Development of a Selective Agar for the Detection of Probiotic Strain Ligilactobacillus animalis NP51 and Other Lactic Acid Bacteria in Cattle Feed
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria, Culture, and Growth Parameters
2.2. Samples and Analysis of Microorganisms in Cattle Feed
2.3. Screening for Selective Chemical Agents
2.4. Fermentation Profile of Ligilactobacillus animalis Probiotic Strain NP51
2.5. Bile Salt Hydrolase Activity Assay
2.6. Real-Time PCR Detection and Enumeration of Probiotic Strain NP51
2.7. Determining the Sensitivity of L. animalis Probiotic Strain NP51 Real-Time qPCR
2.8. Whole Genome Sequencing to Confirm Media Selectivity for LAB
2.9. Statistical Analysis
3. Results
3.1. Growth Conditions for Cultivating Ligilactobacillus animalis Probiotic Strain NP51
3.2. Microorganisms Isolated from Cattle Feed on Commercial MRS Agar
3.3. Identification of Inhibitors That Improve Selectivity of MRS Agar for LAB and Specifically Probiotic Strain NP51 in Cattle Feed and Silage
3.4. Effort to Improve Probiotic Strain Specific Detection
3.5. Modified MRS Medium Selects for LAB Present in Cattle Feed
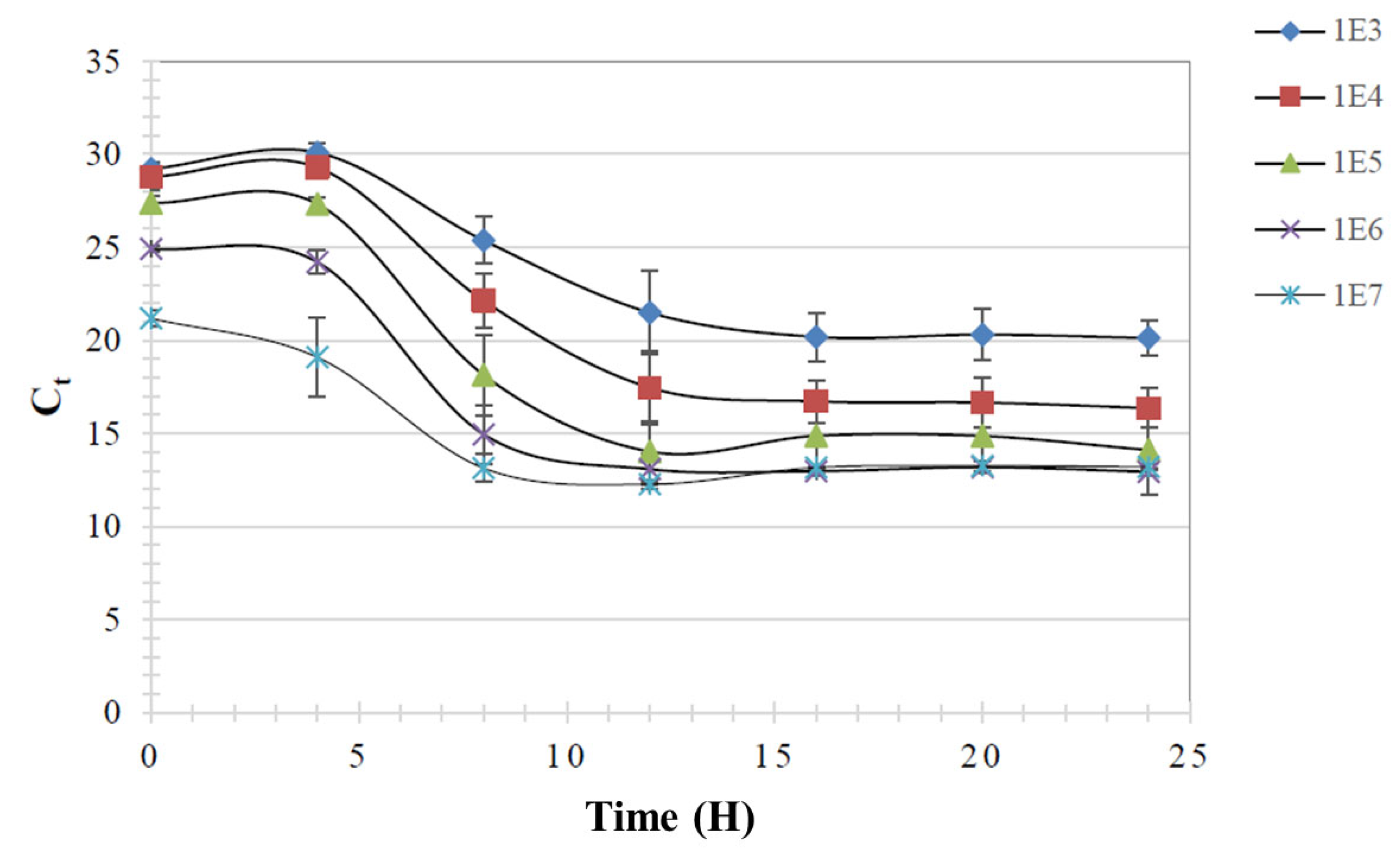
3.6. Detection of Ligilactobacillus NP51 Abundance by qPCR
3.7. Assessing the Antimicrobial Activity of Cattle Feed in Culture of LAB Strain NP51
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Metchnikoff, E. The Prolongation of Life: Optimistic Studies; G. P. Putnam’s Sons: New York, NY, USA; London, UK, 1908. [Google Scholar]
- Lilly, D.M.; Stillwell, R.H. Probiotics: Growth-Promoting Factors Produced by Microorganisms. Science 1965, 147, 747–748. [Google Scholar] [CrossRef] [PubMed]
- Parker, R.B. Probiotics, the other half of the antibiotic story. Anim. Nutr. Health 1974, 29, 4–8. [Google Scholar]
- Fuller, R. Probiotics in man and animals. J. Appl. Bacteriol. 1989, 66, 365–378. [Google Scholar] [PubMed]
- Mack, D.R. Probiotics-mixed messages. Can. Fam. Physician 2005, 51, 1455–1457, 1462–1464. [Google Scholar] [PubMed Central]
- Anonymous. Direct Fed Microbial Products. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/cpg-sec-689100-direct-fed-microbial-products (accessed on 23 October 2015).
- Gilliland, S.E. Health and nutritional benefits from lactic acid bacteria. FEMS Microbiol. Rev. 1990, 7, 175–188. [Google Scholar] [CrossRef]
- Bajagai, Y.S.; Klieve, A.V.; Dart, P.J.; Bryden, W.L. Probiotics in Animal Nutrition: Production, Impact and Regulation; FAO: Rome, Italy, 2016.
- Silva, D.R.; Sardi, J.d.C.O.; de Souza Pitangui, N.; Roque, S.M.; da Silva, A.C.B.; Rosalen, P.L. Probiotics as an alternative antimicrobial therapy: Current reality and future directions. J. Funct. Foods 2020, 73, 104080. [Google Scholar] [CrossRef]
- Chen, X.; Xu, J.; Shuai, J.; Chen, J.; Zhang, Z.; Fang, W. The S-layer proteins of Lactobacillus crispatus strain ZJ001 is responsible for competitive exclusion against Escherichia coli O157:H7 and Salmonella typhimurium. Int. J. Food Microbiol. 2007, 115, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Klaenhammer, T.R. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 1993, 12, 39–85. [Google Scholar] [CrossRef]
