Genome-Wide Identification and Expression Analysis of the PIN Auxin Transporter Gene Family in Zanthoxylum armatum DC
Abstract
:1. Introduction
2. Materials and Methods
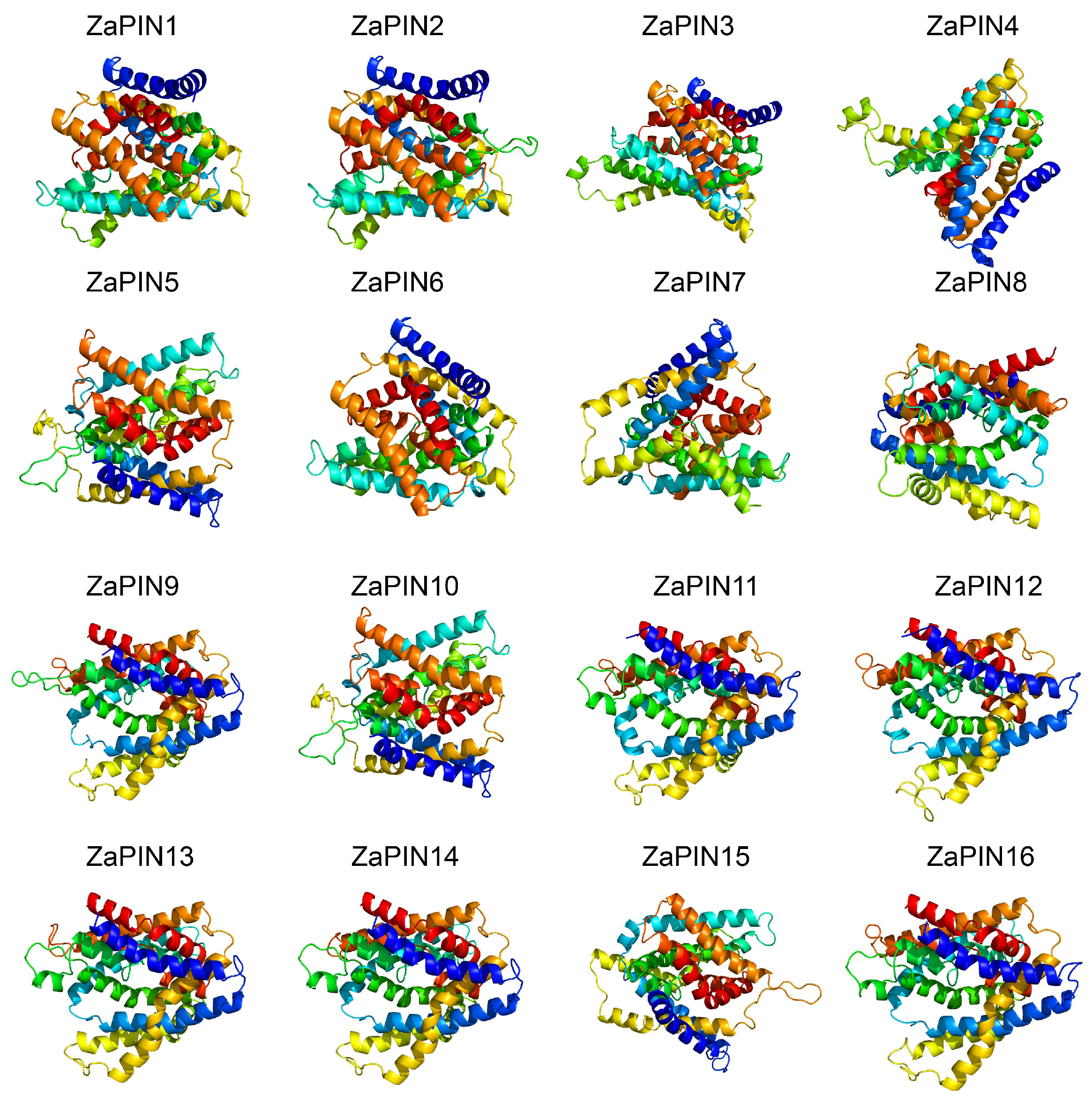
2.1. Physicochemical Characteristics, Subcellular Localization, and Three-Dimensional Structure Analysis
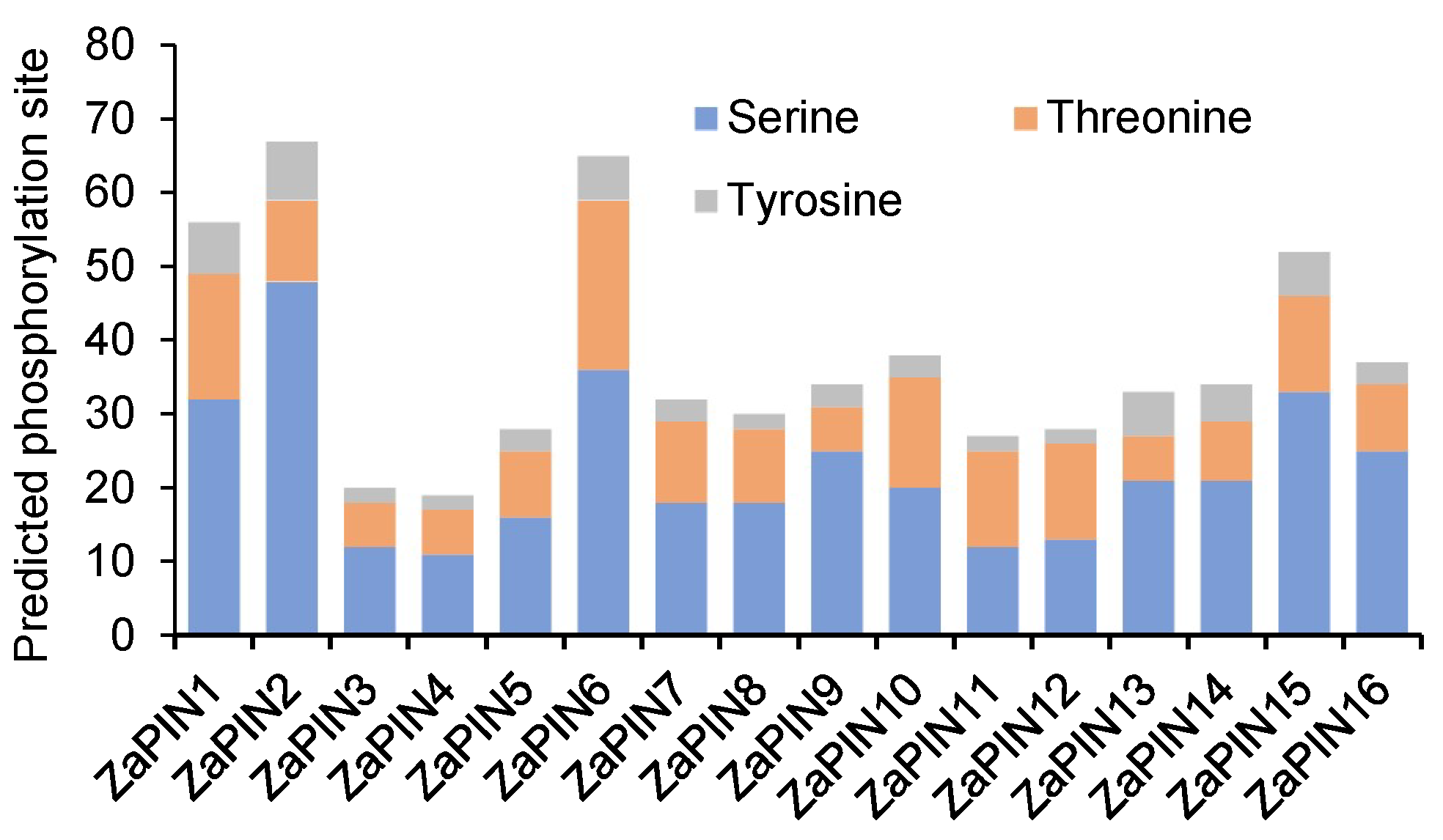
2.2. Prediction of Phosphorylation Modification Sites
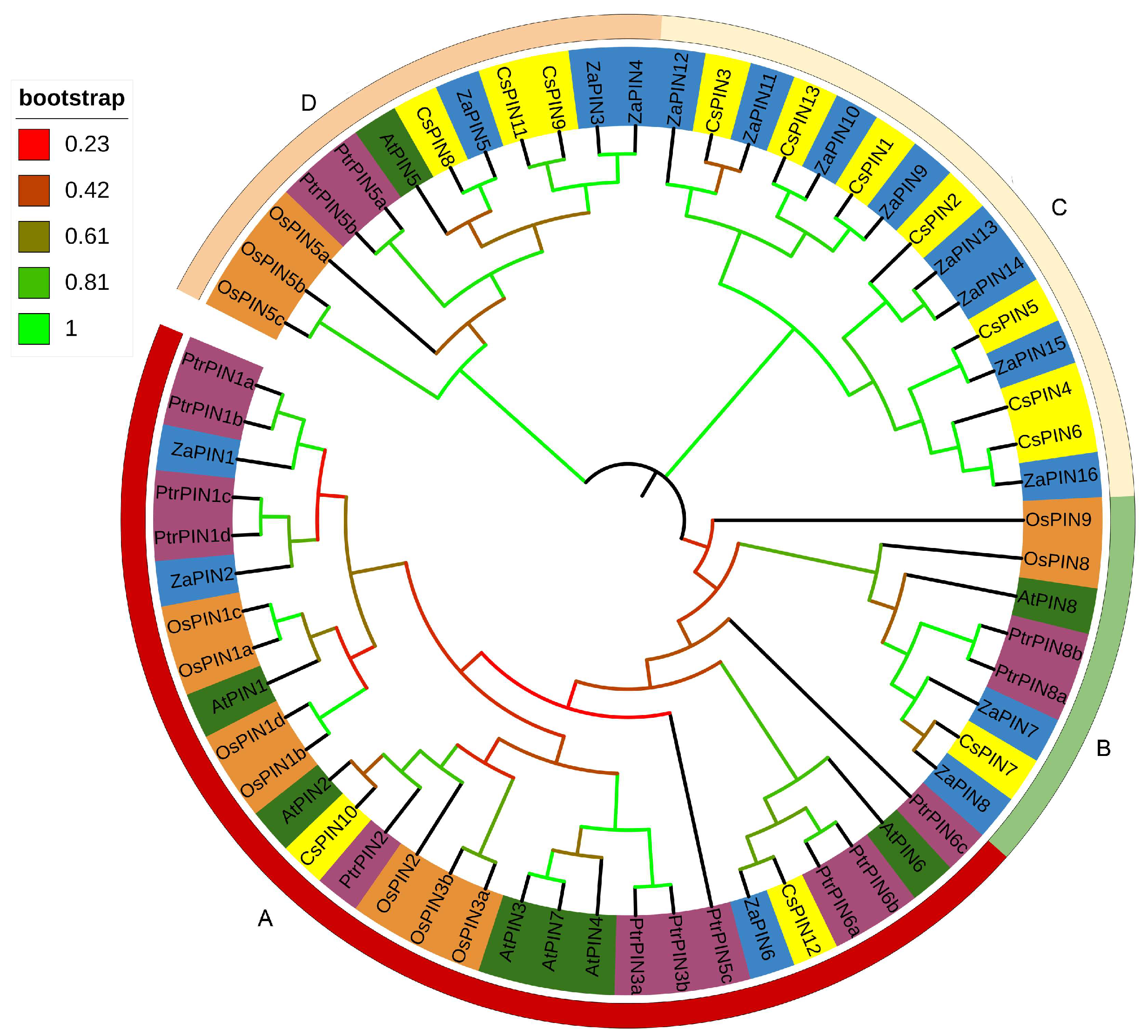
2.3. Phylogenetic Analysis
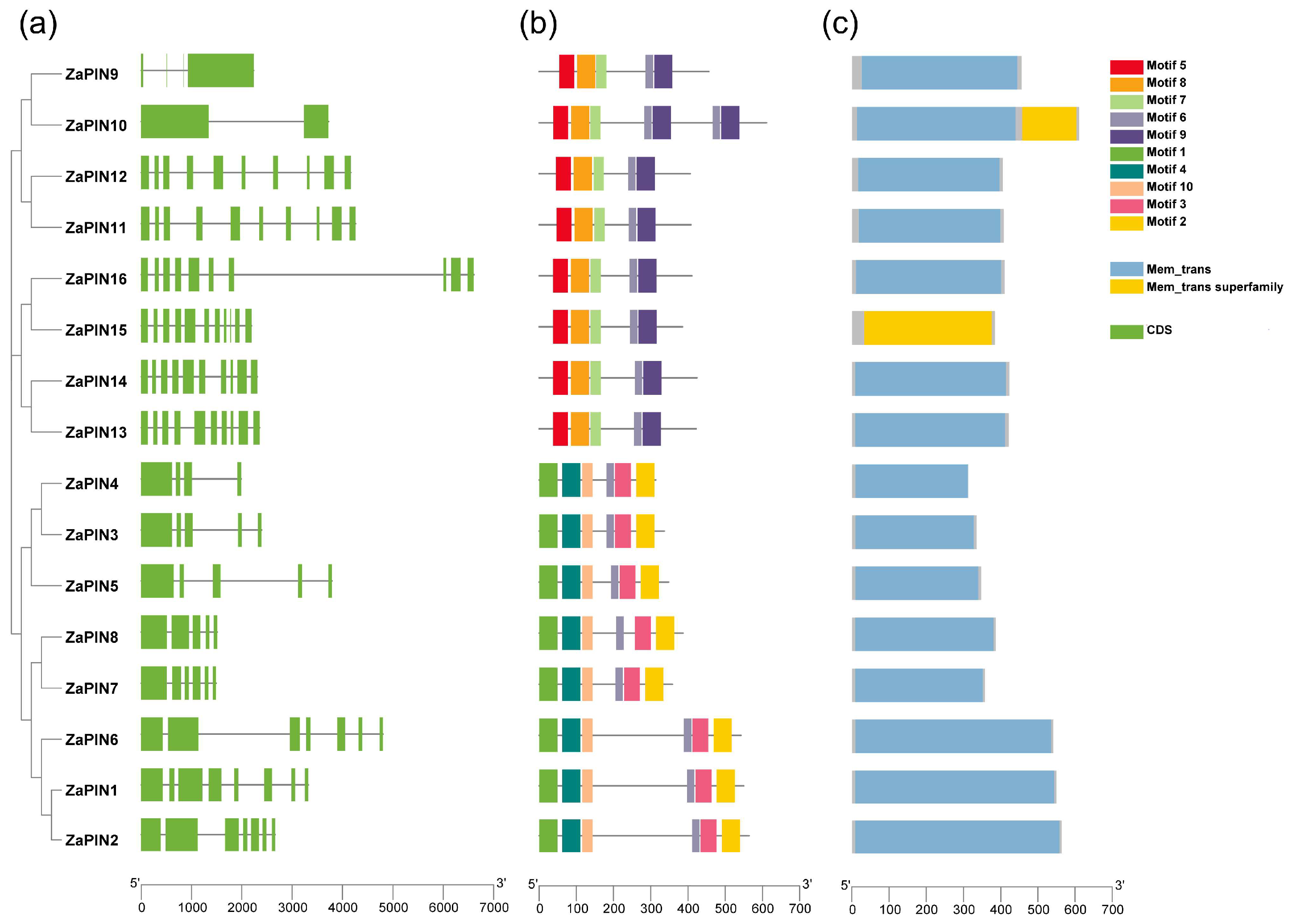
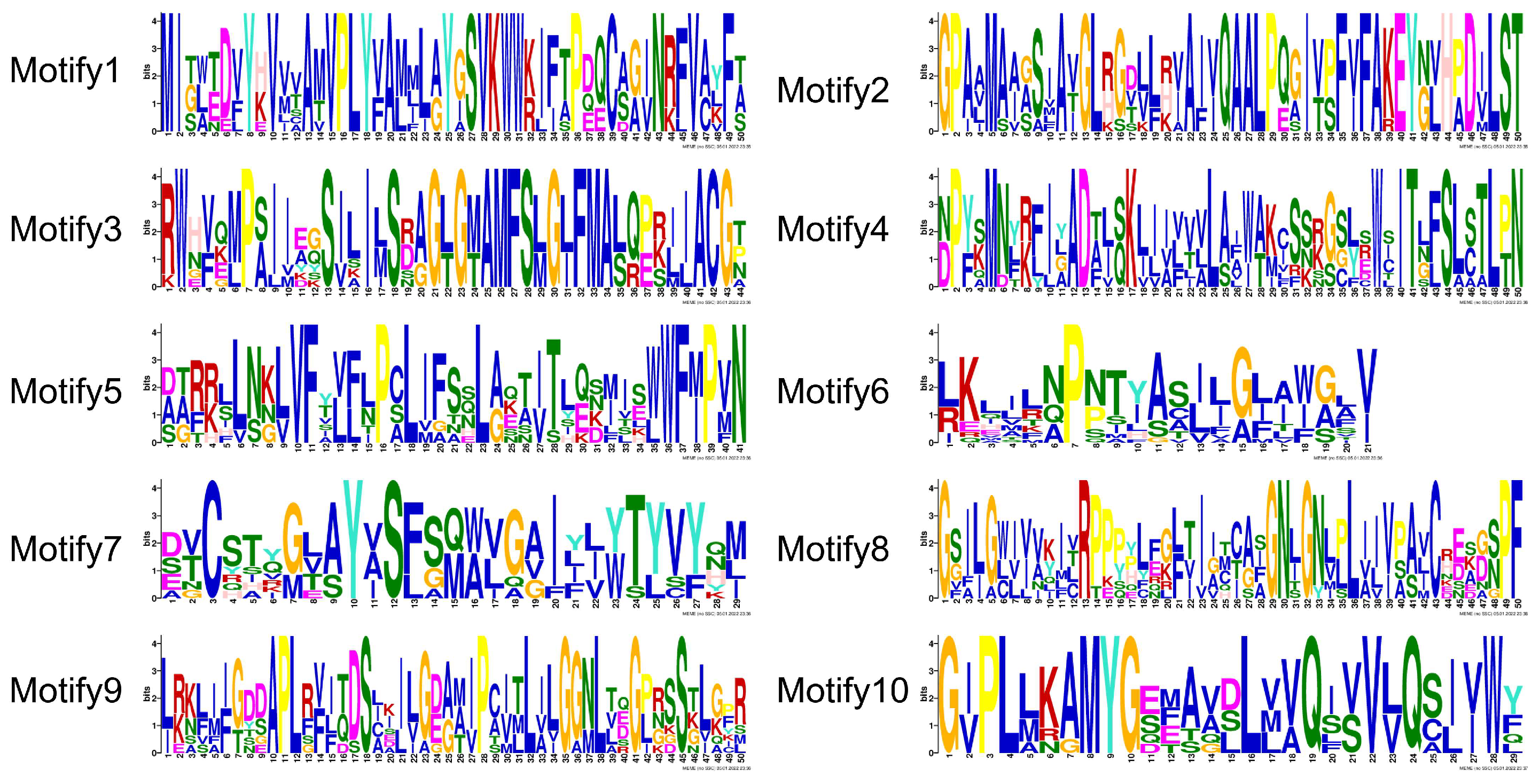
2.4. Gene Structure and Conserved Motif Analysis
2.5. Chromosomal Location, Gene Duplication, and Collinearity Analysis
2.6. Cis-Acting Regulatory Elements (CAREs) Analysis
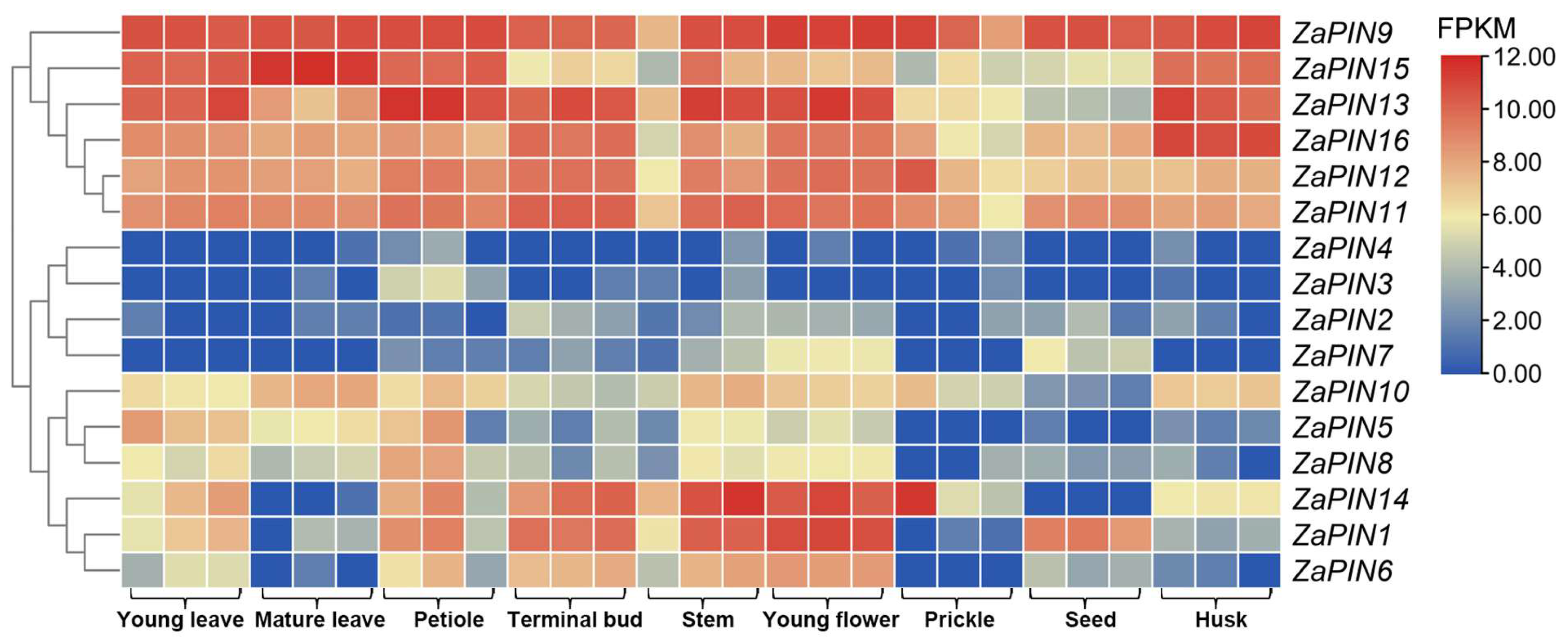
2.7. Gene Expression Analysis
2.8. Plant Treatments, RNA Extraction, and qRT-PCR Analysis
3. Results
3.1. Identification of PIN Family Members in Z. armatum
3.2. Prediction of Phosphorylation Modification Sites in ZaPINs
3.3. Phylogenetic Analysis of ZaPIN Family
3.4. Gene Structure and Conserved Motif Analysis of ZaPIN Family
3.5. Chromosome Locations and Gene Duplication Analysis of ZaPINs
3.6. Cis-Acting Elements in ZaPIN Promoters
3.7. Tissue-Specific Transcript Profiles of ZaPINs
3.8. Expression of ZaPINs in Response to Auxin and Gibberellin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Enders, T.A.; Strader, L.C. Auxin Activity: Past, Present, And Future. Am. J. Bot. 2015, 102, 180–196. [Google Scholar] [CrossRef]
- Li, D.; Pan, C.; Lu, J.; Zaman, W.; Zhao, H.; Zhang, J.; Lü, S. Lupeol Accumulation Correlates with Auxin in the Epidermis of Castor. Molecules 2021, 26, 2978. [Google Scholar] [CrossRef] [PubMed]
- Weijers, D.; Wagner, D. Transcriptional Responses to the Auxin Hormone. In Annual Review of Plant Biology; Merchant, S.S., Ed.; Annual Reviews: Palo Alto, CA, USA, 2016; Volume 67, pp. 539–574. [Google Scholar]
- Hayashi, K.I.; Nakamura, S.; Fukunaga, S.; Nishimura, T.; Jenness, M.K.; Murphy, A.S.; Motose, H.; Nozaki, H.; Furutani, M.; Aoyama, T. Auxin transport sites are visualized in planta using fluorescent auxin analogs. Proc. Natl. Acad. Sci. USA 2014, 111, 11557–11562. [Google Scholar] [CrossRef]
- Swarup, R.; Peret, B. AUX/LAX family of auxin influx carriers-an overview. Front. Plant Sci. 2012, 3, 225. [Google Scholar] [CrossRef] [PubMed]
