Phytoremediation of Secondary Salinity in Greenhouse Soil with Astragalus sinicus, Spinacea oleracea and Lolium perenne
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Site Setup, Management and Sampling
2.2. Crop Yield and Nutrient Accumulation
2.3. Determination of Soil Physicochemical Properties
2.4. Soil DNA Extraction and Microbial Community Analysis
2.5. Real-Time Quantitative Polymerase Chain Reaction (qPCR)
2.6. Data Analysis
3. Results
3.1. Response of Soil Biochemical Properties to Green Manure Crops
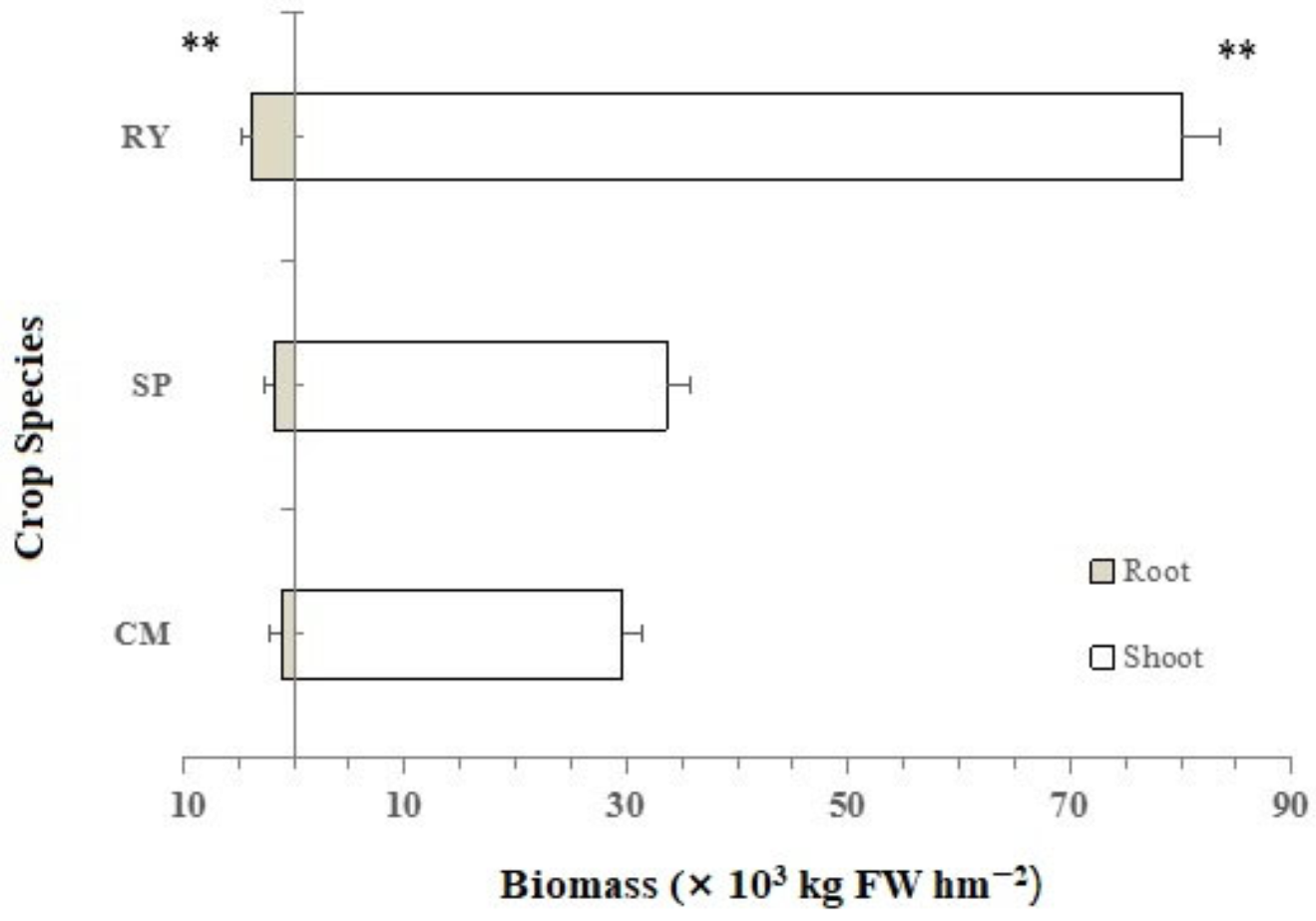
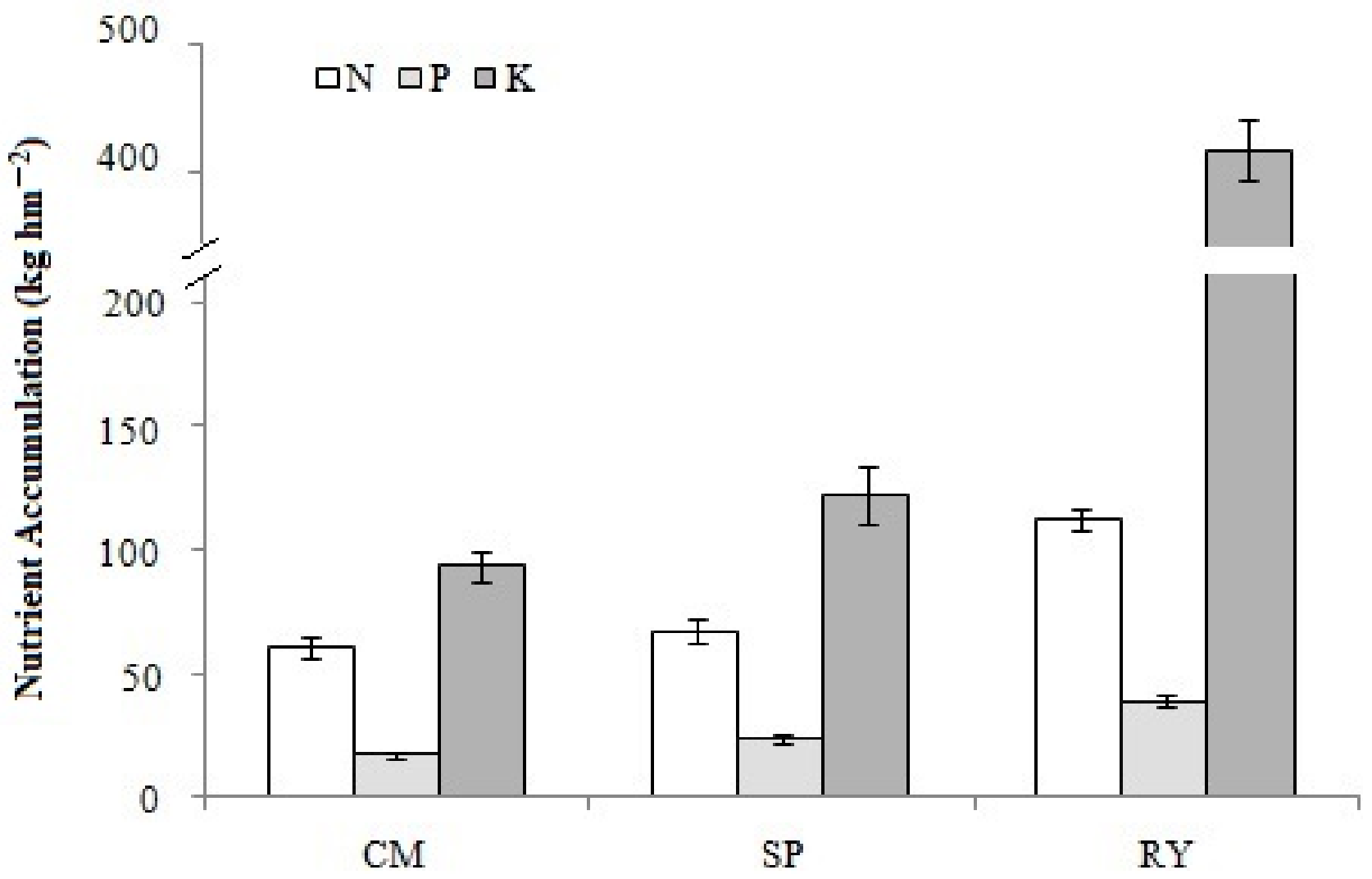
3.2. The Yield and Nutrient Uptake and Accumulation of the Different Crop Species
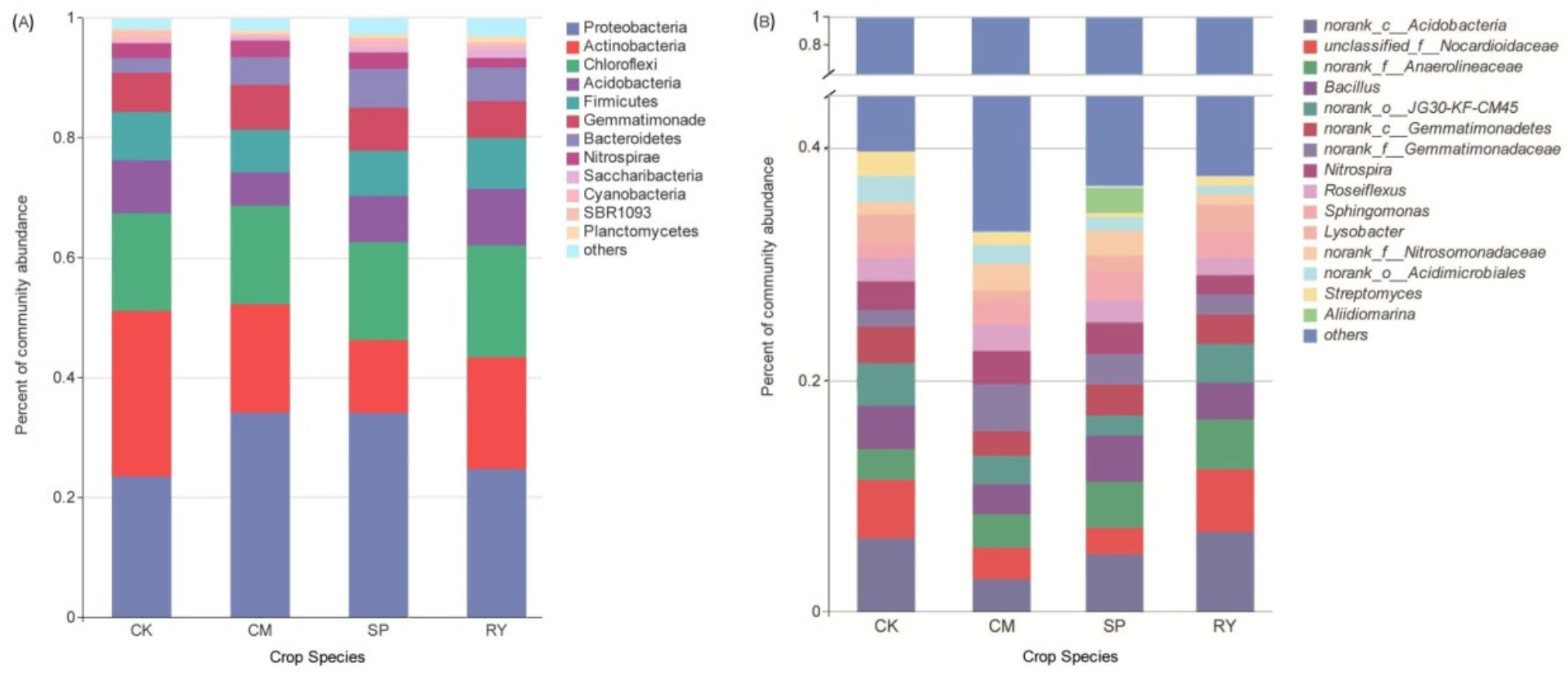
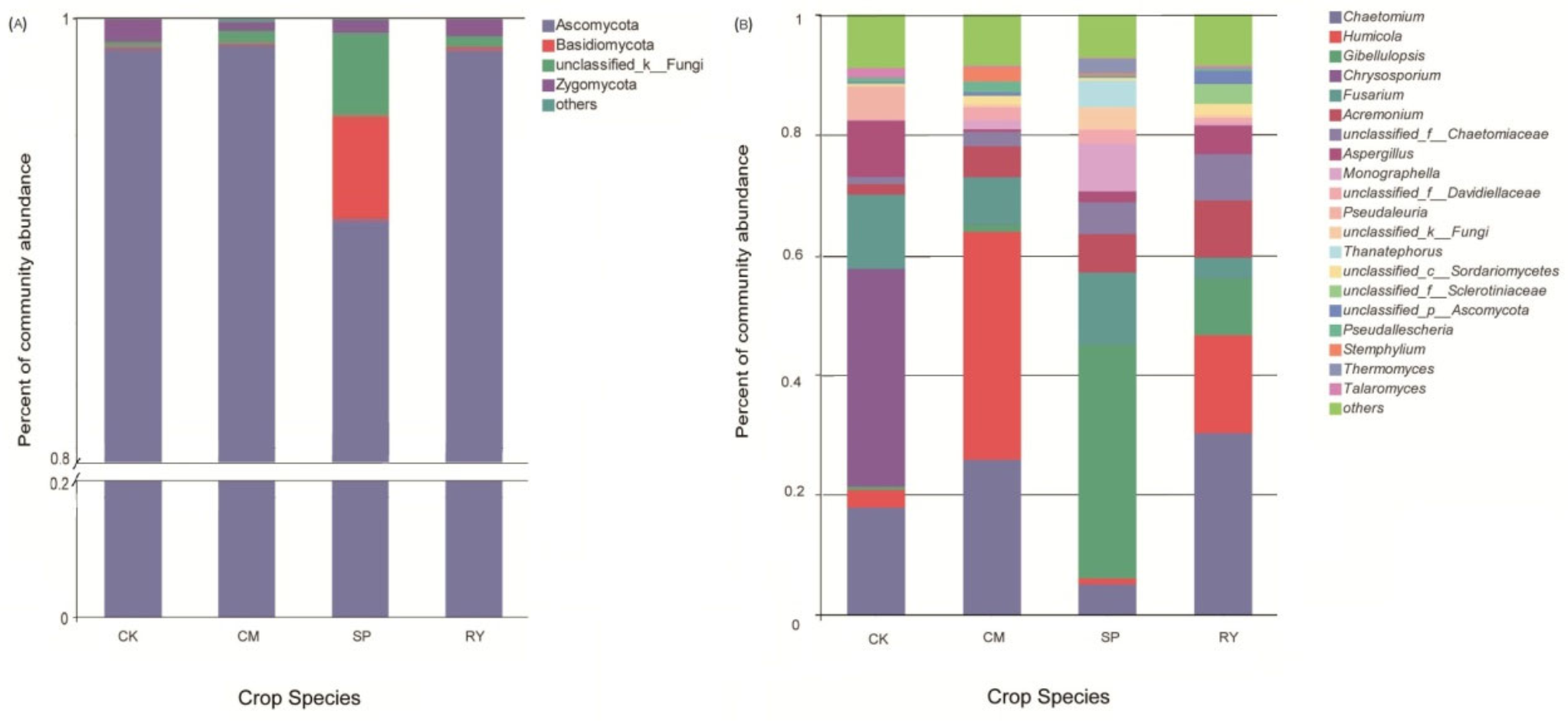
3.3. Abundance and Diversity of Doil Microbial Communities
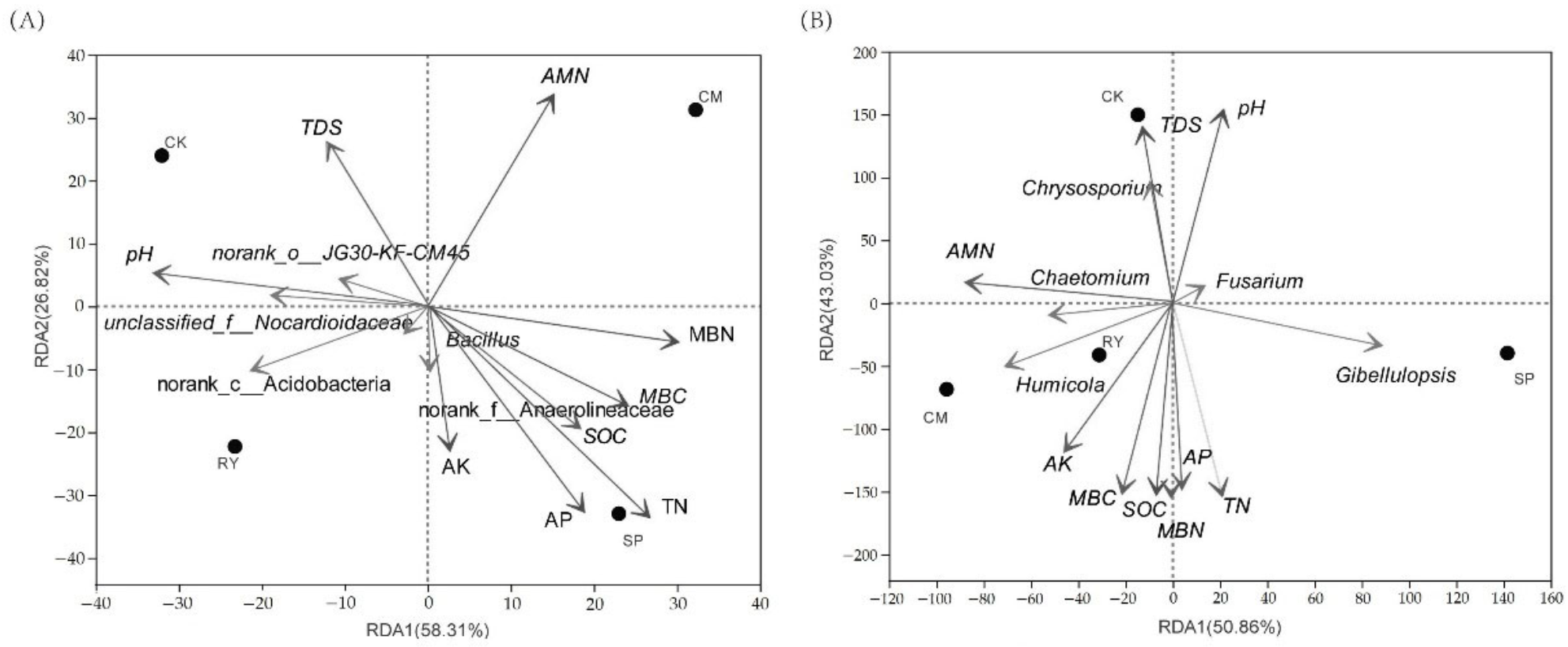
3.4. Relationships between the Relative Abundance of Soil Microorganisms and Crop Nutrient Accumulation
4. Discussion and Conclusions
4.1. Crop Biomass Accumulation and Nutrient Absorption in Secondary Salinized Soil
4.2. Plant–Soil Feedback in Soil Subjected to Secondary Salinization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Chu, S.H.; Zhang, D.; Wang, D.X.; Zhi, Y.; Zhou, P. Heterologous expression and biochemical characterization of assimilatory nitrate and nitrite reductase reveals adaption and potential of Bacillus megaterium NCT-2 in secondary salinization soil. Int. J. Biol. Macromol. 