Real-Time Corn Variety Recognition Using an Efficient DenXt Architecture with Lightweight Optimizations
Abstract
1. Introduction
- Data collection and pre-processing: We collected images of leaves, staminates, and root caps of 40 corn varieties in Gansu Province at the nodulation stage and ensured the quality and usability of the data through image pre-processing and screening techniques, which provided high-quality data support for model training.
- Model optimization: We introduced the Representative Batch Normalization (RBN) structure into the DenseNet121 network model, which improves the generalization ability of the model under different data distributions and batch sizes.
- Structure optimization and feature extraction: Combining the advantages of the SE module and deep separable convolution improves the feature expression ability of the model, while reducing the computational cost, decreasing the model complexity, and ensuring high efficiency.
- Regularization and generalization ability: By introducing dropout regularization, the risk of overfitting of the model is reduced and the robustness on new data is improved.
2. Materials and Methods
2.1. Experimental Materials and Processing
2.1.1. Image Acquisition
2.1.2. Image Processing
2.2. Basic Methodology and Test Environment
2.2.1. Contrast Model
2.2.2. Evaluation Metrics
2.2.3. Test Environment
3. Model Improvements
3.1. Improving the DenseNet Model
3.1.1. Representative BatchNorm (RBN)
3.1.2. SE Attention Mechanism
3.1.3. Depth Separable Convolution
3.1.4. Dropout
3.2. The DenXt Model
4. Results and Discussion
4.1. Ablation Experiments and Comparative Analysis
4.2. Analysis of Classification Results
4.3. Comparison with Other Models
4.4. Network Visualization
5. Conclusions and Outlook
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations (FAO). FAOSTAT Database. 2023. Available online: https://www.fao.org/faostat/en/#home (accessed on 20 August 2024).
- Edmeades, G.O.; Trevisan, W.; Prasanna, B.M.; Campos, H. Tropical maize (Zea mays L.). In Genetic Improvement of Tropical Crops; Springer: Cham, Switzerland, 2017; pp. 57–109. [Google Scholar]
- Erenstein, O.; Jaleta, M.; Sonder, K.; Mottaleb, K.; Prasanna, B.M. Global maize production, consumption and trade: Trends and R&D implications. Food Secur. 2022, 14, 1295–1319. [Google Scholar]
- Guerra, A.; Scremin-Dias, E. Leaf traits, sclerophylly and growth habits in plant species of a semiarid environment. Braz. J. Bot. 2018, 41, 131–144. [Google Scholar] [CrossRef]