- Krouk, G.; Lacombe, B.; Bielach, A.; Perrine-Walker, F.; Malinska, K.; Mounier, E.; Hoyerova, K.; Tillard, P.; Leon, S.; Ljung, K.; et al. Nitrate-Regulated Auxin Transport by NRT1.1 Defines a Mechanism for Nutrient Sensing in Plants. Dev. Cell 2010, 18, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Swarup, K.; Benkova, E.; Swarup, R.; Casimiro, I.; Peret, B.; Yang, Y.; Parry, G.; Nielsen, E.; De Smet, I.; Vanneste, S.; et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 2008, 10, 946–954. [Google Scholar] [CrossRef]
- Peret, B.; Swarup, K.; Ferguson, A.; Seth, M.; Yang, Y.D.; Dhondt, S.; James, N.; Casimiro, I.; Perry, P.; Syed, A.; et al. AUX/LAX Genes Encode a Family of Auxin Influx Transporters That Perform Distinct Functions during Arabidopsis Development. Plant Cell 2012, 24, 2874–2885. [Google Scholar] [CrossRef]
- Okada, K.; Ueda, J.; Komaki, M.K.; Bell, C.J.; Shimura, Y. Requirement of the Auxin Polar Transport System in Early Stages of Arabidopsis Floral Bud Formation. Plant Cell 1991, 3, 677–684. [Google Scholar] [CrossRef]
- Wisniewska, J.; Xu, J.; Seifertova, D.; Brewer, P.B.; Ruzicka, K.; Blilou, I.; Rouquie, D.; Benkova, E.; Scheres, B.; Friml, J. Polar PIN localization directs auxin flow in plants. Science 2006, 312, 883. [Google Scholar] [CrossRef]
- Mravec, J.; Skupa, P.; Bailly, A.; Hoyerova, K.; Krecek, P.; Bielach, A.; Petrasek, J.; Zhang, J.; Gaykova, V.; Stierhof, Y.-D.; et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 2009, 459, U1136–U1140. [Google Scholar] [CrossRef]
- Galweiler, L.; Guan, C.; Muller, A.; Wisman, E.; Mendgen, K.; Yephremov, A.; Palme, K. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 1998, 282, 2226–2230. [Google Scholar] [CrossRef] [PubMed]
- Vanneste, S.; Friml, J. Auxin: A Trigger for Change in Plant Development. Cell 2009, 136, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Forestan, C.; Farinati, S.; Varotto, S. The maize PIN gene family of auxin transporters. Front. Plant Sci. 2012, 3, 16. [Google Scholar] [CrossRef]
- Kumar, M.; Kherawat, B.S.; Dey, P.; Saha, D.; Singh, A.; Bhatia, S.K.; Ghodake, G.S.; Kadam, A.A.; Kim, H.U.; Chung, S.M.; et al. Genome-Wide Identification and Characterization of PIN-FORMED (PIN) Gene Family Reveals Role in Developmental and Various Stress Conditions in Triticum aestivum L. Int. J. Mol. Sci. 2021, 22, 7396. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhang, J.; Wang, L.; Li, J.; Zheng, H.; Chen, J.; Lu, M. A survey of Populus PIN-FORMED family genes reveals their diversified expression patterns. J. Exp. Bot. 2014, 65, 2437–2448. [Google Scholar] [CrossRef]