2017, 101, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Liu, L.J.; Zuo, S.F.; Ali, M.; Wang, Z.L. Soil salinity control and cauliflower quality promotion by intercropping with five turfgrass species. J. Clean. Prod. 2020, 266, 121991. [Google Scholar] [CrossRef]
- Shen, W.S.; Ni, Y.Y.; Gao, N.; Bian, B.Y.; Zheng, S.N.; Lin, X.G.; Chu, H.Y. Bacterial community composition is shaped by soil secondary salinization and acidification brought on by high nitrogen fertilization rates. Appl. Soil Ecol. 2016, 108, 76–83. [Google Scholar] [CrossRef]
- Fan, Y.N.; Zhang, Y.X.; Wan, M.X.; Hu, W.Y.; Chen, Z.K.; Huang, B. Plastic shed production intensified secondary soil salinization in perennial fruit production systems. Agr. Ecosyst. Environ. 2021, 316, 107469. [Google Scholar] [CrossRef]
- Gavrichkova, O.; Brykova, R.A.; Brugnoli, E.; Calfapietra, C.; Cheng, Z.Q.; Kuzyakov, Y.; Liberati, D.; Moscatelli, M.C.; Pallozzi, E.; Vasenev, V.I. Secondary soil salinization in urban lawns: Microbial functioning, vegetation state, and implications for carbon balance. Land Degrad. Dev. 2020, 31, 2591–2604. [Google Scholar] [CrossRef]
- Wu, R.Y.; Sun, H.W.; Xue, J.; Yan, D.; Liu, Y.; Gui, D.W.; Wang, X.G.; Yang, J.Z. Acceleration of soil salinity accumulation and soil degradation due to greenhouse cultivation: A survey of farmers’ practices in China. Environ. Monitor. Assess. 2020, 192, 399. [Google Scholar] [CrossRef]