- Ramiah, K.; van Reenen, C.A.; Dicks, L.M. Surface-bound proteins of Lactobacillus plantarum 423 that contribute to adhesion of Caco-2 cells and their role in competitive exclusion and displacement of Clostridium sporogenes and Enterococcus faecalis. Res. Microbiol. 2008, 159, 470–475. [Google Scholar] [CrossRef]
- Sakai, F.; Hosoya, T.; Ono-Ohmachi, A.; Ukibe, K.; Ogawa, A.; Moriya, T.; Kadooka, Y.; Shiozaki, T.; Nakagawa, H.; Nakayama, Y.; et al. Lactobacillus gasseri SBT2055 induces TGF-beta expression in dendritic cells and activates TLR2 signal to produce IgA in the small intestine. PLoS ONE 2014, 9, e105370. [Google Scholar] [CrossRef]
- Elshaghabee, F.M.; Rokana, N.; Gulhane, R.D.; Sharma, C.; Panwar, H. Bacillus as potential probiotics: Status, concerns, and future perspectives. Front. Microbiol. 2017, 8, 1490. [Google Scholar] [CrossRef] [PubMed]
- Fuller, R.; Tannock, G.W. Probiotics from Farm Animals. In Probiotics: A Critical Review; Horizon Scientific Press Ltd.: Poole, UK, 1999. [Google Scholar]
- Desnoyers, M.; Giger-Reverdin, S.; Bertin, G.; Duvaux-Ponter, C.; Sauvant, D. Meta-analysis of the influence of Saccharomyces cerevisiae supplementation on ruminal parameters and milk production of ruminants. J. Dairy Sci. 2009, 92, 1620–1632. [Google Scholar] [CrossRef]
- Kung, L., Jr.; Kreck, E.M.; Tung, R.S.; Hession, A.O.; Sheperd, A.C.; Cohen, M.A.; Swain, H.E.; Leedle, J.A. Effects of a live yeast culture and enzymes on in vitro ruminal fermentation and milk production of dairy cows. J. Dairy Sci. 1997, 80, 2045–2051. [Google Scholar] [CrossRef] [PubMed]
- Lan, P.T.; Binh le, T.; Benno, Y. Impact of two probiotic Lactobacillus strains feeding on fecal lactobacilli and weight gains in chicken. J. Gen. Appl. Microbiol. 2003, 49, 29–36. [Google Scholar] [CrossRef]
- Wang, A.; Ran, C.; Wang, Y.; Zhang, Z.; Ding, Q.; Yang, Y.; Olsen, R.E.; Ringo, E.; Bindelle, J.; Zhou, Z. Use of probiotics in aquaculture of China-a review of the past decade. Fish Shellfish Immunol. 2019, 86, 734–755. [Google Scholar] [CrossRef]
- Markowiak, P.; Slizewska, K. The role of probiotics, prebiotics and synbiotics in animal nutrition. Gut Pathog. 2018, 10, 21. [Google Scholar] [CrossRef]
- Al-Khalaifah, H.S. Benefits of probiotics and/or prebiotics for antibiotic-reduced poultry. Poult. Sci. 2018, 97, 3807–3815. [Google Scholar] [CrossRef]
- Jackson, S.A.; Schoeni, J.L.; Vegge, C.; Pane, M.; Stahl, B.; Bradley, M.; Goldman, V.S.; Burguiere, P.; Atwater, J.B.; Sanders, M.E. Improving End-User Trust in the Quality of Commercial Probiotic Products. Front. Microbiol. 2019, 10, 739. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Gruning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Lu, J.; Idris, U.; Harmon, B.; Hofacre, C.; Maurer, J.J.; Lee, M.D. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 2003, 69, 6816–6824. [Google Scholar] [CrossRef]
- Pryde, S.E.; Richardson, A.J.; Stewart, C.S.; Flint, H.J. Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig. Appl. Environ. Microbiol. 1999, 65, 5372–5377. [Google Scholar] [CrossRef] [PubMed]
