1. Introduction
Melon (
Cucumis melo L.) is one of the most economically important horticultural crops, widely grown in many countries. The quality of melon is intrinsically associated with its variety. Various regions, climatic conditions, and soil types impose specific requirements on the cultivated melon varieties. Due to their unique taste, aroma, and nutrients, numerous melon varieties have been developed to satisfy the different demands of growers and consumers. In addition to being used for the propagation of melon plants, melon seeds can also serve as traditional Chinese medicine [
1] and can be processed to produce melon seed oils [
2]. Considering these factors, the identification of melon seed varieties can help to ensure the use of appropriate melon varieties.
The large number of melon varieties, along with the significant morphological similarities among seeds from different varieties, complicates the accurate identification of distinct varieties through visual inspection alone. Gene-based methods can be used to identify melon seed varieties accurately [
3,
4]. However, these methods are destructive, inefficient, and expensive, requiring professional operation skills; thus, they are not suitable for the large-scale analysis of each individual seed. Various non-destructive techniques have been used for seed variety identification [
5,
6], including melon seeds [
7]. Among these techniques, hyperspectral imaging has been proven to be an efficient and promising technique for seed variety identification [
8]. Being capable of acquiring spectral and spatial information simultaneously, hyperspectral imaging has been studied in various fields. As for seed quality inspection, hyperspectral imaging can be used for both batch samples and single-kernel seeds [
8].
The application of hyperspectral imaging in seed variety identification depends on the analysis of the hyperspectral images. Hyperspectral images can provide one-dimensional (1D) spectra, two-dimensional (2D) images, and three-dimensional (3D) hyperspectral data cubes. Seed variety identification has been successfully achieved based on the multi-dimensional data. Conventional data analysis methods have shown great success, including data preprocessing, feature selection/extraction, and modeling methods and strategies [
8]. In addition to these data analysis methods, deep learning has emerged as one of the most prominent approaches for analyzing hyperspectral images [
9,
10]. Multi-dimensional data can be directly processed using deep learning models. Deep learning, primarily known for its remarkable feature learning and mining ability, has now been extended to various fields. For seed variety identification, deep learning-based methods have demonstrated strong performance. The combination of deep learning and hyperspectral imaging has emerged as a potential alternative for real-world applications of seed variety identification. The spectral analysis of hyperspectral images, which is primarily a 1D data analysis issue, is the most commonly used data analysis strategy in hyperspectral imaging. Spectral features can reflect the physical and chemical properties of the samples. Deep learning models using 1D spectral data have demonstrated great success across various tasks, including seed variety classification [
5].
Moreover, image features can also provide useful information for seed quality inspection [
11,
12]. These features can provide information regarding morphology, color, and texture [
11]. Image feature-based seed variety classification has also been reported for seeds exhibiting external differences, such as variations in morphology, color, and texture [
11,
12,
13]. For certain seed varieties with differences in internal and external features, external features may strengthen classification performance. For image feature extraction, the manually defined features are extracted in advance using conventional approaches and then these features are used for classification [
11]. Some studies have demonstrated that image feature-based classification obtained good performance [
5,
11,
12,
13]. When the seeds exhibit quite similar external features, image feature-based analysis might not achieve satisfactory results. Thus, attempts to fuse the image features and spectral features to enhance classification performance have been widely explored [
14,
15]. In the case of hyperspectral imaging, image features are generally obtained through a dimension reduction approach to reduce the amount of data and explore informative features.
It is a matter of fact that hyperspectral imaging instruments are significantly more expensive, and, consequently, some researchers have combined spectral features with the features of RGB images [
14,
15]. For both hyperspectral images and RGB images, these features are predefined and must be extracted prior to further analysis [
14,
15,
16,
17,
18].
Due to the excellent feature learning abilities, deep learning-based analysis can be performed in an end-to-end manner, eliminating the need for the prior extraction of predefined features. However, the calculation time for manually extracted features is short, whereas the feature learning process of deep learning might be relatively longer. It is worth investigating the performance of manually extracted features (image features extracted manually based on the equations defined and validated by previous studies) with that of features automatically extracted by deep learning models. As for hyperspectral imaging, deep learning models using the 3D data cube can mine both spectral and image features. Generally, image features of hyperspectral imaging are extracted from gray-scale images at selected wavelengths. Currently, improving seed variety classification performance is still challenging.
In this research, hyperspectral imaging with deep learning was used to identify melon seed varieties. The fusion of spectral features and image features was explored. The specific objectives were to (1) compare the classification performance of LR, SVC, XGBoost, and CNN models for melon seed variety identification using 1D spectra data; (2) identify significant wavelengths and important image features for melon seed variety classification using Grad-CAM++; (3) establish 1D CNN models using image features extracted from RGB images and gray-scale images at selected feature wavelengths and 2D CNN models using RGB and spectral images at selected wavelengths, and compare the performance of these models; and (4) establish end-to-end CNN models to fuse spectral features and image features, and compare these models with the established models.
