Genomic Prediction for Inbred and Hybrid Polysomic Tetraploid Potato Offspring
Abstract
1. Introduction
2. Materials and Methods
2.1. Statistical Models
Cross-Validation
2.2. Software
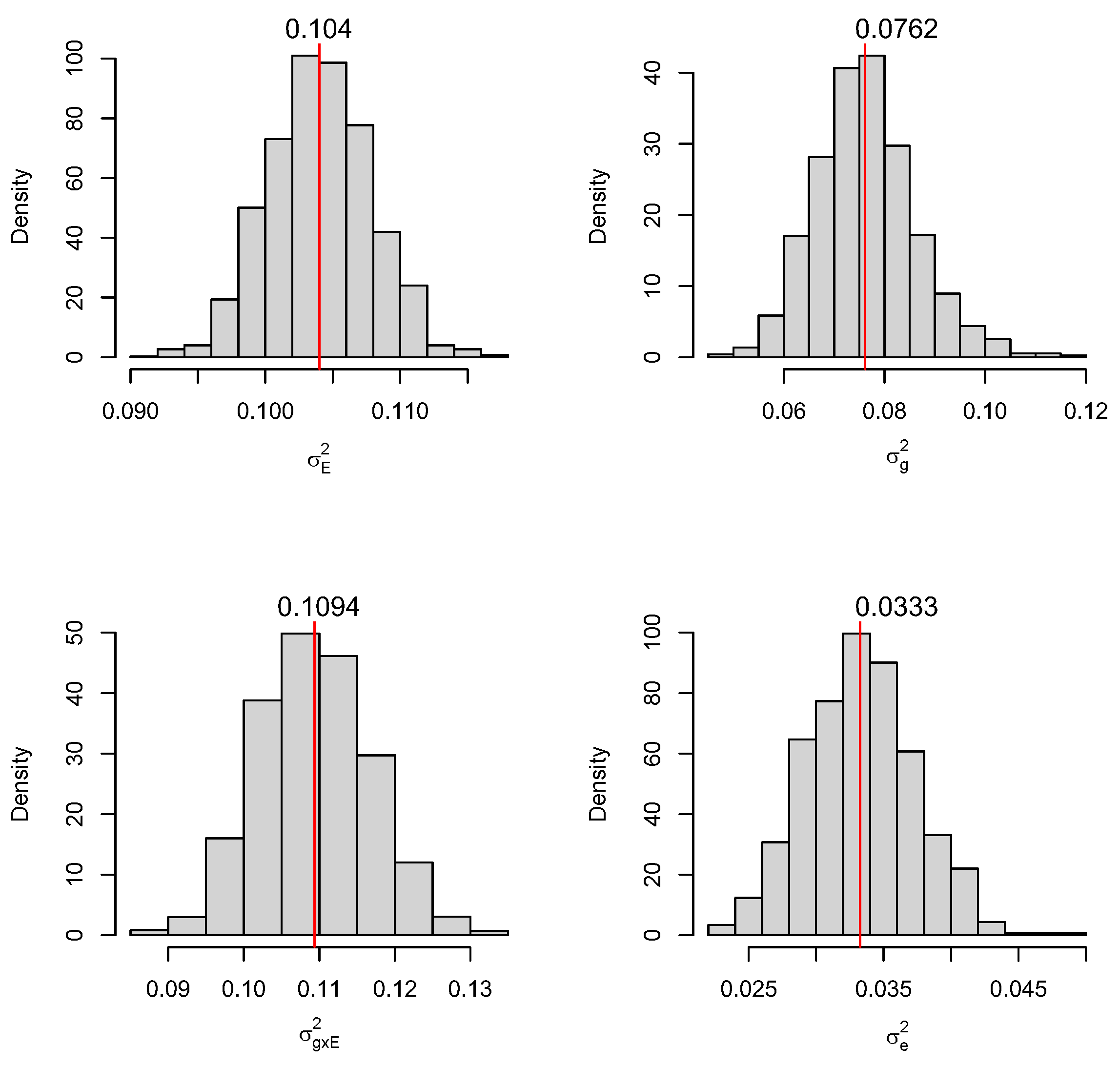
3. Results
4. Discussion
4.1. Inbred vs. Hybrid Offspring Performance
4.2. Genotype Influence on Predictive Ability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eriksson, D.; Carlson-Nilsson, U.; Ortíz, R.; Andreasson, E. Overview and breeding strategies of table potato production in Sweden and the Fennoscandian Region. Potato Res. 2016, 59, 279–294. [Google Scholar] [CrossRef]
- Ortiz, R.; Selga, C.; Reslow, F.; Carlson-Nilsson, U. Svensk potatisförädling: Breeding the new table and crisp potatoes. Sveriges Utsädesförenings Tidskrif 2020, 2020, 16–26. [Google Scholar]
- Brown, J.; Caligari, P.D.S.; Mackay, G.R.; Swan, G.E.L. The efficiency of visual selection in early generations of a potato breeding programme. Ann. Appl. Biol. 1987, 110, 357–363. [Google Scholar] [CrossRef]
- Gopal, J. Progeny selection for agronomic characters in early generations of potato breeding programme. Theor. Appl. Genet. 1997, 95, 307–311. [Google Scholar] [CrossRef]
- Tarn, T.R.; Tai, G.C.C.; Jong, H.D.; Murphy, A.M.; Seabrook, J.E.A. Breeding potatoes for long-day, temperate climates. Plant Breed. Rev. 1992, 9, 219–332. [Google Scholar]
- Brown, J.; Caligari, P.D.S. The efficiency of seedling selection for yield and yield components in a potato breeding programme. Pflanzenzucht 1986, 96, 53–62. [Google Scholar]
- Neele, A.E.F.; Nab, H.J.; Louwes, K.K. Identification of superior parents in a potato breeding programme. Theor. Appl. Genet. 1991, 82, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues de Figueiredo, I.C.; Pinto, C.A.B.P.; Ribeiro, G.H.M.R.; de Oliveira Lino, L.; Lyra, D.H.; Moreira, C.M. Efficiency of selection in early generations of potato families with a view toward heat tolerance. Crop Breed. Appl. Biotechnol. 2015, 15, 210–217. [Google Scholar] [CrossRef]
- Bradshaw, J.E.; Dale, M.F.B.; Swan, G.E.L.; Todd, D.; Wilson, R.N. Early-generation selection between and within pair crosses in a potato (Solanum tuberosum subsp. tuberosum) breeding programme. Theor. Appl. Genet. 1998, 97, 1331–1339. [Google Scholar] [CrossRef]
- Diniz, M.C.D.R.; Pinto, C.A.B.P.; Lambert, E.S. Sample size for family evaluation in potato breeding programs. Ciência e Agrotecnologia 2006, 30, 277–282. [Google Scholar] [CrossRef]
- Simmonds, N.W. Family selection in plant breeding. Euphytica 1996, 90, 201–208. [Google Scholar] [CrossRef]
- Simon, G.A.; Pinto, C.A.B.P.; Benites, F.R.G. Seleção de familias clonais de batata em diferentes ambientes. Ciência e Agrotecnologia 2009, 33, 164–169. [Google Scholar] [CrossRef]