- Busconi, M.; Reggi, S.; Fogher, C. Evaluation of biodiversity of lactic acid bacteria microbiota in the calf intestinal tracts. Antonie Van Leeuwenhoek 2008, 94, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Nawaz, M.S.; Robertson, L.; Khan, S.A.; Cerniglia, C.E. Identification of predominant human and animal anaerobic intestinal bacterial species by terminal restriction fragment patterns (TRFPs): A rapid, PCR-based method. Mol. Cell. Probes 2001, 15, 349–355. [Google Scholar] [CrossRef]
- Balcazar, J.L.; de Blas, I.; Ruiz-Zarzuela, I.; Vendrell, D.; Girones, O.; Muzquiz, J.L. Sequencing of variable regions of the 16S rRNA gene for identification of lactic acid bacteria isolated from the intestinal microbiota of healthy salmonids. Comp. Immunol. Microbiol. Infect. Dis. 2007, 30, 111–118. [Google Scholar] [CrossRef]
- George, F.; Daniel, C.; Thomas, M.; Singer, E.; Guilbaud, A.; Tessier, F.J.; Revol-Junelles, A.M.; Borges, F.; Foligne, B. Occurrence and Dynamism of Lactic Acid Bacteria in Distinct Ecological Niches: A Multifaceted Functional Health Perspective. Front. Microbiol. 2018, 9, 2899. [Google Scholar] [CrossRef]
- Tannock, G.W.; Tilsala-Timisjarvi, A.; Rodtong, S.; Ng, J.; Munro, K.; Alatossava, T. Identification of Lactobacillus isolates from the gastrointestinal tract, silage, and yoghurt by 16S-23S rRNA gene intergenic spacer region sequence comparisons. Appl. Environ. Microbiol. 1999, 65, 4264–4267. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Zhang, Z.; Zhu, K.; Xue, Y.; Xie, F.; Mao, S. Comprehensive Understanding of the Bacterial Populations and Metabolites Profile of Fermented Feed by 16S rRNA Gene Sequencing and Liquid Chromatography-Mass Spectrometry. Metabolites 2019, 9, 239. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Brashears, M.M.; Jaroni, D.; Trimble, J. Isolation, Selection, and Characterization of Lactic Acid Bacteria for a Competitive Exclusion Product to Reduce Shedding of Escherichia coli O157:H7 in Cattle. J. Food Prot. 2003, 66, 355–363. [Google Scholar] [CrossRef]
- Peterson, R.E.; Klopfenstein, T.J.; Erickson, G.E.; Folmer, J.; Hinkley, S.; Moxley, R.A.; Smith, D.R. Effect of Lactobacillus acidophilus strain NP51 on Escherichia coli O157:H7 fecal shedding and finishing performance in beef feedlot cattle. J. Food Prot. 2007, 70, 287–291. [Google Scholar] [CrossRef]
- Flach, M.G.; Dogan, O.B.; Kreikemeier, W.M.; Nightingale, K.K.; Brashears, M.M. Reduction of Pathogens in Feces and Lymph Nodes Collected from Beef Cattle Fed Lactobacillus salivarius (L28), Lactobacillus acidophilus (NP51) and Propionibacterium freudenreichii (NP28), Commercially Available Direct-Fed Microbials. Foods 2022, 11, 3834. [Google Scholar] [CrossRef] [PubMed]
- Karunasena, E.; Kurkure, P.C.; Lackey, R.D.; McMahon, K.W.; Kiernan, E.P.; Graham, S.; Alabady, M.S.; Campos, D.L.; Tatum, O.L.; Brashears, M.M. Effects of the probiotic Lactobacillus animalis in murine Mycobacterium avium subspecies paratuberculosis infection. BMC Microbiol. 2013, 13, 8. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, F.E.; Chien, C.C.; Paster, B.J.; Ericson, R.L.; Orcutt, R.P.; Schauer, D.B.; Fox, J.G. Phylogeny of the defined murine microbiota: Altered Schaedler flora. Appl. Environ. Microbiol. 1999, 65, 3287–3292. [Google Scholar] [CrossRef] [PubMed]