2. Materials and Methods
2.1. Sample Preparation
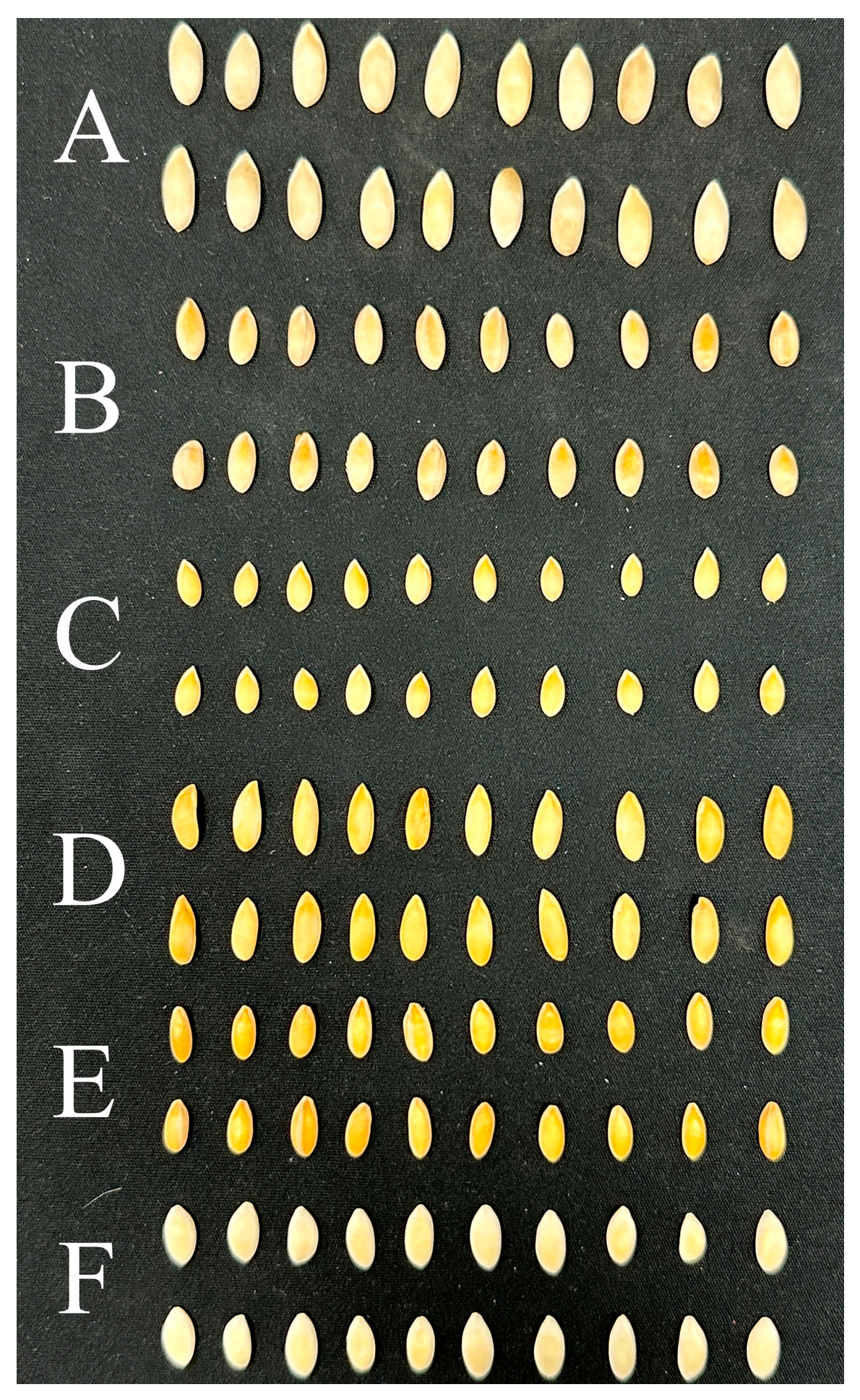
Melon seeds of six varieties (2A-234, CX-264, DFM-268, Zhetian103, Zhetian105, and Zhetian501) were collected from Zhejiang University, Hangzhou, Zhejiang Province, China, in 2023. These six melon varieties exhibit close genetic relationships. All seeds were intact and clean, and then used for hyperspectral image acquisition. Representative images of seeds of the six melon varieties are shown in
Figure 1. Detailed seed information is listed in
Table 1. The melon seeds of 2A-234, CX-264, DFM-268, Zhetian103, Zhetian105, and Zhetian501 were assigned the category values of 0, 1, 2, 3, 4, and 5, respectively.
2.2. Hyperspectral Image Acquisition and Correction
A laboratory-based hyperspectral imaging system was used to acquire hyperspectral images. The hyperspectral imaging system consists of an FX10 hyperspectral camera (Spectral Imaging Ltd., Oulu, Finland) at the spectral range of 400–1000 nm with a spectral resolution of 5.5 nm. The effective pixel size was 19.9 × 9.97 μm. A light source with six halogen lamps (OSRAM, Munich, Germany) was used for illumination. The lamps were symmetrically placed on either side of the camera, with three lamps in a row on each side. Each lamp had a power of 35 W. A mobile platform was used to transport the samples. The distance between the lower edge of the light source and the moving plate was 220 mm. The official software LUMO-Scanner 2020 (Spectral Imaging Ltd., Oulu, Finland) was used to control the entire hyperspectral imaging system. During the acquisition of images of melon seeds, the distance between the seeds and the camera lens was set at 300 mm, and the speed of the moving platform was set at 24.70 mm/s. During image acquisition, the seeds were randomly placed on the moving platform, with no seeds being in contact with one another.
Before image acquisition, a dark reference image was collected by covering the image lens with a black cover. During image acquisition, a white Teflon board was placed in front of the samples for white reference image acquisition. After acquiring the raw hyperspectral image, the raw image was corrected as the reflectance image using the dark reference image and white reference image based on the following equation:
where
IR is the raw hyperspectral image,
ID is the dark reference image,
IW is the white reference image, and
IC is the corrected image.