- Verissimo, M.A.A.; Pereira, A.S.; Silva, S.D.A.; Terres, L.R.; Ney, V.G.; Silva, G.O. Expressão de caracteres de tubérculos em função do tamanho de recipiente usado no cultivo de batata na generação de plântulas. Rev. Ceres 2012, 59, 787–793. [Google Scholar] [CrossRef]
- Ticona-Benavente, C.A.; Pinto, C.A.B.P.; Rodrigues de Figueiredo, I.C.; Ribeiro, G.H.M.R. Repeatability of family means in early generations of potato under heat stress. Crop Breed. Appl. Biotechnol. 2011, 11, 330–337. [Google Scholar] [CrossRef]
- Brown, J.; Caligari, P.D.S.; Mackay, G.R. The repeatability of progeny means in the early generations of a potato breeding programme. Ann. Appl. Biol. 1987, 110, 365–370. [Google Scholar] [CrossRef]
- Brown, J.; Caligari, P.D.S. Cross prediction in a potato breeding programme by evaluation of parental material. Theor. Appl. Genet. 1989, 77, 246–252. [Google Scholar] [CrossRef]
- Brown, J.; Caligari, P.D.S.; Mackay, G.R.; Swan, G.E.L. The efficiency of seedling selection by visual preference in a potato breeding programme. J. Agric. Sci. 1984, 103, 339–346. [Google Scholar] [CrossRef]
- Swieżyński, K.M. Selection of individual tubers in potato breeding. Theor. Appl. Genet. 1978, 53, 71–80. [Google Scholar] [CrossRef]
- Brown, J.; Caligari, P.D.S.; Dale, M.F.B.; Swan, G.E.L.; Mackay, G.R. The use of cross prediction methods in a practical potato breeding programme. Theor. Appl. Genet. 1988, 76, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Ticona-Benavente, C.A.; da Silva Filho, D.F. Comparison of BLUE and BLUP/REML in the selection of clones and families of potato (Solanum tuberosum). Genet. Mol. Res. 2015, 14, 18421–18430. [Google Scholar] [CrossRef] [PubMed]
- Henderson, C.R. Applications of Linear Models in Animal Breeding; University of Guelph: Guelph, Canada, 1984. [Google Scholar]
- Slater, A.T.; Wilson, G.M.; Cogan, N.O.; Forster, J.W.; Hayes, B.J. Improving the analysis of low heritability complex traits for enhanced genetic gain in potato. Theor. Appl. Genet. 2014, 127, 809–820. [Google Scholar] [CrossRef]
- Slater, A.T.; Cogan, N.O.; Hayes, B.J.; Schultz, L.; Dale, M.F.; Bryan, G.J.; Forster, J.W. Improving breeding efficiency in potato using molecular and quantitative genetics. Theor. Appl. Genet. 2014, 127, 2279–2292. [Google Scholar] [CrossRef]
- Slater, A.T.; Cogan, N.O.I.; Rodoni, B.C.; Hayes, B.J.; Forster, J.W. Improving the selection efficiency in potato breeding. Acta Hort. 2016, 1127, 237–242. [Google Scholar] [CrossRef]
- Slater, A.T.; Cogan, N.O.; Forster, J.W.; Hayes, B.J.; Daetwyler, H.D. Improving genetic gain with genomic selection in autotetraploid potato. Plant Genome 2016, 9, 3. [Google Scholar] [CrossRef]