- Chen, F.; Liu, J.; Liu, Z.; Chen, Z.; Ren, W.; Gong, X.; Wang, L.; Cai, H.; Pan, Q.; Yuan, L.; et al. Breeding for high-yield and nitrogen use efficiency in maize: Lessons from comparison between Chinese and US cultivars. Adv. Agron. 2021, 166, 251–275. [Google Scholar]
- Ganesh, A.; Shukla, V.; Mohapatra, A.; George, A.P.; Bhukya DP, N.; Das, K.K.; Kola, V.S.R.; Suresh, A.; Ramireddy, E. Root cap to soil interface: A driving force toward plant adaptation and development. Plant Cell Physiol. 2022, 63, 1038–1051. [Google Scholar] [CrossRef] [PubMed]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Tiwari, V.; Joshi, R.C.; Dutta, M.K. Dense convolutional neural networks based multiclass plant disease detection and classification using leaf images. Ecol. Inform. 2021, 63, 101289. [Google Scholar] [CrossRef]
- Laabassi, K.; Belarbi, M.A.; Mahmoudi, S.; Mahmoudi, S.A.; Ferhat, K. Wheat varieties identification based on a deep learning approach. J. Saudi Soc. Agric. Sci. 2021, 20, 281–289. [Google Scholar] [CrossRef]
- Oikonomidis, A.; Catal, C.; Kassahun, A. Deep learning for crop yield prediction:a systematic literature review. N. Zeal. J. Crop Hortic. Sci. 2023, 51, 1–26. [Google Scholar] [CrossRef]
- Wang, Z.; Huang, W.; Tian, X.; Long, Y.; Li, L.; Fan, S. Rapid and non-destructive classification of new and aged maize seeds using hyperspectral image and chemometric methods. Front. Plant Sci. 2022, 13, 849495. [Google Scholar] [CrossRef]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef]
- Rasti, S.; Bleakley, C.J.; Silvestre, G.C.; Holden, N.M.; Langton, D.; O’Hare, G.M. Crop growth stage estimation prior to canopy closure using deep learning algorithms. Neural Comput. Appl. 2021, 33, 1733–1743. [Google Scholar] [CrossRef]
- Anami, B.S.; Malvade, N.N.; Palaiah, S. Deep learning approach for recognition and classification of yield affecting paddy crop stresses using field images. Artif. Intell. Agric. 2020, 4, 12–20. [Google Scholar] [CrossRef]
- Song, Z.; Wang, P.; Zhang, Z.; Yang, S.; Ning, J. Recognition of sunflower growth period based on deep learning from UAV remote sensing images. Precis. Agric. 2023, 24, 1417–1438. [Google Scholar] [CrossRef]
- Xu, J.; Wang, J.; Xu, X.; Ju, X. Rice growth stage image recognition based on RAdam convolutional neural network. Trans. Chin. Soc. Agric. Eng. 2021, 37, 143–150. [Google Scholar]
- Liu, P.; Liu, L.; Wang, C.; Zhu, Y.; Wang, H.; Li, X. Method for determining the flowering stage of wheat in the field based on machine vision. J. Agric. Mach. 2022, 53, 251–258. [Google Scholar]