2.3. Spectra Extraction
After image acquisition and correction, the hyperspectral images were preprocessed to remove the background by establishing and applying the masks. The hyperspectral images were then cut into sub-images to ensure that each sub-image contained only one intact seed. After obtaining the hyperspectral images of each single melon seed, the spectral information was extracted. To extract the spectral information, each seed was defined as a region of interest (ROI). Each pixel within the ROI (seed) contained a spectrum, and the reflectance values of all pixels within the seed at each wavelength were averaged to represent the reflectance value of the seed at certain wavelengths. The average reflectance value of each wavelength was then combined to form the average spectrum of the seed. For each seed, although the number of pixels was different, the average spectra would help reduce the influence of seed sizes. Due to the fact that the head and tail of the spectra contained noises caused by the sensor responses and environments, spectra at these wavelengths were removed. Only the spectra in the range of 424–987 nm were used for analysis. As for spectral preprocessing, the models were first established using the raw spectra by trials, and good performance was obtained. Thus, no further spectral preprocessing was conducted.
2.4. Image Feature Extraction
During the acquisition of hyperspectral images, the software LUMO-Scanner automatically generated the RGB images of melon seeds with the same spatial size as the hyperspectral images. The RGB image was then cut into sub-images as the same as the sub-images of hyperspectral images. In each sub-image, there were only the seed and the black moving plate without any foreign materials, which were quite simple. There were obvious differences between the seed and the black plate, and a binarization approach was conducted to remove the background. The binarization was achieved as follows: a fixed threshold method was applied to segment the generated sub-image, producing a binary mask to remove the background. The threshold was determined using the Otsu method for each sub-image. Then, the image features were extracted according to the literature [
11]. A total of 47 features were extracted. There were 12 color features from the R, G, and B images, including the mean, standard deviation, maximum value, and minimum value of R, G, and B images. Then, the RGB images were transformed into HSV spaces as HSV images, and 12 similar color features were extracted. The monochrome image was constructed for gray-scale feature extraction, resulting in four features (mean, variance, standard deviation, and population mean). In addition to the color features, the morphological features were also extracted, including area, perimeter, convex hull perimeter, maximum feret diameter, major axis length, minor axis length, aspect ratio, ellipse ratio, thinness ratio, hydraulic radius, and orientation (the definitions of these features can be found in the literature [
11]). The texture features were also extracted from the monochrome image, including contrast, dissimilarity, homogeneity, energy, correlation, and angular second moment (ASM). The extracted features were used to establish the classification models. Moreover, the manually extracted image features were used for data fusion with the spectral features.
The 2D images can be directly processed by deep learning models. Thus, in addition to the manually extracted features, the preprocessed seed images were also used as inputs of the CNN models and the fusion model.
2.5. Data Analysis Methods
In this study, five datasets were constructed, namely, spectral features, manually extracted image features, seed images, fusion of spectral features and manually extracted image features, and fusion of spectral features and images. The classification models were established using the five datasets. The samples were randomly divided into the training, validation, and test sets. The number of samples in the training, validation, and test sets are shown in
Table 1. It should be noted that the orders of the samples in the spectral dataset, the image features dataset, and the fusion dataset were consistent. The conventional machine learning methods Logistic Regression (LR), Support Vector Classification (SVC), and Extreme Gradient Boosting (XGBoost) were used to establish models using spectral features and the manually extracted image features. The Convolutional Neural Network (CNN) was used to establish models based on the five datasets.
2.6. Conventional Machine Learning Methods
2.6.1. Logistic Regression (LR)
LR is a widely used classification method [
19]. LR is primarily a binary class classification method, and it calculates the probability of the sample to be one class based on the input features. The core of LR is the Sigmoid function to calculate the probability. LR can be extended to deal with multi-class classification problems.
2.6.2. Support Vector Classification (SVC)
SVC is the classification version of support vector machine (SVM) [
20]. It deals with both linear and non-linear issues effectively. For linearly separable samples, the goal of SVC is to find a linear classifier to maximumly classify the samples. For linear non-separable samples, SVC first maps the original data into a high-dimension space using kernel functions. The goal of SVC is to construct maximum-margin hyperplanes to maximumly classify the samples in the high-dimensional space. The selection of the kernel function is of importance. In spectral data analysis, the radial basis function (RBF) has shown good performance.
2.6.3. Extreme Gradient Boosting (XGBoost)
XGBoost is an ensemble-based machine learning method [
21]. XGBoost is based on the Gradient Boosting Decision Tree (GBDT), and ensembles several classification and regression trees (CARTs). The goal of XGboost is to minimize the loss function. The loss function of XGboost contains the output results of all CARTs and a regularization term. This type of loss function is used to obtain better prediction performance, reduce complexity, and avoid overfitting.
2.6.4. Convolutional Neural Network (CNN)
The revolutionary development of deep learning has gained great success in various fields. Deep learning-based artificial intelligence has groundbreaking applications. CNN is one of the most used deep learning architectures in data analysis [
22], including hyperspectral imaging [
9,
23]. A CNN model typically contains a number of convolutional layers, pooling layers, and fully connected layers, with batch normalization layers, dropout layers, etc. Due to the extraordinary feature learning abilities, CNN can deal with 1D, 2D, and 3D data in an end-to-end manner, which makes it quite suitable for hyperspectral image analysis. In this study, a shallow CNN was used to deal with the 1D spectra, 1D extracted image features, and 2D seed images to classify the varieties of melon seeds.