- Ripamonti, B.; Agazzi, A.; Bersani, C.; De Dea, P.; Pecorini, C.; Pirani, S.; Rebucci, R.; Savoini, G.; Stella, S.; Stenico, A.; et al. Screening of species-specific lactic acid bacteria for veal calves multi-strain probiotic adjuncts. Anaerobe 2011, 17, 97–105. [Google Scholar] [CrossRef]
- Rezvani, M.; Mendoza, M.; Koci, M.D.; Daron, C.; Levy, J.; Hassan, H.M. Draft Genome Sequences of Lactobacillus animalis Strain P38 and Lactobacillus reuteri Strain P43 Isolated from Chicken Cecum. Genome Announc. 2016, 4, e01229-16. [Google Scholar] [CrossRef]
- Ruiz-Moyano, S.; Martín, A.; Benito, M.J.; Nevado, F.P.; de Guía Córdoba, M. Screening of lactic acid bacteria and bifidobacteria for potential probiotic use in Iberian dry fermented sausages. Meat Sci. 2008, 80, 715–721. [Google Scholar] [CrossRef]
- Lee, H.J.; Lee, J.B.; Park, S.Y.; Choi, I.S.; Lee, S.W. Antimicrobial activity of dominant Ligilactobacillus animalis strains in healthy canine feces and their probiotic potential. FEMS Microbiol. Lett. 2022, 369, fnac115. [Google Scholar] [CrossRef]
- De Man, J.C.; Rogosa, M.; Sharpe, M.E. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 1960, 23, 130–135. [Google Scholar] [CrossRef]
- Chopin, A. Organization and regulation of genes for amino acid biosynthesis in lactic acid bacteria. FEMS Microbiol. Rev. 1993, 12, 21–37. [Google Scholar] [CrossRef]
- Makarova, K.; Slesarev, A.; Wolf, Y.; Sorokin, A.; Mirkin, B.; Koonin, E.; Pavlov, A.; Pavlova, N.; Karamychev, V.; Polouchine, N.; et al. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 2006, 103, 15611–15616. [Google Scholar] [CrossRef]
- Jeffries, L. Novobiocin-tetrathionate broth: A medium of improved selectivity for the isolation of Salmonellae from faeces. J. Clin. Pathol. 1959, 12, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Goossens, H.; De Boeck, M.; Butzler, J.P. A new selective medium for the isolation of Campylobacter jejuni from human faeces. Eur. J. Clin. Microbiol. 1983, 2, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Livingston, S.J.; Kominos, S.D.; Yee, R.B. New medium for selection and presumptive identification of the Bacteroides fragilis group. J. Clin. Microbiol. 1978, 7, 448–453. [Google Scholar] [CrossRef]
- Krumwiede, C.; Pratt, J.S. Observations on the Growth of Bacteria on Media Containing Various Anilin Dyes. J. Exp. Med. 1914, 19, 20–27. [Google Scholar] [CrossRef][Green Version]
- Stokes, J.L.; Osborne, W.W. A selenite brilliant green medium for the isolation of Salmonella. Appl. Microbiol. 1955, 3, 217–220. [Google Scholar] [CrossRef] [PubMed]
- Macconkey, A.T. Bile Salt Media and their advantages in some Bacteriological Examinations. J. Hyg. 1908, 8, 322–334. [Google Scholar] [CrossRef]
- Miller, R.G.; Tate, C.R.; Mallinson, E.T.; Scherrer, J.A. Xylose-lysine-tergitol 4: An improved selective agar medium for the isolation of Salmonella. Poult. Sci. 1991, 70, 2429–2432. [Google Scholar] [CrossRef]