- Wang, Y.; Chai, C.; Valliyodan, B.; Maupin, C.; Annen, B.; Nguyen, H.T. Genome-wide analysis and expression profiling of the PIN auxin transporter gene family in soybean (Glycine max). BMC Genom. 2015, 16, 951. [Google Scholar] [CrossRef]
- He, P.; Zhao, P.; Wang, L.; Zhang, Y.; Wang, X.; Xiao, H.; Yu, J.; Xiao, G. The PIN gene family in cotton (Gossypium hirsutum): Genome-wide identification and gene expression analyses during root development and abiotic stress responses. BMC Genom. 2017, 18, 507. [Google Scholar] [CrossRef]
- Xie, X.; Qin, G.; Si, P.; Luo, Z.; Gao, J.; Chen, X.; Zhang, J.; Wei, P.; Xia, Q.; Lin, F.; et al. Analysis of Nicotiana tabacum PIN genes identifies NtPIN4 as a key regulator of axillary bud growth. Physiol. Plant. 2017, 160, 222–239. [Google Scholar] [CrossRef]
- Yang, C.H.; Wang, D.D.; Zhang, C.; Kong, N.N.; Ma, H.L.; Chen, Q. Comparative Analysis of the PIN Auxin Transporter Gene Family in Different Plant Species: A Focus on Structural and Expression Profiling of PINs in Solanum tuberosum. Int. J. Mol. Sci. 2019, 20, 3270. [Google Scholar] [CrossRef]
- Barkoulas, M.; Hay, A.; Kougioumoutzi, E.; Tsiantis, M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 2008, 40, 1136–1141. [Google Scholar] [CrossRef]
- Wang, L.; Guo, M.; Li, Y.; Ruan, W.; Mo, X.; Wu, Z.; Sturrock, C.J.; Yu, H.; Lu, C.; Peng, J.; et al. LARGE ROOT ANGLE1, encoding OsPIN2, is involved in root system architecture in rice. J. Exp. Bot. 2018, 69, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Jiang, Y.; Han, X.; Liu, X.; Cao, R.; Qi, M.; Xu, T.; Li, T. SlPIN1 regulates auxin efflux to affect flower abscission process. Sci. Rep. 2017, 7, 14919. [Google Scholar] [CrossRef] [PubMed]
- Phuyal, N.; Jha, P.K.; Raturi, P.P.; Rajbhandary, S. Zanthoxylum armatum DC.: Current knowledge, gaps and opportunities in Nepal. J. Ethnopharmacol. 2019, 229, 326–341. [Google Scholar] [CrossRef] [PubMed]
- Alam, F.; Saqib, Q.N.U.; Ashraf, M. Zanthoxylum armatum DC extracts from fruit, bark and leaf induce hypolipidemic and hypoglycemic effects in mice- in vivo and in vitro study. BMC Complementary Altern. Med. 2018, 18, 68. [Google Scholar] [CrossRef]