- Ortiz, R.; Crossa, J.; Reslow, F.; Perez-Rodriguez, P.; Cuevas, J. Genome-based genotype environment prediction enhances potato (Solanum tuberosum L.) improvement using pseudo-diploid and polysomic tetraploid modeling. Front. Plant Sci. 2022, 13, 785196. [Google Scholar] [CrossRef]
- Jinks, J.L.; Lawrence, M.J. The Genetic Basis of Inbreeding Depression and Heterosis: Its Implications for Plant and Animal Breeding; Palm Oil Research Institute of Malaysia: Kuala Lumpur, Malaysia, 1983. [Google Scholar]
- Jinks, J.L. The genetic framework of plant breeding. Phil. Trans. R. Soc. Lond. B 1981, 292, 407–419. [Google Scholar]
- Jinks, J.L. Biometrical genetics of heterosis. In Heterosis; Frankel, R., Ed.; Monograph Theoretical and Applied Genetics 6; Springer: Berlin, Germany, 1983; pp. 1–46. [Google Scholar]
- Ortiz, R.; Reslow, F.; Vetukuri, R.; García-Gil, M.R.; Pérez-Rodríguez, P.; Crossa, J. Inbreeding effects on the performance and genomic prediction for polysomic tetraploid potato offspring grown at high Nordic latitudes. Genes 2024, 14, 1302. [Google Scholar] [CrossRef]
- Jarquín, D.; Crossa, J.; Lacaze, X.; Cheyron, P.D.; Daucourt, J.; Lorgeou, J.; Piraux, F.; Guerreiro, L.; Pérez, P.; Calus, M.; et al. A reaction norm model for genomic selection using high-dimensional genomic and environmental data. Theor. Appl. Genet. 2014, 127, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Burgueño, J.; Cadena, A.; Crossa, J.; Banziger, M.; Gilmour, A.R.; Cullis, B. User’s Guide for Spatial Analysis of Field Variety Trials Using ASREML; Centro Internacional de Mejoramiento de Maíz y Trigo: Texcoco, Mexico, 2000. [Google Scholar]
- Cullis, B.R.; Lill, W.J.; Fisher, J.A.; Read, B.J.; Gleeson, A.C. A new procedure for the analysis of early generation variety trials. Appl. Stat. 1989, 38, 361–375. [Google Scholar] [CrossRef]
- Gleeson, A.C.; Cullis, B.R. Residual maximum likelihood (REML) estimation of a neighbour model for field experiments. Biometrics 1987, 43, 277–288. [Google Scholar] [CrossRef]
- Hill, J.; Becker, H.C.; Tigerstedt, P.M.A. Quantitative and Ecological Aspects of Plant Breeding; Springer: Dordrecht, The Netherlands, 1998. [Google Scholar]
- Endelman, J.B.; Kante, M.; Lindqvist-Kreuze, H.; Kilian, A.; Shannon, L.M.; Caraza-Harter, M.V.; Vaillancourt, B.; Mailloux, K.; Hamilton, P.P.; Buell, C.R. Targeted genotyping-by-sequencing of potato and software for imputation. bioRiv 2024. [Google Scholar] [CrossRef]
- Amadeu, R.R.; Ferrão, L.F.V.; Oliveira, I.B.; Benevenuto, J.; Endelman, J.B.; Munoz, P.R. Impact of dominance effects on autotetraploid genomic prediction. Crop Sci. 2020, 260, 656–665. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 24 May 2023).
