd | |
| 2019 | 10,822 | 24.34 ± 0.04 Bb | |
| 2020 | 7705 | 25.27 ± 0.04 Cc | |
| 2021 | 16,524 | 25.51 ± 0.03 Aa | |
| 2022 | 10,625 | 25.51 ± 0.04 Aa | |
| Calving month | 1 | 5974 | 24.59 ± 0.05 DEde |
| 2 | 5566 | 24.72 ± 0.05 BCDcd | |
| 3 | 5337 | 24.91 ± 0.05 ABab | |
| 4 | 4396 | 24.67 ± 0.05 CDEcd | |
| 5 | 5223 | 24.79 ± 0.05 ABCDabcd | |
| 6 | 6435 | 24.46 ± 0.04 Ee | |
| 7 | 8664 | 24.23 ± 0.04 Ff | |
| 8 | 9130 | 24.17 ± 0.04 Ff | |
| 9 | 6333 | 24.73 ± 0.04 BCDbcd | |
| 10 | 6323 | 24.85 ± 0.04 ACac | |
| 11 | 5964 | 24.95 ± 0.05 Aa | |
| 12 | 5888 | 24.86 ± 0.05 ABCabc | |
| Parity | 1 | 28,624 | 24.24 ± 0.03 Bb |
| 2 | 27,688 | 24.84 ± 0.03 Aa | |
| 3 | 18,921 | 24.90 ± 0.03 Aa | |
| Interval between calving and colostrum collection (in hours) | 0–2 | 42,776 | 24.82 ± 0.03 Aa |
| 2–6 | 32,457 | 24.50 ± 0.03 Bb | |
| Farm area scale 2 | Group 1 | 13,859 | 23.59 ± 0.03 Dd |
| Group 2 | 13,557 | 23.18 ± 0.03 Ee | |
| Group 3 | 24,310 | 25.36 ± 0.04 Bb | |
| Group 4 | 15,188 | 24.54 ± 0.04 Cc | |
| Group 5 | 8319 | 26.63 ± 0.04 Aa |
| Trait a | N b | ± SE b | ± SE b | ± SE b |
|---|---|---|---|---|
| Brix1 | 24,108 | 2.68 ± 0.20 | 6.69 ± 0.16 | 0.29 ± 0.02 |
| Brix2 | 25,685 | 2.54 ± 0.19 | 5.82 ± 0.17 | 0.30 ± 0.02 |
| Brix3 | 17,413 | 1.80 ± 0.02 | 6.91 ± 0.24 | 0.21 ± 0.03 |
| Trait 1 | Brix1 | Brix2 | Brix3 |
|---|---|---|---|
| Brix1 | 0.57 ± 0.07 * | 0.37 ± 0.14 * | |
| Brix2 | 0.11 ± 0.01 * | 0.81 ± 0.13 * | |
| Brix3 | 0.04 ± 0.02 * | 0.16 ± 0.01 * |
| Chromosome | Regions, Mb | Proportion of the Total Additive Genetic Variance Explained, % | Candidate Genes | ||
|---|---|---|---|---|---|
| Trait 1 | |||||
| Brix1 | Brix2 | Brix3 | |||
| BTA6 | 38.23–38.25 | 0.24 | 0.35 | - | |
| BTA9 | 61.69–61.88 | 0.13 | - | 0.12 | CNR1, SPACA1 |
| BTA19 | 12.81–13.04 | - | 0.14 | 0.19 | CA4, ZNHIT3, MYO19, PIGW, GGNBP2, DHRS11, MRM1 |
| BTA19 | 13.07–13.26 | - | 0.13 | 0.14 | |
| BTA22 | 60.46–60.58 | - | 0.11 | 0.17 | CHST13, UROC1, ZXDC, SLC41A3, ALDH1L1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Brito, L.F.; An, T.; Zhang, H.; Chang, Y.; Chen, S.; Wang, X.; Bai, L.; Guo, G.; Wang, Y. Estimation of Genetic Parameters and Weighted Single-Step Genome-Wide Association Study for Indicators of Colostrum Quality in Chinese Holstein Cattle. Agriculture 2025, 15, 1763. https://doi.org/10.3390/agriculture15161763
Ma Y, Brito LF, An T, Zhang H, Chang Y, Chen S, Wang X, Bai L, Guo G, Wang Y. Estimation of Genetic Parameters and Weighted Single-Step Genome-Wide Association Study for Indicators of Colostrum Quality in Chinese Holstein Cattle. Agriculture. 2025; 15(16):1763. https://doi.org/10.3390/agriculture15161763
Chicago/Turabian StyleMa, Yehua, Luiz F. Brito, Tao An, Hailiang Zhang, Yao Chang, Shaohu Chen, Xin Wang, Libing Bai, Gang Guo, and Yachun Wang. 2025. "Estimation of Genetic Parameters and Weighted Single-Step Genome-Wide Association Study for Indicators of Colostrum Quality in Chinese Holstein Cattle" Agriculture 15, no. 16: 1763. https://doi.org/10.3390/agriculture15161763
APA StyleMa, Y., Brito, L. F., An, T., Zhang, H., Chang, Y., Chen, S., Wang, X., Bai, L., Guo, G., & Wang, Y. (2025). Estimation of Genetic Parameters and Weighted Single-Step Genome-Wide Association Study for Indicators of Colostrum Quality in Chinese Holstein Cattle. Agriculture, 15(16), 1763. https://doi.org/10.3390/agriculture15161763