- Han, Y.; Xing, H.; Jin, H. Design of an automatic detection system for maize seedling emergence and three-leaf stage based on OpenCV. J. Electron. Meas. Instrum. 2017, 31, 1574–1581. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, R.; Liu, M.; Gong, Y. Recognition of maize growth stages based on deep convolutional features. Electron. Meas. Technol. 2018, 41, 79–84. [Google Scholar] [CrossRef]
- Shi, L.; Lei, J.; Wang, J.; Yang, C.; Liu, Z.; Lei, X.; Xiong, S. A lightweight wheat growth stage recognition model based on improved FasterNet. J. Agric. Mach. 2024, 55, 226–234. [Google Scholar]
- Zheng, G.; Wei, J.; Ren, Y.; Liu, H.; Lei, X. Research on a lightweight wheat growth monitoring model based on deep separable and dilated convolutions. Jiangsu J. Agric. Sci. 2022, 50, 226–232. [Google Scholar] [CrossRef]
- Sheng, R.T.-C.; Huang, Y.-H.; Chan, P.-C.; Bhat, S.A.; Wu, Y.-C.; Huang, N.-F. Rice growth stage classification via RF-based machine learning and image processing. Agriculture 2022, 12, 2137. [Google Scholar] [CrossRef]
- Mo, H.; Wei, L. SA-ConvNeXt: A hybrid approach for flower image classification using selective attention mechanism. Mathematics 2024, 12, 2151. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Yang, H.; Ni, J.; Gao, J.; Han, Z.; Luan, T. A novel method for peanut variety identification and classification by Improved VGG16. Sci. Rep. 2021, 11, 15756. [Google Scholar] [CrossRef]
- Koonce, B.; Koonce, B. MobileNetV3. In Convolutional Neural Networks with Swift for Tensorflow: Image Recognition and Dataset Categorization; Apress: New York, NY, USA, 2021; pp. 125–144. [Google Scholar]
- Mukti, I.Z.; Biswas, D. Transfer learning based plant diseases detection using ResNet50. In Proceedings of the 2019 4th International Conference on Electrical Information and Communication Technology (EICT), Khulna, Bangladesh, 20–22 December 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–6. [Google Scholar]
- Feng, J.; Tan, H.; Li, W.; Xie, M. Conv2NeXt: Reconsidering Conv NeXt Network Design for Image Recognition. In Proceedings of the 2022 International Conference on Computers and Artificial Intelligence Technologies (CAIT), Quzhou, China, 4–6 November 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 53–60. [Google Scholar]
- Xing, X.; Liu, C.; Han, J.; Feng, Q.; Lu, Q.; Feng, Y. Wheat-seed variety recognition based on the GC_DRNet model. Agriculture 2023, 13, 2056. [Google Scholar] [CrossRef]
- Gao, S.H.; Han, Q.; Li, D.; Cheng, M.M.; Peng, P. Representative batch normalization with feature calibration. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 8669–8679. [Google Scholar]