- Berrah, G.; Konetzka, W.A. Selective and reversible inhibition of the synthesis of bacterial deoxyribonucleic acid by phenethyl alcohol. J. Bacteriol. 1962, 83, 738–744. [Google Scholar] [CrossRef]
- Hunt, D.E.; Sandham, H.J. Improved agar gradient-plate technique. Appl. Microbiol. 1969, 17, 329–330. [Google Scholar] [CrossRef]
- Randhawa, S.; Brashears, M.; McMahon, K.; Fokar, M.; Karunasena, E. Comparison of Phenotypic and Genotypic Methods Used for the Species Identification of Lactobacillus NP51 and Development of a Strain-Specific PCR Assay. Probiotics Antimicrob. Proteins 2010, 2, 274–283. [Google Scholar] [CrossRef]
- Keegan, K.P.; Glass, E.M.; Meyer, F. MG-RAST, a Metagenomics Service for Analysis of Microbial Community Structure and Function. Methods Mol. Biol. 2016, 1399, 207–233. [Google Scholar] [CrossRef]
- Ekmekci, B.; McAnany, C.E.; Mura, C. An Introduction to Programming for Bioscientists: A Python-Based Primer. PLoS Comput. Biol. 2016, 12, e1004867. [Google Scholar] [CrossRef]
- Kluyver, T.; Ragan-Kelley, B.; Pérez, F.; Granger, B.E.; Bussonnier, M.; Frederic, J.; Kelley, K.; Hamrick, J.B.; Grout, J.; Corlay, S. Jupyter Notebooks-a publishing format for reproducible computational workflows. Elpub 2016, 2016, 87–90. [Google Scholar]
- Hammes, W.P.; Hertel, C. Bergey’s manual of systematic bacteriology. Volume 3, The firmicutes; In Genus I. Lactobacillus, 2nd ed.; online resource; Springer: Berlin/Heidelberg, Germany, 2009; Volume 2, p. 1. [Google Scholar]
- Schaeffer, A.B.; Fulton, M.D. A Simplified Method of Staining Endospores. Science 1933, 77, 194. [Google Scholar] [CrossRef]
- Sigma. Nystatin dihydrate Cell Culture Tested. 2007. Available online: https://www.sigmaaldrich.com/deepweb/assets/sigmaaldrich/product/documents/305/915/n4014pis.pdf?srsltid=AfmBOoqbeVgCF1wtEJ0gJLdlHL55suonZq2xL5u1vC2yUVWFHjS3ReZ4 (accessed on 3 July 2014).
- Fung, D.Y.; Miller, R.D. Effect of dyes on bacterial growth. Appl. Microbiol. 1973, 25, 793–799. [Google Scholar] [CrossRef]
- Baker, Z.; Harrison, R.; Miller, B.F. The bactericidal action of synthetic detergents. J. Exp. Med. 1941, 74, 611. [Google Scholar] [CrossRef]
- Xu, F.F.; Imlay, J.A. Silver (I), mercury (II), cadmium (II), and zinc (II) target exposed enzymic iron-sulfur clusters when they toxify Escherichia coli. Appl. Environ. Microbiol. 2012, 78, 3614–3621. [Google Scholar] [CrossRef]
- Macomber, L.; Imlay, J.A. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 8344–8349. [Google Scholar] [CrossRef]
- Whitman, W.B.; Parte, A.C. Systematic Bacteriology; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Cunningham, A.; Smith, A. The microbiology of silage made by the addition of mineral acids to crops rich in protein. II. The microflora. J. Dairy. Res. 1940, 2, 243. [Google Scholar] [CrossRef]
- Arslan, S. A review: Chemical, microbiological and nutritional characteristics of kefir. CyTA J. Food 2015, 13, 340–345. [Google Scholar] [CrossRef]
- FDA. Approved Animal Drug Products (Green Book); US Food and Drug Administration: Silver Spring, MD, USA, 2018.