- Jesus, J.M.; Danko, A.S.; Fiúza, A.; Borges, M.T. Comparison of phytoremediation and chemical amendments for non-calcareous highly saline-sodic soil remediation. Water Air Soil Pollut. 2018, 229, 1–10. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, H.T.; Wang, Y.J.; Li, S.; Cao, Z.; Ji, S.; He, Y.; Zhang, H. Effect of dietary forage to concentrate ratios on dynamic profile changes and interactions of ruminal microbiota and metabolites in holstein heifers. Front. Microbiol. 2017, 8, 2206. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Chen, J.; Xing, Y.X.; Xie, X.Y.; Wang, L.C. Influence of intercropping Chinese milk vetch on the soil microbial community in rhizosphere of rape. Plant Soil 2019, 440, 85–96. [Google Scholar] [CrossRef]
- He, H.B.; Li, W.X.; Zhang, Y.W.; Cheng, J.K.; Jia, X.; Li, S.; Yang, H.; Chen, B.; Xin, G.R. Effects of Italian ryegrass residues as green manure on soil properties and bacterial communities under an Italian ryegrass (Lolium multiflorum L.)-rice (Oryza sativa L.) rotation. Soil Till. Res. 2020, 196, 104487. [Google Scholar] [CrossRef]
- Guo, J.J.; Liu, W.B.; Zhu, C.; Luo, G.W.; Kong, Y.L.; Ling, N.; Wang, M.; Dai, J.Y.; Shen, Q.R.; Guo, S.W. Bacterial rather than fungal community composition is associated with microbial activities and nutrient-use efficiencies in a paddy soil with short-term organic amendments. Plant Soil 2017, 424, 335–349. [Google Scholar] [CrossRef]