- Wang, M.C.; Tong, S.F.; Ma, T.; Xi, Z.X.; Liu, J.Q. Chromosome-level genome assembly of Sichuan pepper provides insights into apomixis, drought tolerance, and alkaloid biosynthesis. Mol. Ecol. Resour. 2021, 21, 2533–2545. [Google Scholar] [CrossRef]
- Miller, D.R.; Leek, T.; Schwartz, R.M. A hidden Markov model information retrieval system. In Proceedings of the 22nd Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, Berkeley, CA, USA, 15–19 August 1999; pp. 214–221. [Google Scholar]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Springer: New York, NY, USA, 2005; pp. 571–607. [Google Scholar]
- Nakai, K. PSORT: A program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem. Sci 1999, 24, 34–35. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Nugent, T.; Ward, S.; Jones, D.T. The MEMPACK alpha-helical transmembrane protein structure prediction server. Bioinformatics 2011, 27, 1438–1439. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Blom, N.; Sicheritz-Ponten, T.; Gupta, R.; Gammeltoft, S.; Brunak, S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004, 4, 1633–1649. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; He, Y.; Xia, R. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018, 289660, 289660. [Google Scholar]
- Leinonen, R.; Sugawara, H.; Shumway, M.; Collaboration, I.N.S.D. The sequence read archive. Nucleic Acids Res. 2010, 39, D19–D21. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Janssens, V.; Goris, J. Protein phosphatase 2A: A highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 2001, 353, 417–439. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Nielsen, R. Mutation-selection models of codon substitution and their use to estimate selective strengths on codon usage. Mol. Biol. Evol. 2008, 25, 568–579. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.-J.; Luo, J. The PIN-FORMED Auxin Efflux Carriers in Plants. Int. J. Mol. Sci. 2018, 19, 2759. [Google Scholar] [CrossRef]
- Pahari, S.; Cormark, R.D.; Blackshaw, M.T.; Liu, C.; Erickson, J.L.; Schultz, E.A. Arabidopsis UNHINGED encodes a VPS51 homolog and reveals a role for the GARP complex in leaf shape and vein patterning. Development 2014, 141, 1894–1905. [Google Scholar] [CrossRef]
- Balzan, S.; Johal, G.S.; Carraro, N. The role of auxin transporters in monocots development. Front. Plant Sci. 2014, 5, 393. [Google Scholar] [CrossRef]