2.6.5. Efficient Channel Attention (ECA)
ECA is a lightweight attention mechanism optimized for Convolutional Neural Networks (CNNs) [
24]. ECA is designed to enhance performance with minimal model complexity. It avoids traditional dimensionality reduction after global average pooling to preserve the integrity of channel features, and employs a size-adaptive 1D convolution kernel to efficiently capture local cross-channel interactions between each channel and its k neighbors, thereby generating precise channel weights. Compared to methods like SENet, ECA significantly reduces parameter count and computational overhead while achieving an excellent balance of performance and efficiency.
2.6.6. Data Fusion Using CNN
The fusion of spectral features and image features has been widely explored in hyperspectral image analysis [
25,
26]. To explore the performance of fusing spectral and image features, CNN models for data fusion were designed. The data fusion consisted in two ways. The first way was to fuse the spectral features with the manually extracted image features. The second way was to fuse the spectral features and the seed images. In both ways, two-branch CNN models were established as the end-to-end deep fusion networks.
2.7. Software and Model Performance Evaluation Metrics
The hardware used in this research was a computer with 16 GB RAM, NVIDIA GeForce RTX 4060Ti GPU and INTEL i5-12400 CPU. Spectral extraction, image feature extraction, and data processing were carried out with Opencv-Python (version: 4.9.0) and Pycharm (2021.1.1) software based on Python (version: 3.9). LR, SVC, and XGboost models were developed using Scikit-Learn (version: 1.4.2) and CNN models using Pytorch (version: 2.2.2). The performance of the models was evaluated by classification accuracy.
3. Results
3.1. Spectral Profiles
Figure 2 shows the reflectance spectra of the six varieties of melon seeds. The colored shadow indicates the standard deviation of the reflectance of each wavelength, which shows the variation in the reflectance of each wavelength of the samples in the corresponding melon seed variety. The general trends of the spectra of the six varieties of melon seeds were similar, and overlaps could be observed. Hoverer, differences could also be observed among these spectral profiles. There was a crossing of the spectra of different varieties of melon seeds between 600 nm and 650 nm. These differences would help to differentiate melon seed varieties.
3.2. Analysis of Image Features
In this study, a total of 45 features were manually extracted from the seed RGB images. The statistical analysis of the 45 image features can be found in
Table S1 (in the Supplementary Materials). As shown in
Table S1, there were differences in the manually extracted image features. Although the differences in some image features of some varieties might not be significant, at least three varieties had significant differences in these image features. Moreover, it should be noted that some of the image features showed significant differences among all six varieties of melon seeds, such as hydraulic radius and feret diameter. All these differences in the image features of the six varieties of sweet melon seeds laid the foundation for the good performance of the models using these image features.
3.2.1. Model Development Using Spectral Features
LR, SVC, XGBoost, and CNN models were established using the 1D spectral features.
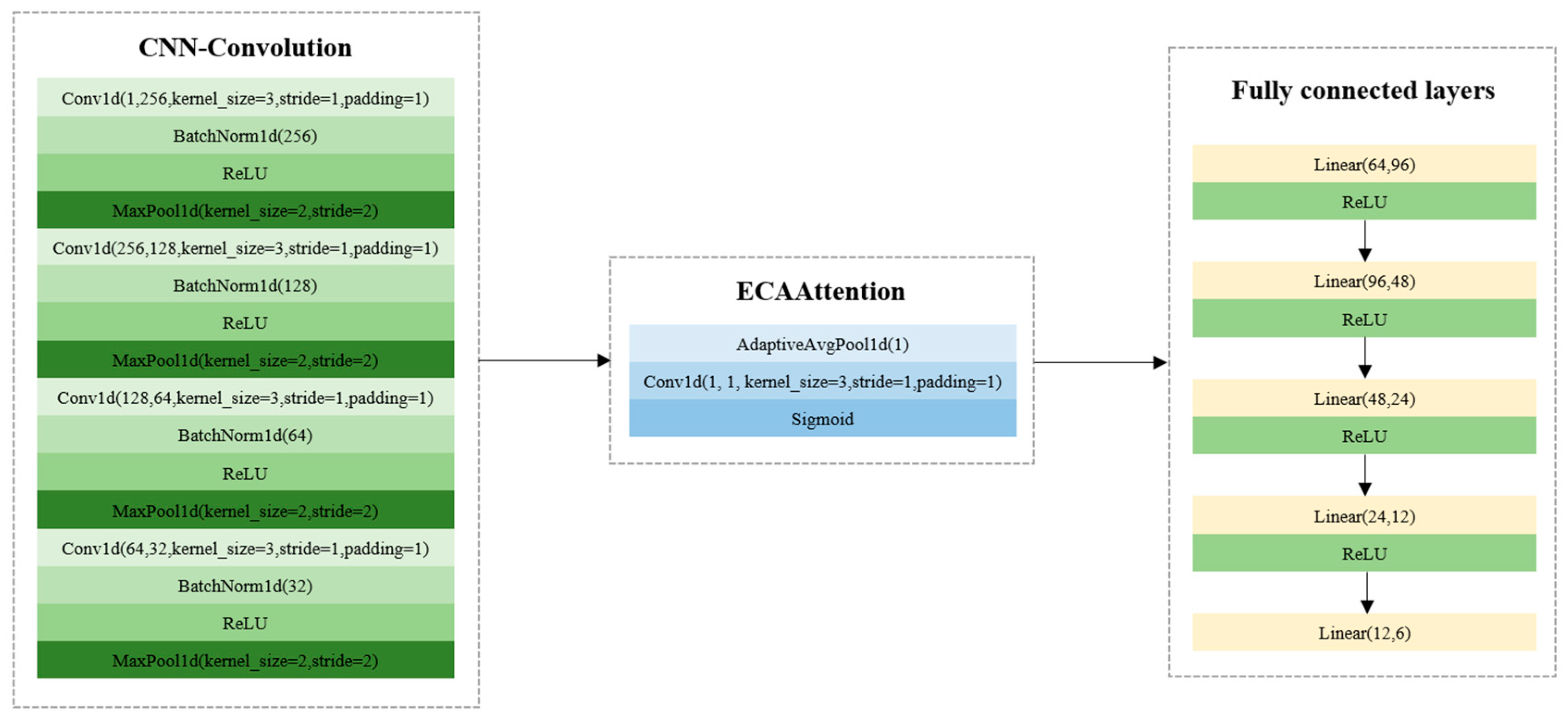