- Perez, P.; de los Campos, G. Genome-wide regression and prediction with the BGLR statistical package. Genetics 2014, 198, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rodríguez, P.; Crossa, J.; Bondalapati, K.; De Meyer, G.; Pita, F.; Campos, G.d.l. A pedigree-based reaction norm model for prediction of cotton yield in multienvironment trials. Crop Sci. 2015, 55, 1143–1151. [Google Scholar] [CrossRef]
- Lian, Q.; Tang, D.; Bai, Z.; Qi, J.; Lu, F.; Huang, S.; Zhang, C. Acquisition of deleterious mutations during potato polyploidization. J. Integr. Plant Biol. 2019, 61, 7–11. [Google Scholar] [CrossRef]
- Charlesworth, D.; Willis, J. The genetics of inbreeding depression. Nat. Rev. Genet. 2009, 10, 783–796. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, Z.; Tang, D.; Zhu, Y.; Wang, P.; Li, D.; Zhu, G.; Xiong, X.; Shang, Y.; Li, C.; et al. Genome design of hybrid potato. Cell 2021, 84, 3873–3883. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, P.; Tang, D.; Yang, Z.; Lu, F.; Qi, J.; Tawari, N.R.; Shang, Y.; Li, C.; Huang, S. The genetic basis of inbreeding depression in potato. Nat. Genet. 2019, 51, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Selga, S.; Reslow, R.; Pérez-Rodríguez, P.; Ortiz, R. The power of genomic estimated breeding values for selection when using a finite population size in genetic improvement of tetraploid potato. G3 2022, 12, jkab362. [Google Scholar] [CrossRef] [PubMed]
- Endelman, J.B.; Carley, C.A.S.; Bethke, P.C.; Coombs, J.J.; Clough, M.E.; da Silva, W.L.; De Jong, W.S.; Douches, D.S.; Frederick, C.M.; Haynes, K.G.; et al. Genetic variance partitioning and genome-wide prediction with allele dosage information in autotetraploid potato. Genetics 2018, 209, 77–87. [Google Scholar] [CrossRef]
- Gemenet, D.C.; Lindqvist-Kreuze, H.; De Boeck, B.; da Silva Pereira, G.; Mollinari, M.; Zeng, Z.B.; Yencho, G.C.; Campos, H. Sequencing depth and genotype quality: Accuracy and breeding operation considerations for genomic selection applications in autopolyploid crops. Theor. Appl. Genet. 2020, 133, 3345–3363. [Google Scholar] [CrossRef]
- Sood, S.; Lin, Z.; Caruana, B.; Slater, A.T.; Daetwyler, H.D. Making the most of all data: Combining non-genotyped and genotyped potato individuals with HBLUP. Plant Genome 2020, 13, e20056. [Google Scholar] [CrossRef] [PubMed]
- Stich, B.; Van Inghelandt, D. Prospects and potential uses of genomic prediction of key performance traits in tetraploid potato. Front. Plant Sci. 2018, 9, 159. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.; Zheng, C.; Maliepaard, C.; Mulder, H.A.; Visser, R.G.F.; van der Burgt, A.; van Eeuwijk, F. Understanding the effectiveness of genomic prediction in tetraploid potato. Front. Plant Sci. 2021, 12, 672417. [Google Scholar] [CrossRef] [PubMed]
- Batista, L.G.; Mello, V.H.; Souza, A.P.; Margarido, G.R.A. Genomic prediction with allele dosage information in highly polyploid species. Theor. Appl. Genet. 2022, 135, 723–739. [Google Scholar] [CrossRef]