- Zhang, Y.; Tao, X.; Liu, Q.; Zhang, Y.J.; Xu, J.; Zhang, W.; Wang, J.; Zhang, D.; Li, B.; Wang, L.; et al. Succession changes of fermentation parameters, nutrient components and bacterial community of sorghum stalk silage. Front. Microbiol. 2022, 13, 982489. [Google Scholar] [CrossRef] [PubMed]
- Agarussi, M.C.N.; Pereira, O.G.; Pimentel, F.E.; Azevedo, C.F.; da Silva, V.P.; FF, E.S. Microbiome of rehydrated corn and sorghum grain silages treated with microbial inoculants in different fermentation periods. Sci. Rep. 2022, 12, 16864. [Google Scholar] [CrossRef]
- Drouin, P.; Tremblay, J.; da Silva É., B.; Apper, E. Changes to the microbiome of alfalfa during the growing season and after ensiling with Lentilactobacillus buchneri and Lentilactobacillus hilgardii inoculant. J. Appl. Microbiol. 2022, 133, 2331–2347. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, J.; Shi, W.; Sun, J.; Xia, T.; Huang, F.; Liu, Y.; Li, H.; Teng, K.; Zhong, J. Dynamic Changes in Fermentation Quality and Structure and Function of the Microbiome during Mixed Silage of Sesbania cannabina and Sweet Sorghum Grown on Saline-Alkaline Land. Microbiol. Spectr. 2022, 10, e0248322. [Google Scholar] [CrossRef] [PubMed]
- Ghannoum, M.A.; Rice, L.B. Antifungal agents: Mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 1999, 12, 501–517. [Google Scholar] [CrossRef]
- Simpson, P.J.; Fitzgerald, G.F.; Stanton, C.; Ross, R.P. Enumeration and identification of pediococci in powder-based products using selective media and rapid PFGE. J. Microbiol. Methods 2006, 64, 120–125. [Google Scholar] [CrossRef]
- Leuschner, R.G.; Bew, J.; Simpson, P.J.; Ross, P.R.; Stanton, C. Enumeration of probiotic pediococci in animal feed: Interlaboratory study. J. AOAC Int. 2003, 86, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Vera, A.; Rigobello, V.; Demarigny, Y. Comparative study of culture media used for sourdough lactobacilli. Food Microbiol. 2009, 26, 728–733. [Google Scholar] [CrossRef]
- Duniere, L.; Jin, L.; Smiley, B.; Qi, M.; Rutherford, W.; Wang, Y.; McAllister, T. Impact of adding Saccharomyces strains on fermentation, aerobic stability, nutritive value, and select lactobacilli populations in corn silage. J. Anim. Sci. 2015, 93, 2322–2335. [Google Scholar] [CrossRef]
- Gomes, A.R.; Varela, C.L.; Pires, A.S.; Tavares-da-Silva, E.J.; Roleira, F.M.F. Synthetic and natural guanidine derivatives as antitumor and antimicrobial agents: A review. Bioorganic Chem. 2023, 138, 106600. [Google Scholar] [CrossRef]