Figure 3 shows the structures of CNN models. LR, SVC, and XGBoost models were developed based on Scikit-Learn (version: 1.4.2), and the model parameters could be found and optimized in Scikit-Learn (version: 1.4.2). After trials, the optimal model parameters were obtained. For LR, the solver was selected as ‘liblinear’, the max_iter was set as 20,000, and the C value was set as 1.0. For SVC, the kernel function was selected as ‘linear’, and the C value was 1.0. For XGBoost, the max_depth was set as 4, the min_child_weight was set as 12, the learning_rate was set as 0.008, the n_estimators was set as 1200, the subsample was set as 0.8, and the colsample_bytree was set as 0.8. To train the CNN model, the batch size, the learning rate, and the number of epochs were set as 64, 0.01, and 500. The results of the LR, SVC, XGBoost, and CNN models are shown in
Table 2. As for the conventional machine learning models, XGBoost and SVC showed close results in the validation and test sets. The LR model obtained relatively worse results. The CNN model had much better performance, with the classification accuracy over 95% in the training, validation, and test sets. The overall results indicated that the spectral features could be used for the classification of the six varieties of melon seeds.
3.2.2. Model Development Using Extracted Image Features
In addition to the spectral features, manually extracted features were also used to build classification models to explore the feasibility of using the external image features to identify melon seed varieties. For LR, the solver was selected as ‘liblinear’, the max_iter was set as 20,000, and the C value was set as 1.0. For SVC, the kernel function was selected as ‘linear’, and the C value was 1.0. For XGBoost, the max_depth was set as 4, the min_child_weight was set as 12, the learning_rate was set as 0.008, the n_estimators was set as 1200, the subsample was set as 0.8, and the colsample_bytree was set as 0.8. To train the CNN model, the batch size, the learning rate, and the number of epochs were set as 64, 0.01, and 500. The classification results are shown in
Table 3. Excellent performance was obtained for melon seed variety identification by different models. The classification accuracy of the training, validation, and test sets was all over 90%. Slight differences in the performance of machine learning and deep learning models could be found. The results indicated that manually extracted 1D images could be used for the classification of the six varieties of melon seeds (
Figure 4).
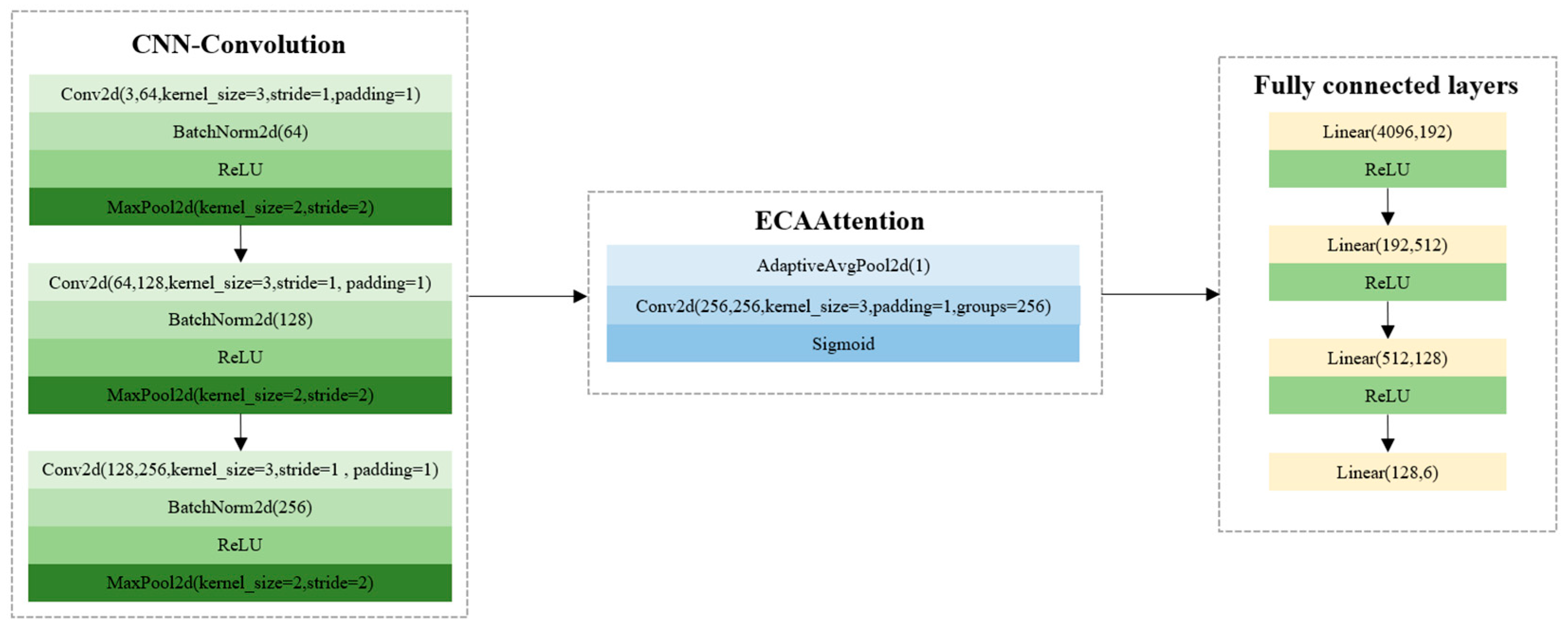
3.2.3. CNN Model Using Seed Images
In addition to the CNN model using 1D image features, the 2D images of melon seeds were also used to develop CNN models for variety identification. To train the CNN model, the batch size, the learning rate, and the number of epochs were set as 64, 0.01, and 500. To avoid the interference of the background, the background was removed for each seed. The seed images were then fed into a CNN model. The CNN model using the seed images as inputs obtained good performance, with the classification accuracy of the training, validation, and test sets at 100%, 94.49%, and 95.08%, respectively. The F1-scores of the training, validation, and test sets were 1.0000, 0.9444, and 0.9504, respectively. The training time of the CNN model was 1089.5620 s. The results indicated that 2D seed images could be used to identify the six varieties of melon seeds (
Figure 5).