- Vasil, V.; Marcotte, W.R., Jr.; Rosenkrans, L.; Cocciolone, S.M.; Vasil, I.K.; Quatrano, R.S.; McCarty, D.R. Overlap of Viviparous1 (VP1) and abscisic acid response elements in the Em promoter: G-box elements are sufficient but not necessary for VP1 transactivation. Plant Cell 1995, 7, 1511–1518. [Google Scholar]
- Ren, R.; Wang, H.; Guo, C.; Zhang, N.; Zeng, L.; Chen, Y.; Ma, H.; Qi, J. Widespread Whole Genome Duplications Contribute to Genome Complexity and Species Diversity in Angiosperms. Mol. Plant 2018, 11, 414–428. [Google Scholar] [CrossRef]
- Nekrutenko, A.; Makova, K.D.; Li, W.-H. The K(A)/K(S) ratio test for assessing the protein-coding potential of genomic regions: An empirical and simulation study. Genome Res. 2002, 12, 198–202. [Google Scholar] [CrossRef]
- Luschnig, C.; Gaxiola, R.A.; Grisafi, P.; Fink, G.R. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 1998, 12, 2175–2187. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, I.C.R.; Hammes, U.Z.; Schwechheimer, C. Activation and Polarity Control of PIN-FORMED Auxin Transporters by Phosphorylation. Trends Plant Sci. 2018, 23, 523–538. [Google Scholar] [CrossRef] [PubMed]
- Adamowski, M.; Friml, J. PIN-Dependent Auxin Transport: Action, Regulation, and Evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Weller, B.; Zourelidou, M.; Frank, L.; Barbosa, I.C.R.; Fastner, A.; Richter, S.; Juergens, G.; Hammes, U.Z.; Schwechheimer, C. Dynamic PIN-FORMED auxin efflux carrier phosphorylation at the plasma membrane controls auxin efflux-dependent growth. Proc. Natl. Acad. Sci. USA 2017, 114, E887–E896. [Google Scholar] [CrossRef]
- Huang, F.; Zago, M.K.; Abas, L.; van Marion, A.; Galvan-Ampudia, C.S.; Offringa, R. Phosphorylation of Conserved PIN Motifs Directs Arabidopsis PIN1 Polarity and Auxin Transport. Plant Cell 2010, 22, 1129–1142. [Google Scholar] [CrossRef] [PubMed]
- Grones, P.; Abas, M.; Hajny, J.; Jones, A.; Waidmann, S.; Kleine-Vehn, J.; Friml, J. PID/WAG-mediated phosphorylation of the Arabidopsis nnnPIN3 auxin transporter mediates polarity switches during gravitropism. Sci. Rep. 2018, 8, 10279. [Google Scholar] [CrossRef]
- Zažímalová, E.; Křeček, P.; Skůpa, P.; Hoyerova, K.; Petrášek, J. Polar transport of the plant hormone auxin–the role of PIN-FORMED (PIN) proteins. Cell. Mol. Life Sci. 2007, 64, 1621–1637. [Google Scholar] [CrossRef]