- Lu, R.K. Soil Argrochemistry Analysis Protocoes; China Agriculture Science Press: Beijing, China, 1999. [Google Scholar]
- Moore, J.M.; Klose, S.; Tabatabai, M.A. Soil microbial biomass carbon and nitrogen as affected by cropping systems. Biol. Fertil. Soils 2020, 31, 200–210. [Google Scholar] [CrossRef]
- Li, Z.Q.; Zhang, X.; Xu, J.X.; Cao, K.; Wang, J.H.; Xu, C.X.; Cao, W.D. Green manure incorporation with reductions in chemical fertilizer inputs improves rice yield and soil organic matter accumulation. J. Soil Sediment 2020, 20, 2784–2793. [Google Scholar] [CrossRef]
- Cai, S.M.; Lv, W.G.; Zhu, H.T.; Zhang, D.S.; Fu, Z.S.; Zhang, H.L.; Xu, S.X. Effect of nitrogen application rate on soil fungi community structure in a rice-fish mutualistic system. Sci. Rep. 2019, 9, 16188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.T.; Long, M.; Gatesoupe, F.J.; Zhang, Q.; Li, A.; Gong, X. Comparative analysis of the intestinal bacterial communities in different species of carp by pyrosequencing. Microb. Ecol. 2014, 69, 25–36. [Google Scholar] [CrossRef]
- Kotula, L.; Kwa, H.Y.; Nichols, P.G.; Colmer, T.D. Tolerance and recovery of the annual pasture legumes Melilotus siculus, Trifolium michelianum and Medicago polymorpha to soil salinity, soil waterlogging and the combination of these stresses. Plant Soil 2019, 444, 267–280. [Google Scholar] [CrossRef]