3.2.4. CNN Fusion Models Using Spectral Features and Image Features
As mentioned above, both spectral features and image features could be used for melon seed variety identification. Attempts to fuse the spectral features and image features were also explored. Hyperspectral imaging can also provide the 3D data cube containing both spectral and image features. However, the training of CNN models using 3D data required more powerful computation capability and higher computation complexity. Considering that both spectral features and image features obtained good classification performance, the use of the 3D data cube for melon seed variety identification was not utilized.
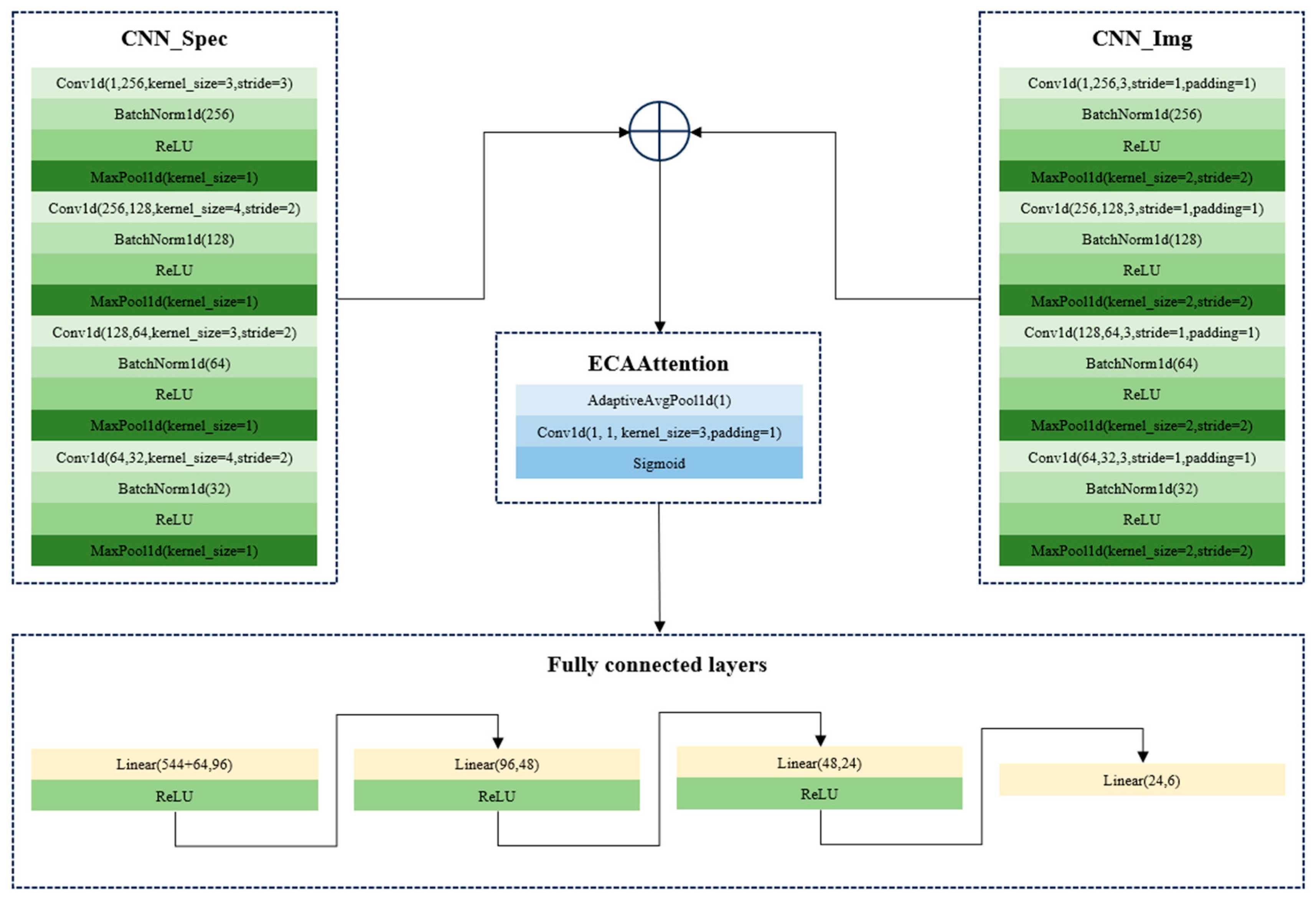
In this study, the fusion of spectral and image features was conducted using two strategies. First, the spectral features (1D) and the manually extracted image features (1D) were used as inputs of a two-branch CNN model for end-to-end fusion. The CNN architectures for the fusion of 1D spectral features and 1D manually extracted image features are shown in
Figure 6. Second, the spectral features (1D) and the processed images (2D) were used as inputs of a two-branch CNN model for end-to-end fusion. The CNN architectures for the fusion of 1D spectral features and 2D seed images are shown in
Figure 7. In both strategies, the features were firstly learned by one branch, and the learned features were then also concatenated for the next steps. During the training of CNN models using the fusion set, the batch size, the learning rate, and the number of epochs were set as 64, 0.01, and 500 for the two CNN models. The results are shown in
Table 4.