- Bradshaw, J.E. Review and a nalysis of limitations in way to improve conventional potato breeding. Potato Res. 2017, 60, 171–193. [Google Scholar] [CrossRef]
- Wu, P.Y.; Stich, B.; Renner, J.M.; Muders, K.; Prigge, V.; van Inghelandt, D. Optimal implementation of genomic selection in clone breeding programs–Exemplified in potato: I. Effect of selection strategy, implementation stage, and selection intensity on short-term genetic gain. Plant Genome 2023, 16, e20327. [Google Scholar] [CrossRef]
| S1 or F1 | Offspring Number |
|---|---|
| Colleen S1 (C S1) | 162 |
| Melody S1 (M S1) | 94 |
| Queen Anne S1 (QA S1) | 177 |
| Rudolph S1 (R S1) | 173 |
| Queen Anne × Colleen (QA × C) | 36 |
| Queen Anne × Melody (QA × M) | 272 |
| Genotype | Umeå | Helgegården | |
|---|---|---|---|
| 2021 | 2022 | ||
| Colleen | 0.693 | 0.867 | 1.594 |
| Colleen S1 | 0.181 | 0.601 | 0.733 |
| Melody | 0.826 | 1.128 | 1.746 |
| Melody S1 | 0.195 | 0.739 | 1.040 |
| Queen Anne | 0.562 | 1.145 | 1.585 |
| Queen Anne S1 | 0.124 | 0.392 | 0.807 |
| Queen Anne × Colleen | 0.290 | 0.857 | 1.189 |
| Queen Anne × Melody | 0.386 | 0.981 | 2.058 |
| Rudolph | N/A | 1.100 | 1.472 |
| Rudolph S1 | 0.222 | 0.759 | 0.619 |
| LSD0.05 | 0.094 | 0.125 | 0.279 |
| Breeding T2 population (30% of the records for a given population) in Helgegården 2022 | (QA × C) | (QA × M) | (C S1) | (M S1) | (QA S1) | (R S1) |
| Training T1 population in Umeå 2021 and 70% of that population evaluated in Helgegården 2022 | (QA × C) | (QA × M) | (C S1) | (M S1) | (QA S1) | (R S1) |
| r | 0.503 | 0.141 | 0.262 | 0.392 | 0.683 | 0.389 |
| SD | 0.210 | 0.129 | 0.113 | 0.162 | 0.087 | 0.122 |
| Breeding T2 population (30% of the records for a given population) in Umeå 2022 | (QA × C) | (QA × M) | (C S1) | (M S1) | (QA S1) | (R S1) |
| Training T1 population in Umeå 2021 and 70% of that population evaluated in Umeä 2022 | (QA × C) | (QA × M) | (C S1) | (M S1) | (QA S1) | (R S1) |
| r | 0.607 | 0.428 | 0.514 | 0.448 | 0.769 | 0.517 |
| SD | 0.23 | 0.116 | 0.092 | 0.172 | 0.058 | 0.109 |
| Training Population | Breeding T2 Population | r |
|---|---|---|
| U2021 C S1 + U2021 QA × M + H2022 C S1 | H2022 QA × C | 0.6183 |
| U2021 M S1 + U2021 QA × M + H2022 M S1 | H2022 QA × M | 0.0994 |
| U 2021 QA S1 + U2021 QA × C + H2022 QA S1 | H2022 QA × C | 0.5345 |
| U2021 QA S1 + U2021 QA × M+ H2022 QA S1 | H2022 QA × M | 0.1089 |
| U2021 QA × C + U2021 QA × M + H2022 QA × C | H2022 Q × M | 0.1229 |
| U2021 QA × C + U2021 QA × M + H2022 QA × M | H2022 QA × C | 0.4650 |
| U2021 R S1 + U2021 Queen Anne × C + H2022 R S1 | H 2022 QA × C | 0.5137 |
| U2021 RS1 + U2021 QA × M + H2022 R S1 | H2022 QA × M | 0.1139 |
| U2021 C S1 + U2021 R S1 + H2022 R S1 | H2022 C S1 | 0.2636 |
| U2021 M S1 + Umeå 2021 R S1 + H2022 R S1 | H2022 M S1 | 0.4154 |
| U2021 QAS1 + U2021 R S1 + H2022 R S1 | H2022 QA S1 | 0.6570 |