- Mi, Z.; Zhang, X.; Su, J.; Han, D.; Su, B. Wheat stripe rust grading by deep learning with attention mechanism and images from mobile devices. Front. Plant Sci. 2020, 11, 558126. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
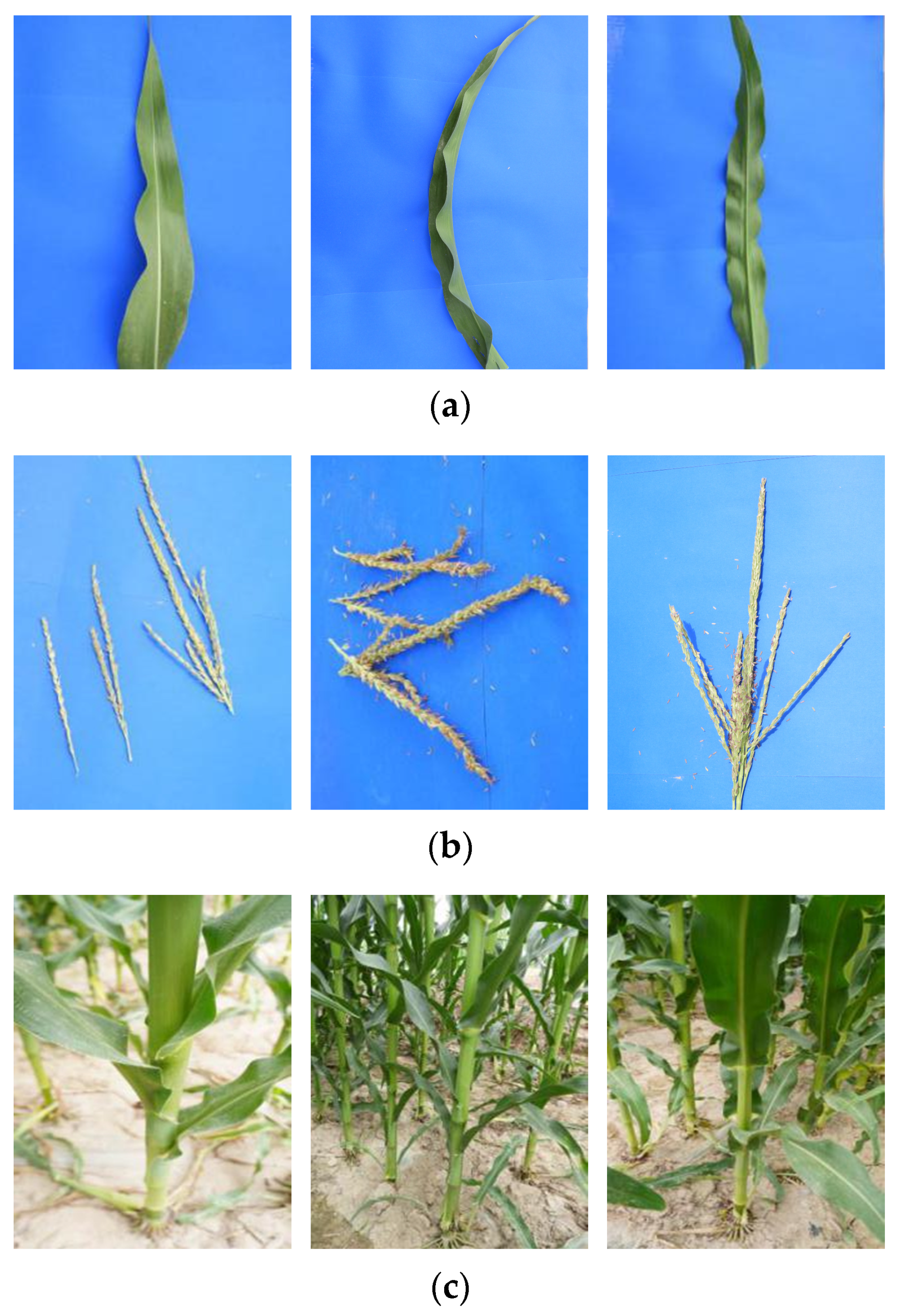








| Range | Breed (Line) | Blade Image | Staminate Image | Root Cap Image | ||
|---|---|---|---|---|---|---|
| Hybrids | LD632 | LD633 | LD635 | 450 | 150 | 300 |
| LD635 | LD655 | LD656 | ||||
| LD657 | LD659 | LD636 | ||||
| LD2463 | LD24159 | LD634 | ||||
| XY1483 | XY335 | XY698 | ||||
| XY1620 | XY1516 | R1831 | ||||
| RP909 | DF899 | |||||
| Parent | Parent 1 | Parent 2 | Parent 3 | 300 | 150 | 200 |
| Parent 4 | Parent 5 | Parent 6 | ||||
| Parent 7 | Parent 8 | Parent 9 | ||||
| Parent 10 | Parent 11 | Parent 12 | ||||
| Parent 13 | Parent 14 | Parent 15 | ||||
| Parent 16 | Parent 17 | Parent 18 | ||||
| Parent 19 | Parent 20 |
| Indicator | Formula |
|---|---|
| Accuracy (A) | |
| Precision (P) | |
| Recall (R) | |
| F1 |
| Attention Mechanism | Accuracy (%) | Precision (%) | Recall (%) | F1 Score (%) |
|---|---|---|---|---|
| ECA | 89.89 | 90.68 | 89.88 | 89.95 |
| CBAM | 91.71 | 91.84 | 91.51 | 91.45 |
| SE | 94.81 | 95.04 | 94.66 | 94.67 |
| Model | Total Parameters | Trainable Parameters | Model Parameters (MB) |
|---|---|---|---|
| Densenet 121 | 6,994,856 | 6,994,856 | 20.68 |
| DenXt | 5,276,664 | 5,276,664 | 20.13 |
| Layer Type | DenseNet121 | DenXt | Improvement |
|---|---|---|---|
| Conv Layer | 7 × 7 Conv, stride = 2, BN-ReLU | 7 × 7Conv, stride = 2, RBN | RBN combines ReLU and BatchNorm to enhance stability and speed up training. |
| Pooling | Maxpool, 3 × 3, stride = 2 | Maxpool, 3 × 3, stride = 2 | - |
| Dense Block1 | BN-ReLU Conv1, BN-ReLU Conv2 (6x) | ReLU-RBN Conv1, ReLu-RBN-SE, ReLU-RBN Conv2 (6x) | The introduction of RBN and SEBlock for feature recalibration helps to learn important features. |
| Transition Layer1 | BN-ReLU 1 × 1 Conv, Maxpool 2 × 2 | RBN-ReLU 1 × 1 Conv, Maxpool 2 × 2 | Apply RBN to 1 × 1 convolution to stabilize training. |
| Dense Block2 | BN-ReLU Conv1, BN-ReLU Conv2 (12x) | ReLU-RBN Conv1, ReLu-RBN-SE, ReLU-RBN Conv2 (12x) | The introduction of RBN and SEBlock for feature recalibration helps to learn important features. |
| Transition Layer2 | BN-ReLU 1 × 1 Conv, Maxpool 2 × 2 | RBN-ReLU 1 × 1 Conv, Maxpool 2 × 2 | Apply RBN to 1 × 1 convolution to stabilize training. |
| Dense Block3 | BN-ReLU Conv1, BN-ReLU Conv2 (24x) | ReLU-RBN Conv1, ReLu-RBN-SE, ReLU-RBN Conv2 (24x) | The introduction of RBN and SEBlock for feature recalibration helps to learn important features. |