- Oberg, C.J.; Moyes, L.V.; Domek, M.J.; Brothersen, C.; McMahon, D.J. Survival of probiotic adjunct cultures in cheese and challenges in their enumeration using selective media. J. Dairy Sci. 2011, 94, 2220–2230. [Google Scholar] [CrossRef]
- Farahmand, N.; Ouoba, L.I.I.; Naghizadeh Raeisi, S.; Sutherland, J.; Ghoddusi, H.B. Probiotic Lactobacilli in Fermented Dairy Products: Selective Detection, Enumeration and Identification Scheme. Microorganisms 2021, 9, 1600. [Google Scholar] [CrossRef]
- Colombo, M.; de Oliveira, A.E.; de Carvalho, A.F.; Nero, L.A. Development of an alternative culture medium for the selective enumeration of Lactobacillus casei in fermented milk. Food Microbiol. 2014, 39, 89–95. [Google Scholar] [CrossRef]
- Ingham, S.C. Use of modified Lactobacillus selective medium and Bifidobacterium iodoacetate medium for differential enumeration of Lactobacillus acidophilus and Bifidobacterium spp. in powdered nutritional products. J. Food Prot. 1999, 62, 77–80. [Google Scholar] [CrossRef]
- Suzuki, M.T.; Taylor, L.T.; DeLong, E.F. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 2000, 66, 4605–4614. [Google Scholar] [CrossRef]
- Polz, M.F.; Cavanaugh, C.M. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 1998, 64, 3724–3730. [Google Scholar] [CrossRef]
- Wali, A.; Hou, J.; Tsuruta, T.; Nishino, N. Bacterial and fungal microbiota of total mixed ration silage stored at various temperatures. J. Appl. Microbiol. 2022, 133, 579–590. [Google Scholar] [CrossRef]
- Ridwan, R.; Abdelbagi, M.; Sofyan, A.; Fidriyanto, R.; Astuti, W.D.; Fitri, A.; Sholikin, M.M.; Rohmatussolihat; Sarwono, K.A.; Jayanegara, A.; et al. A meta-analysis to observe silage microbiome differentiated by the use of inoculant and type of raw material. Front. Microbiol. 2023, 14, 1063333. [Google Scholar] [CrossRef]
- Xu, D.; Ding, W.; Ke, W.; Li, F.; Zhang, P.; Guo, X. Modulation of Metabolome and Bacterial Community in Whole Crop Corn Silage by Inoculating Homofermentative Lactobacillus plantarum and Heterofermentative Lactobacillus buchneri. Front. Microbiol. 2018, 9, 3299. [Google Scholar] [CrossRef]
- Dennis, S.M.; Nagaraja, T.G.; Bartley, E.E. Effects of lasalocid or monensin on lactate-producing or -using rumen bacteria. J. Anim. Sci. 1981, 52, 418–426. [Google Scholar] [CrossRef]



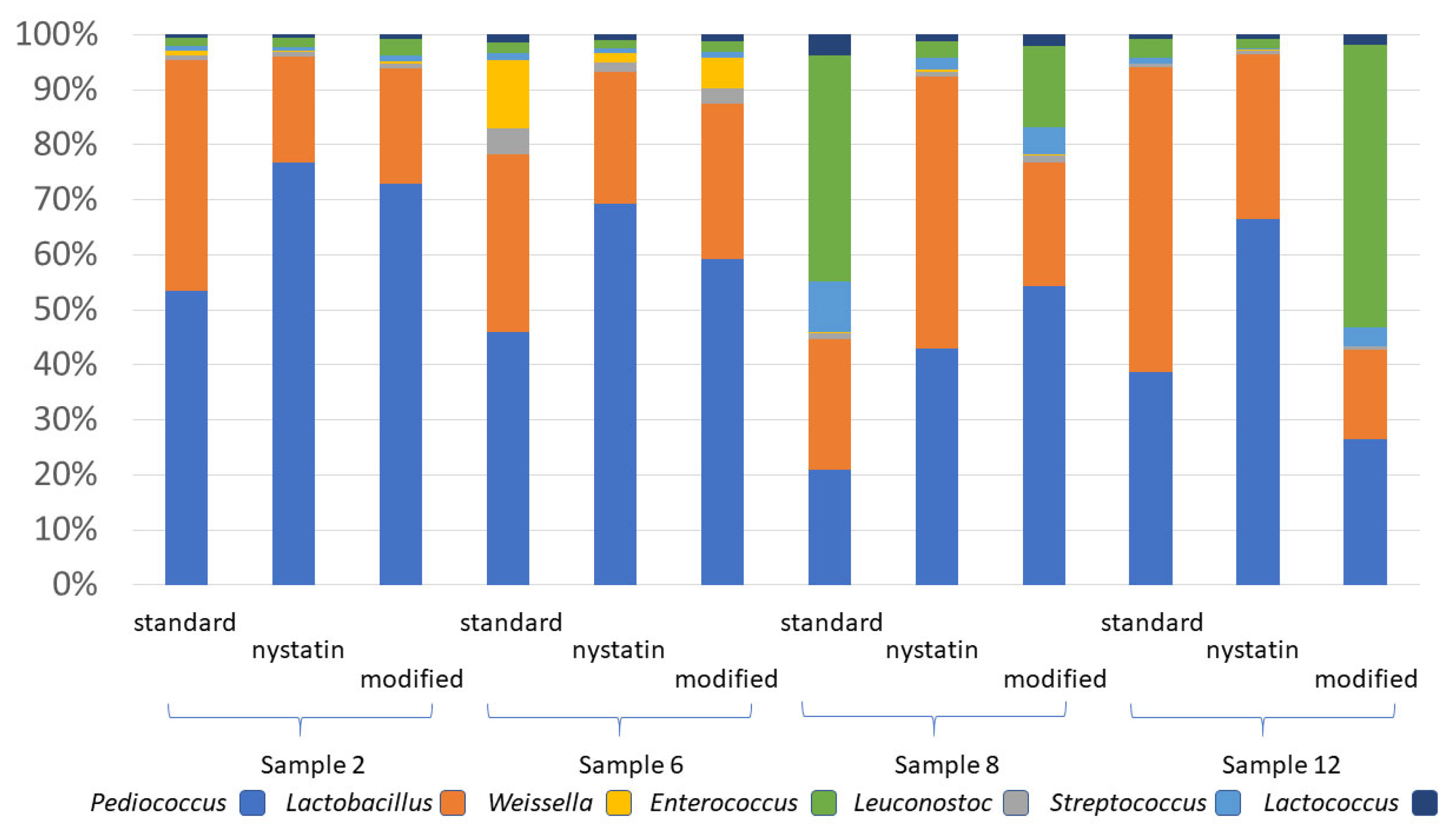


| Sample Name 1 | Project ID 2 | Sample Name 1 | Project ID 2 |
|---|---|---|---|
| MRS 2 | mgp83932 | MRS 6 | mgp84598 |
| MRS 8 | mgp84599 | MRS 12 | mgp86311 |
| MRS plus Nys 2 | mgp83479 | MRS plus Nys 6 | mgp83478 |
| MRS plus Nys 8 | mgp83480 | MRS plus Nys 12 | mgp84597 |
| Modified MRS 2 | mgp83308 | Modified MRS 6 | mgp83442 |
| Modified MRS 8 | mgp83443 | Modified MRS 12 | mgp83477 |