| U2021 C S1 + U2021 M S1 + H2022 C S1 | H2022 M S1 | 0.1891 |
| U2021 QA S1 + U2021 C S1 + H2022 C S1 | H2022 QA S1 | 0.6342 |
| U2021 C S1 + U2021 M S1 + H 2022 S1 | H2022 C S1 | 0.1359 |
| U2021 QA S1 + U2021 M S1 + H2022 M S1 | H2022 QA S1 | 0.6656 |
| U2021 QA S1 + U2021 C S1 + H2022 QA S1 | H2022 C S1 | 0.2635 |
| U2021 QA S1 + U2021 M S1 + H2022 QA S1 | H2022 M S1 | 0.4059 |
| U2021 M S1 + U2021 QA × C + H2022 M S1 | H2022 QA × C | 0.4965 |
| U2021 C S1 + U2021 QA × M + H2022 C S1 | H2022 QA × M | 0.1137 |
| Training Population | Breeding T2 Population | r |
|---|---|---|
| U2021 C S1 + U2021 QA × C + U2022 C S1 | U2022 QA × C | 0.5870 |
| U2021 M S1 + U2021 QA × M + U2022 M S1 | U2022 QA × M | 0.4363 |
| U2021 QA S1 + U2021 QA × C + U022 QA S1 | U2022 QA × C | 0.5720 |
| U2021 QA S1 + U2021 QA × M + U2022 QA S1 | U2022 QA × M | 0.4363 |
| U2021 QA × C + U2021 QA × M + U2022 QA × C | U2022 QA × M | 0.4358 |
| U2021 QA × C + U2021 QA× M + U2022 QA × M | U2022 QA × C | 0.5863 |
| U2021 R S1 + U2021 QA × C + U2022 R S1 | U2022 QA × C | 0.5700 |
| U2021 R S1+ U2021 QA × M + U2022 R S1 | U2022 QA × M | 0.4406 |
| U2021 C S1 + U2021 R S1 + U2022 R S1 | U2022 C S1 | 0.4944 |
| U2021 M S1 + U2021 R S1 + U2022 R S1 | U2022 M S1 | 0.4231 |
| U2021 Q S1 + U2021 R S1 + U2022 R S1 | U2022 Q S1 | 0.7815 |
| U2021 C S1 + U2021 M S1 + U2022 C S1 | U2022 M S1 | 0.4324 |
| U2021 QA S1 + U2021 C S1 + U2022 C S1 | U2022 QA S1 | 0.7528 |
| U2021 C S1 + U2021 M S1 + U2022 M S1 | U2022 C S1 | 0.4713 |
| U2021 QA S1 + U2021 M S1 + U2022 M S1 | U2022 Q S1 | 0.7577 |
| U2021 QA S1 + U2021 C S1 + U2022 QA S1 | U2022 C S1 | 0.5095 |
| U2021 QA S1 + U2021 M S1+ U2022 QA S1 | U2022 M S1 | 0.4629 |
| U2021 M S1 + U2021 QA × C + U2022 M S1 | U022 QA × C | 0.5698 |
| U2021 C S1 + U2021 QA × M + U2022 CS1 | U2022 QA × M | 0.4350 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortiz, R.; Reslow, F.; Vetukuri, R.; García-Gil, M.R.; Pérez-Rodríguez, P.; Crossa, J. Genomic Prediction for Inbred and Hybrid Polysomic Tetraploid Potato Offspring. Agriculture 2024, 14, 455. https://doi.org/10.3390/agriculture14030455
Ortiz R, Reslow F, Vetukuri R, García-Gil MR, Pérez-Rodríguez P, Crossa J. Genomic Prediction for Inbred and Hybrid Polysomic Tetraploid Potato Offspring. Agriculture. 2024; 14(3):455. https://doi.org/10.3390/agriculture14030455
Chicago/Turabian StyleOrtiz, Rodomiro, Fredrik Reslow, Ramesh Vetukuri, M. Rosario García-Gil, Paulino Pérez-Rodríguez, and José Crossa. 2024. "Genomic Prediction for Inbred and Hybrid Polysomic Tetraploid Potato Offspring" Agriculture 14, no. 3: 455. https://doi.org/10.3390/agriculture14030455
APA StyleOrtiz, R., Reslow, F., Vetukuri, R., García-Gil, M. R., Pérez-Rodríguez, P., & Crossa, J. (2024). Genomic Prediction for Inbred and Hybrid Polysomic Tetraploid Potato Offspring. Agriculture, 14(3), 455. https://doi.org/10.3390/agriculture14030455