- Habets, M.E.; Offringa, R. PIN-driven polar auxin transport in plant developmental plasticity: A key target for environmental and endogenous signals. New Phytol. 2014, 203, 362–377. [Google Scholar] [CrossRef]
- Wang, J.-R.; Hu, H.; Wang, G.-H.; Li, J.; Chen, J.-Y.; Wu, P. Expression of PIN genes in rice (Oryza sativa L.): Tissue specificity and regulation by hormones. Mol. Plant 2009, 2, 823–831. [Google Scholar] [CrossRef]
- Matthes, M.S.; Best, N.B.; Robil, J.M.; Malcomber, S.; Gallavotti, A.; McSteen, P. Auxin EvoDevo: Conservation and diversification of genes regulating auxin biosynthesis, transport, and signaling. Mol. Plant 2019, 12, 298–320. [Google Scholar] [CrossRef] [Green Version]




| Primer | Primer Sequence (5′-3′) |
|---|---|
| ZaACT-F | GTGAGCCACACAGTACCCAT |
| ZaACT-R | GGTGAAAGAGTACCCACGCT |
| ZaPIN1-F | ACTTTACCCAACACGCTGGT |
| ZaPIN1-R | AGCTTGAGCCTCTTGGTGTT |
| ZaPIN2-F | CTCAAGGTGAACCCAAGCCA |
| ZaPIN2-R | ACGTGCCACCTGTAGGAAAC |
| ZaPIN3-F | GGACTGCATGCCGATGTTCT |
| ZaPIN3-R | CAATCAGCACAGGAAGCGAA |
| ZaPIN4-F | GTTGTTAGCATGTGGCCCTG |
| ZaPIN4-R | TGGAAGAGCTGCCTGAATGA |
| ZaPIN5-F | TGGGCCAAGTGTAGTAGCAAG |
| ZaPIN5-R | GGTTCCTGCCCTCCGAATTT |
| ZaPIN6-F | GCAATGCTCCGGAATCAACC |
| ZaPIN6-R | GATAGCCCAAACGGAGAGCA |
| ZaPIN7-F | GGCGCCACAAGAATCACAAG |
| ZaPIN7-R | CGCGACGCCATAAAAAGACC |
| ZaPIN8-F | GGCATCGCGACCTAGCATAA |
| ZaPIN8-R | CCCCTTAGTCCGATGGCAAA |
| ZaPIN9-F | TGGCCGGGTATTGAGGAAAA |
| ZaPIN9-R | CCTTCCGCCGAGAGATCAAG |
| ZaPIN10-F | GCCTTCCGCTATGCTTGTTC |
| ZaPIN10-R | CTGCCAAAATAGCAGTGCCG |
| ZaPIN11-F | AGGACTTGGCATCGTCACTT |
| ZaPIN11-R | CCACAACCGCGTAAACTGGA |
| ZaPIN12-F | TTACTGGCGTTAGGAGGCAA |
| ZaPIN12-R | TACCGCAACCCCGTAAACTG |
| ZaPIN13-F | ATTGCTGTGGTTTGCGTTCG |
| ZaPIN13-R | GCACTCCTCTTGAGCCACAT |
| ZaPIN14-F | CAGCAGCCATCCGCAAAAAG |
| ZaPIN14-R | TCTCAGCCACACAACAGCTC |
| ZaPIN15-F | TACTGCTCTTGGCCTGCTTC |
| ZaPIN15-R | TTGCTACCGATGAGTGCTGG |
| ZaPIN16-F | CTGAGGTGAAAGGTTGGCGT |
| ZaPIN16-R | GGATTCCTGAACCTCGCAGA |
| Gene | Gene ID | Chromosome Location | No. of AA | Molecular Weight (kD) | Theoretical pI | Instability Index | GRAVY | Aliphatic Index |
|---|---|---|---|---|---|---|---|---|
| ZaPIN1 | Zardc43630.t1 | Chr33:18461572~18464892 | 549 | 59.82 | 9.31 | 33.73 | 0.074 | 87.58 |
| ZaPIN2 | Zardc11981.t1 | Chr7:52941759~52944422 | 563 | 60.62 | 8.54 | 33.14 | 0.180 | 93.69 |