As shown in
Table 4, both fusion strategies obtained good and close performance, with the classification accuracy of the training, validation, and test sets over 97%. The overall results illustrated the effectiveness of the fusion strategies and the fact that spectral features and image features can provide complementary information to each other. However, the training of the CNN model using the fusion of the 1D spectral features and 2D seed images required more computation resources. Assuming that 1D manually extracted features can be used for the classification of the six varieties of melon seeds, the fusion of 1D spectral features and 1D manually extracted image features might be more practical.
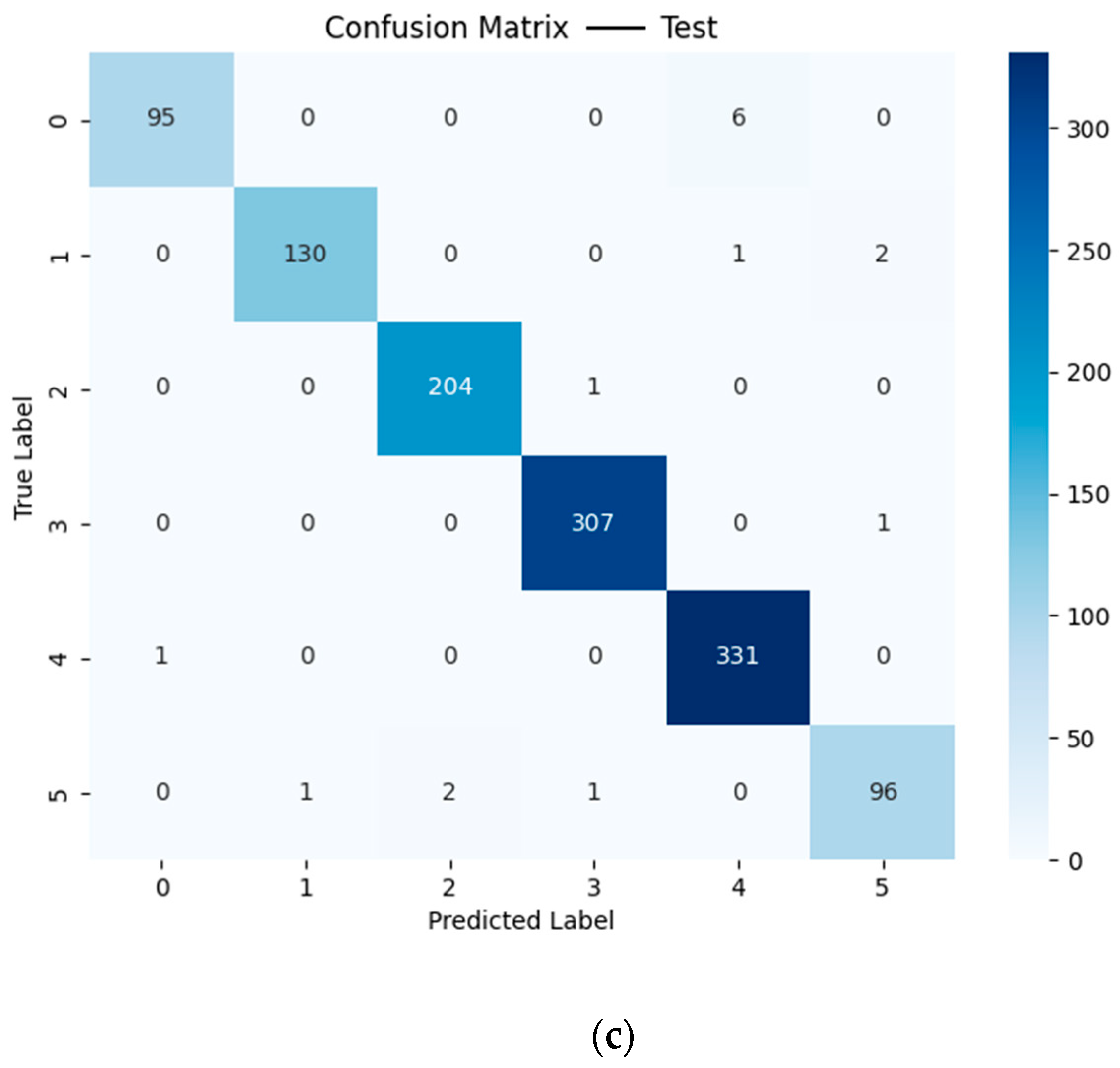
Figure 8 shows the confusion matrix of the training, validation, and test sets based on the CNN model using the fusion of 1D spectral features and 1D manually extracted image features. It should be noted that no particular pattern was found for the misclassification.