- Jeromela, A.M.; Mikic, A.M.; Vujic, S.; Cupina, B.; Krstic, D.; Dimitrijevic, A.; Vasiljevic, S.; Mihailovic, V.; Cvejic, S.; Miladinovic, D. Potential of legume-brassica intercrops for forage production and green manure: Encouragements from a temperate southeast european environment. Front. Plant Sci. 2017, 8, 312. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Wang, Y.; Zou, C.B.; Wang, Z.L. Aboveground biomass invariance masks significant belowground productivity changes in response to salinization and nitrogen loading in reed marshes. Wetlands 2017, 37, 985–995. [Google Scholar] [CrossRef]
- Stagg, C.L.; Schoolmaster, D.R.; Piazza, S.C.; Snedden, G.; Steyer, G.D.; Fischenich, C.J.; McComas, R.W. A landscape-scale assessment of above- and belowground primary production in coastal wetlands: Implications for climate change-induced community shifts. Estuar. Coast 2016, 40, 856–879. [Google Scholar] [CrossRef]
- Rane, N.R.; Tapase, S.; Kanojia, A.; Watharkar, A.; Salama, E.S.; Jang, M.; Yadav, K.K.; Amin, M.A.; Cabral-Pinto, M.M.; Jadhav, J.P.; et al. Molecular insights into plant–microbe interactions for sustainable remediation of contaminated environment. Bioresour. Technol. 2022, 344, 126246. [Google Scholar] [CrossRef]
- Thakur, M.P.; Putten, W.; Wilschut, R.A.; Veen, C.; Bezemer, T.M. Plant-soil feedbacks and temporal dynamics of plant diversity-productivity relationships. Trends Ecol. Evol. 2021, 36, 651–661. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.; Sandhu, D.; Liu, X.; Halvorson, J. Spinach (Spinacea oleracea L.) response to salinity: Nutritional value, physiological parameters, antioxidant capacity, and gene expression. Agriculture 2018, 8, 163. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.; Pallavi; Negi, R.; Rani, A.; Parul. Fungal isolation and characterization from spoiled vegetables Lycopersicon esculentum, Brassica oleracea, Spinacia oleracea. Indo Am. J. P. Sc. 2016, 3, 1271–1275. [Google Scholar]
- Zhang, X.Y.; Zha, L.N.; Jiang, P.Y.; Wang, X.Y.; Lu, K.W.; He, S.B.; Huang, J.C.; Zhou, W.L. Comparative study on nitrogen removal and functional genes response between surface flow constructed wetland and floating treatment wetland planted with Iris pseudacorus. Environ. Sci. Pollut. R. 2019, 26, 23696–23706. [Google Scholar] [CrossRef]
- Hung, P.M.; Wattanachai, P.; Kasem, S.; Poeaim, S. Efficacy of chaetomium species as biological control agents against phytophthora nicotianae root rot in citrus. Mycobiology 2015, 43, 288–296. [Google Scholar] [CrossRef] [Green Version]

| Treatments | pH | TDS (g kg−1) | SOC (g kg−1) | AMN (mg kg−1) | AP (mg kg−1) | AK (mg kg−1) | MBC (mg kg−1) | MBN (mg kg−1) |
|---|---|---|---|---|---|---|---|---|
| CK | 4.98 c | 3.25 a | 11.70 b | 120.58 b | 45.56 c | 205.20 a | 108.45 c | 38.70 c |
| CM | 5.29 b | 2.81 b | 12.58 ab | 139.33 a | 90.47 b | 336.92 c | 192.94 ab | 62.60 a |
| SP | 5.33 b | 2.74 b | 11.79 b | 104.06 c | 92.44 b | 282.98 b | 188.36 b | 52.56 b |