| ZaPIN3 | Zardc29606.t1 | Chr20:60577488~60579880 | 335 | 37.12 | 7.61 | 35.32 | 0.818 | 115.25 |
| ZaPIN4 | Zardc29009.t1 | Chr20:20274558~20276547 | 312 | 34.54 | 8.13 | 35.72 | 0.734 | 110.03 |
| ZaPIN5 | Zardc28594.t1 | Chr19:59627359~59631149 | 347 | 38.15 | 7.57 | 30.26 | 0.805 | 115.24 |
| ZaPIN6 | Zardc52636.t1 | unanchor10525:28196~32999 | 551 | 59.57 | 8.74 | 33.45 | 0.334 | 100.43 |
| ZaPIN7 | Zardc45630.t1 | unanchor489:82629~84119 | 357 | 38.94 | 9.67 | 34.67 | 0.677 | 124.96 |
| ZaPIN8 | Zardc46880.t1 | unanchor1921:45537~47048 | 386 | 42.36 | 9.25 | 36.35 | 0.714 | 124.12 |
| ZaPIN9 | Zardc01636.t1 | Chr1:86981515~86983753 | 455 | 50.07 | 6.32 | 40.45 | 0.638 | 126.42 |
| ZaPIN10 | Zardc28240.t1 | Chr19:42027501~42031224 | 610 | 66.83 | 5.56 | 33.69 | 0.716 | 125.28 |
| ZaPIN11 | Zardc29440.t1 | Chr20:44357827~44362085 | 407 | 44.07 | 8.59 | 34.47 | 0.779 | 125.55 |
| ZaPIN12 | Zardc27357.t1 | Chr19:26142114~26146278 | 405 | 43.99 | 7.51 | 33.93 | 0.713 | 121.11 |
| ZaPIN13 | Zardc30903.t1 | Chr22:2924357~2926710 | 421 | 46.15 | 5.70 | 42.44 | 0.635 | 123.23 |
| ZaPIN14 | Zardc26161.t1 | Chr18:67606835~67609143 | 423 | 46.72 | 6.10 | 45.80 | 0.571 | 118.51 |
| ZaPIN15 | Zardc09344.t1 | Chr6:95154~97348 | 384 | 51.23 | 4.85 | 45.64 | 0.730 | 126.48 |
| ZaPIN16 | Zardc24304.t1 | Chr17:24324787~24331393 | 510 | 44.29 | 8.61 | 40.25 | 0.661 | 123.93 |
| Protein | Alpha Helix (aa) (Proportion (%)) | Extended Strand (aa) (Proportion (%)) | Beta Turn (aa) (Proportion (%) | Random Coil (aa) (Proportion (%)) | Predicted Subcellular Location |
|---|---|---|---|---|---|
| ZaPIN1 | 173 (31.51%) | 77 (14.03%) | 30 (5.46%) | 269 (49.00%) | Plasma Membrane (13) |
| ZaPIN2 | 161 (28.60%) | 81 (14.39%) | 24 (4.26%) | 297 (52.75%) | Plasma Membrane (11) |
| ZaPIN3 | 199 (59.40%) | 42 (12.54%) | 18 (5.37%) | 76 (22.69%) | Vacuolar Membrane (11) |
| ZaPIN4 | 172 (55.13%) | 47 (15.06%) | 15 (4.81%) | 78 (25.00%) | Vacuolar Membrane (12) |
| ZaPIN5 | 187 (53.89%) | 56 (16.14%) | 18 (5.19%) | 86 (24.78%) | Vacuolar Membrane (7) |
| ZaPIN6 | 186 (34.38%) | 88 (16.27%) | 29 (5.36%) | 238 (43.99%) | Plasma Membrane (11) |
| ZaPIN7 | 187 (52.38%) | 56 (15.69%) | 17 (4.76%) | 97 (27.17%) | Plasma Membrane (7) |
| ZaPIN8 | 216 (55.96%) | 55 (14.25%) | 20 (5.18%) | 95 (24.61%) | Plasma Membrane (11) |
| ZaPIN9 | 194 (42.64%) | 73 (16.04%) | 14 (3.08%) | 174 (38.24%) | Plasma Membrane (11) |