| Selective Agents 2 | Plate Counts (Log10, CFU/g) 1 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L. animalis NP51 | Cattle Feed | Silage | ||||||||||||
| 1 | 2 | 3 | 4 | 2 | 4 | 6 | 8 | 10 | 12 | 14 | 18 | 1 | 2 | |
| None | 7.64 | 7.80 | 8.00 | 7.91 | TNTC | 7.50 | TNTC | TNTC | 5.31 | 5.50 | 5.48 | 5.92 | 4.31 | 6.96 |
| A | 7.57 | 7.81 | 8.08 | 7.99 | TNTC | 7.33 | TNTC | TNTC | 5.30 | 5.52 | 5.48 | 6.03 | 4.33 | 6.79 |
| B | 7.65 | 7.77 | 7.96 | 7.86 | TNTC | 7.16 | TNTC | 5.48 | 5.31 | 5.67 | NG | 5.79 | 4.32 | 6.52 |
| C | 7.55 | 7.78 | 7.97 | 7.82 | 5.48 | 5.95 | TNTC | NG | NG | NG | NG | 4.48 | 2.90 | 5.48 |
| D | 7.53 | 7.75 | 8.99 | 7.60 | 5.70 | 6.56 | TNTC | 5.32 | 3.48 | 4.65 | NG | 5.18 | 4.17 | 6.26 |
| E | 7.61 | 7.85 | 7.96 | 7.88 | TNTC | 6.97 | TNTC | 5.00 | 3.48 | 4.08 | NG | 5.04 | 3.59 | 6.61 |
| F | 7.55 | 7.83 | 8.00 | 7.70 | 5.18 | 5.78 | TNTC | NG | NG | NG | NG | NG | 3.15 | 5.60 |
| G | 7.43 | 7.66 | 7.90 | 7.24 | 5.18 | 5.90 | TNTC | 3.48 | 3.30 | NG | NG | 4.60 | 3.23 | 6.00 |
| H | 7.45 | 7.81 | 7.98 | 7.84 | TNTC | 6.89 | TNTC | 3.78 | 3.60 | NG | NG | 4.85 | 3.57 | 6.54 |
| I | 7.27 | 7.59 | 7.70 | 7.61 | 5.00 | 5.60 | TNTC | NG | NG | NG | NG | NG | 3.08 | 6.04 |
| J | 7.21 | 7.62 | 7.66 | 7.05 | 4.90 | 5.95 | TNTC | NG | NG | NG | NG | 4.70 | 3.11 | 6.28 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thompson, K.; Akter, S.; Ferguson-Noel, N.; Maurer, J.J.; Lee, M.D. Development of a Selective Agar for the Detection of Probiotic Strain Ligilactobacillus animalis NP51 and Other Lactic Acid Bacteria in Cattle Feed. Agriculture 2025, 15, 1284. https://doi.org/10.3390/agriculture15121284
Thompson K, Akter S, Ferguson-Noel N, Maurer JJ, Lee MD. Development of a Selective Agar for the Detection of Probiotic Strain Ligilactobacillus animalis NP51 and Other Lactic Acid Bacteria in Cattle Feed. Agriculture. 2025; 15(12):1284. https://doi.org/10.3390/agriculture15121284
Chicago/Turabian StyleThompson, Kasey, Shamima Akter, Naola Ferguson-Noel, John J. Maurer, and Margie D. Lee. 2025. "Development of a Selective Agar for the Detection of Probiotic Strain Ligilactobacillus animalis NP51 and Other Lactic Acid Bacteria in Cattle Feed" Agriculture 15, no. 12: 1284. https://doi.org/10.3390/agriculture15121284
APA StyleThompson, K., Akter, S., Ferguson-Noel, N., Maurer, J. J., & Lee, M. D. (2025). Development of a Selective Agar for the Detection of Probiotic Strain Ligilactobacillus animalis NP51 and Other Lactic Acid Bacteria in Cattle Feed. Agriculture, 15(12), 1284. https://doi.org/10.3390/agriculture15121284