| RY | 5.52 a | 2.52 b | 13.09 a | 95.89 c | 113.60 a | 426.08 d | 194.84 a | 54.92 b |
| Treatments | 16S Gene Copy Numbers (Copies × 1010) | ITS Gene Copy Numbers (Copies × 108) | B/F (Bacteria/Fungi 103) |
|---|---|---|---|
| CK | 2.50 d | 1.44 a | 0.17 c |
| CM | 4.21 b | 0.82 b | 0.51 b |
| SP | 2.83 c | 0.26 c | 1.11 a |
| RY | 6.69 a | 1.47 a | 0.46 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, S.; Xu, S.; Zhang, D.; Fu, Z.; Zhang, H.; Zhu, H. Phytoremediation of Secondary Salinity in Greenhouse Soil with Astragalus sinicus, Spinacea oleracea and Lolium perenne. Agriculture 2022, 12, 212. https://doi.org/10.3390/agriculture12020212
Cai S, Xu S, Zhang D, Fu Z, Zhang H, Zhu H. Phytoremediation of Secondary Salinity in Greenhouse Soil with Astragalus sinicus, Spinacea oleracea and Lolium perenne. Agriculture. 2022; 12(2):212. https://doi.org/10.3390/agriculture12020212
Chicago/Turabian StyleCai, Shumei, Sixin Xu, Deshan Zhang, Zishi Fu, Hanlin Zhang, and Haitao Zhu. 2022. "Phytoremediation of Secondary Salinity in Greenhouse Soil with Astragalus sinicus, Spinacea oleracea and Lolium perenne" Agriculture 12, no. 2: 212. https://doi.org/10.3390/agriculture12020212
APA StyleCai, S., Xu, S., Zhang, D., Fu, Z., Zhang, H., & Zhu, H. (2022). Phytoremediation of Secondary Salinity in Greenhouse Soil with Astragalus sinicus, Spinacea oleracea and Lolium perenne. Agriculture, 12(2), 212. https://doi.org/10.3390/agriculture12020212