| ZaPIN10 | 228 (37.38%) | 127 (20.82%) | 24 (3.93%) | 231 (37.87%) | Plasma Membrane (11) |
| ZaPIN11 | 166 (40.79%) | 85 (20.88%) | 16 (3.93%) | 140 (34.40%) | Plasma Membrane (10) |
| ZaPIN12 | 168 (51.48%) | 76 (18.77%) | 15 (3.70%) | 146 (36.05%) | Plasma Membrane (11) |
| ZaPIN13 | 171 (40.62%) | 91 (21.62%) | 13 (3.09%) | 146 (34.68%) | Plasma Membrane (7) |
| ZaPIN14 | 163 (38.53%) | 88 (20.80%) | 14 (3.31%) | 158 (37.35%) | Plasma Membrane (9) |
| ZaPIN15 | 171 (44.53%) | 71 (18.49%) | 13 (3.39%) | 129 (33.59%) | Vacuolar Membrane (8) |
| ZaPIN16 | 170 (51.46%) | 85 (20.73%) | 17 (4.15%) | 138 (33.66%) | Vacuolar Membrane (12) |
| Gene Pair Name | Gene Pair ID | Ka | Ks | Ka/Ks |
|---|---|---|---|---|
| ZaPIN14/ZaPIN13 | Zardc26161.t1/Zardc30903.t1 | 0.078 | 0.177 | 0.438 |
| ZaPIN12/ZaPIN11 | Zardc27357.t1/Zardc29440.t1 | 0.041 | 0.138 | 0.298 |
| ZaPIN10/ZaPIN11 | Zardc28240.t1/Zardc29440.t1 | 0.573 | 2.157 | 0.266 |
| ZaPIN8/ZaPIN7 | Zardc46880.t1/Zardc45630.t1 | 0.043 | 0.260 | 0.166 |
| Zardc27658.t1/ZaPIN2 | Zardc27658.t1/Zardc11981.t1 | 0.032 | 0.297 | 0.106 |
| ZaPIN1/Zardc06555.t1 | Zardc43630.t1/Zardc06555.t1 | 0.038 | 0.236 | 0.162 |
| ZaPIN2/Zardc12357.t1 | Zardc11981.t1/Zardc12357.t1 | 0.005 | 0.035 | 0.156 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, T.; Chen, J.; Huang, Y.; Jin, Z.; Li, J.; Li, Y.; Zeng, X. Genome-Wide Identification and Expression Analysis of the PIN Auxin Transporter Gene Family in Zanthoxylum armatum DC. Agriculture 2022, 12, 1318. https://doi.org/10.3390/agriculture12091318
Zhou T, Chen J, Huang Y, Jin Z, Li J, Li Y, Zeng X. Genome-Wide Identification and Expression Analysis of the PIN Auxin Transporter Gene Family in Zanthoxylum armatum DC. Agriculture. 2022; 12(9):1318. https://doi.org/10.3390/agriculture12091318
Chicago/Turabian StyleZhou, Tao, Jiajia Chen, Yanhui Huang, Zhengyu Jin, Jianrong Li, Yan Li, and Xiaofang Zeng. 2022. "Genome-Wide Identification and Expression Analysis of the PIN Auxin Transporter Gene Family in Zanthoxylum armatum DC" Agriculture 12, no. 9: 1318. https://doi.org/10.3390/agriculture12091318
APA StyleZhou, T., Chen, J., Huang, Y., Jin, Z., Li, J., Li, Y., & Zeng, X. (2022). Genome-Wide Identification and Expression Analysis of the PIN Auxin Transporter Gene Family in Zanthoxylum armatum DC. Agriculture, 12(9), 1318. https://doi.org/10.3390/agriculture12091318