3.3. Comparison of Results of Different Datasets
The classification models using the spectral feature dataset, the manually extracted image feature dataset, the image data, the fusion dataset of spectral features and manually extracted image features, and the fusion dataset of spectral features and the images all obtained good performance. Conventional machine learning algorithms (LR, SVC, and XGBoost) based on image features performed better than those based on spectral features. However, the CNN models using spectral features and image features obtained good and close results, indicating the effectiveness of CNN models in melon seed variety identification in this study. On the other hand, the number of image features was much lower than that of spectral features, indicating that there were differences in the external features of the melon seeds.
The CNN models using the seed images as inputs also obtained good performance, and the performance was similar to that of models using spectral features and image features. However, compared with the 1D features, the 2D images were more difficult for CNN training, requiring more computation power and time.
The CNN models using the two fusion datasets showed better performance than those using the other three datasets. Both spectral and image features contained distinctive features of the six varieties of melon seeds, and the fusion of the two types of features can provide more comprehensive information for melon seed variety identification.
Regarding the training time, it should be noted that the training time of the models using the fusion of 1D spectra and 2D seed image was the longest, followed by the CNN model using the seed images as input. The training times of CNN models using 1D spectra features or manually extracted image features were relatively longer than those of the conventional machine learning models. However, once the models were trained, the optimal model was saved and the saved models could be used for prediction. The prediction time was quite short for all models.
3.4. SHapley Additive exPlanations Analysis
The overall results show that the classification models using spectral features and image features obtained good performance. To further explore the important features contributing more to the melon seed variety identification, SHapley Additive exPlanations (SHAP) analysis was conducted based on the models established using spectral features and manually extracted image features. The features were ranked according to the mean absolute SHAP values of each model for each melon seed variety.
As for spectral features, the top 50 wavelengths with the highest mean absolute SHAP values of each variety in the LR, SVC, XGBoost, and CNN models are listed in
Tables S2–S7. It can be observed that for each variety, there were some common important wavelengths for any two models, but there were quite a few common wavelengths for all four models. As for manually extracted image features, the top 20 image features with the highest mean absolute SHAP values of each variety are listed in
Tables S8–S13. It can be observed that for each variety, there were some common important wavelengths for any two models, but there were only a few common wavelengths for all four models. Differences in feature importance could also be found for different varieties of melon seeds. These differences in the important features of each variety might be attributed to the different principles of the models. Another possible reason might be the limited number of seeds, and more seeds should be studied to further explore the features contributing more to the classification.
4. Discussion
In this research, the utilization of spectral features, image features, and the fusion of spectral and image features to identify melon seed varieties was explored, achieving good performance. The good classification results indicated that hyperspectral imaging combined with CNN can effectively identify the six varieties of melon seeds. The successful application of spectral features, manually extracted image features, seed images, and the fusion of spectral features and images has been demonstrated in various types of seeds [
11,
12,
14,
17,
27].
In this research, the classification performance of conventional machine learning models using manually extracted image features was better than that of models using spectral features, indicating significant differences in the external features of the six varieties of melon seeds. These differences may also be the reason that CNN models using seed images and manually extracted features obtained quite similar and good results.
Although deep learning can directly process 3D hyperspectral images to mine spectral features and image features, the hardware requirements and computation time are relatively high. The fusion of spectral features and image features is widely explored in the data analysis of hyperspectral images by extracting these features separately from the hyperspectral images [
16,
17,
18]. In this research, the models using the fusion of spectral features with manually extracted features and the fusion of spectral features with seed images obtained similar results, which were better than those obtained from models using spectral features and image features, respectively. In previous studies aimed at identifying seed varieties, the models using the fused datasets generally showed better performance than those using spectral features and image features independently [
15,
17,
18].
The good performance of the spectral and image feature-based classification of melon seed varieties indicated that there may be significant differences in both internal and external features among the six varieties of melon seeds. However, there is a wide variety of melon seed types. In future, many more varieties of melon seeds should be studied, and corresponding data analysis strategies should be developed. The spectral features and image features (including manually extracted image features and seed images) demonstrate great potential for identifying melon seed varieties, which is critical for the online real-time automatic detection of melon seed purity and the online real-time automatic sorting of suitable melon seeds. Furthermore, the fusion of spectral features and image features can provide comprehensive information about the seeds, thereby improving classification accuracy. The data analysis strategies employed in this study would be available to identify various plant seed varieties. The use of the spectral features and the image features from seed RGB images also showed the potential to develop low-cost spectrometers and RGB cameras instead of hyperspectral imaging for seed variety identification, via proper and optimized design.
5. Conclusions
Hyperspectral imaging combined with deep learning models was successfully used to identify six varieties of melon seeds: 2A-234, CX-264, DFM-268, Zhetian103, Zhetian105, and Zhetian501. Five datasets were constructed from the hyperspectral images and the RGB images (obtained during hyperspectral image acquisition), namely, seed spectral features, manually extracted features from seed RGB images, seed RGB images, the fusion of seed spectral features and manually extracted features, and the fusion of seed spectral features with seed images. The classification models established using these datasets showed good performance, with CNN models achieving classification accuracy over 90% across the training, validation, and test sets for all five datasets. These results indicated that hyperspectral imaging combined with CNN had great potential for the classification of melon seed varieties. Moreover, the good performance of models using spectral features and image features illustrated that there may be significant differences in chemical compositions and external features among the six varieties of melon seeds. The relatively better performance of the models using fused datasets indicated that the utilization of both spectral features and image features can provide comprehensive information for the identification of the six varieties of melon seeds. The findings of this study could contribute to the development of models that use both spectral features and image features for rapid and accurate melon seed variety identification. In future, more melon seed varieties with relatively minor chemical and physical variations should be studied, and more data analysis strategies should be explored to obtain better performance.