| Transition Layer3 | BN-ReLU 1 × 1 Conv, Maxpool 2 × 2 | RBN-ReLU 1 × 1 Conv, Maxpool 2 × 2 | Apply RBN to 1 × 1 convolution to stabilize training. |
| Dense Block4 | BN-ReLU Conv1, BN-ReLU Conv2 (16x) | ReLU-RBN Conv1, ReLu-RBN-SE, ReLU-RBN Conv2 (16x) | Combining RBN and SEBlock to further refine the features in the final dense block. |
| Classification Layer | 7 × 7 global average pool, 1024D fully conected, softmax | 7 × 7 global average pool, 1024D fully conected, softmax | - |
| Dropout | - | Applied after each Dense Block (6x, 12x, 24x, 16x) | Dropout prevents overfitting by randomly discarding units during training. |
| Model | Improvement Methods | Acc (%) | F1 (%) | |||
|---|---|---|---|---|---|---|
| Representative BatchNorm | Squeeze and Excitation | Depthwise Separable Convolution | Dropout | |||
| Densenet 121 | 94.55 | 94.19 | ||||
| Den-RBN | √ | 95.01 | 94.97 | |||
| Den-SE | √ | 94.81 | 94.67 | |||
| Den-DS | √ | 96.54 | 96.50 | |||
| Den-Drop | √ | 94.89 | 94.66 | |||
| DenXt | √ | √ | √ | √ | 97.79 | 97.75 |
| Model | Accuracy/% | Precision/% | Recall/% | F1 Score/% | Parameters Size (MB) | GPU Memory (MB) | Inference Time (ms) |
|---|---|---|---|---|---|---|---|
| DenseNet 121 | 94.55 | 94.48 | 94.20 | 94.19 | 26.08 | 27.11 | 28,774.34 |
| VGG16 | 89.45 | 89.67 | 89.23 | 89.19 | 512.79 | 513.66 | 13,243.09 |
| MobileNet V3 | 89.49 | 89.34 | 88.99 | 88.91 | 16.22 | 16.40 | 2407.02 |
| ResNet50 | 92.69 | 92.91 | 92.30 | 92.30 | 90.02 | 90.29 | 2661.37 |
| ConvNeXt | 94.49 | 94.46 | 94.23 | 94.23 | 748.79 | 749.67 | 3413.79 |
| DenXt | 97.79 | 97.27 | 97.75 | 97.75 | 20.13 | 20.62 | 2265.95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Liu, C.; Han, J.; Zhou, Y.; Li, Y.; Zhang, L. Real-Time Corn Variety Recognition Using an Efficient DenXt Architecture with Lightweight Optimizations. Agriculture 2025, 15, 79. https://doi.org/10.3390/agriculture15010079
Zhao J, Liu C, Han J, Zhou Y, Li Y, Zhang L. Real-Time Corn Variety Recognition Using an Efficient DenXt Architecture with Lightweight Optimizations. Agriculture. 2025; 15(1):79. https://doi.org/10.3390/agriculture15010079
Chicago/Turabian StyleZhao, Jin, Chengzhong Liu, Junying Han, Yuqian Zhou, Yongsheng Li, and Linzhe Zhang. 2025. "Real-Time Corn Variety Recognition Using an Efficient DenXt Architecture with Lightweight Optimizations" Agriculture 15, no. 1: 79. https://doi.org/10.3390/agriculture15010079
APA StyleZhao, J., Liu, C., Han, J., Zhou, Y., Li, Y., & Zhang, L. (2025). Real-Time Corn Variety Recognition Using an Efficient DenXt Architecture with Lightweight Optimizations. Agriculture, 15(1), 79. https://doi.org/10.3390/agriculture15010079






