Fenlong-Ridging Promotes Microbial Activity in Sugarcane: A Soil and Root Metabarcoding Survey
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Experimental Design
2.1.2. Soil and Root Sampling
2.2. Methods
2.2.1. Estimation of Sugarcane Yield
2.2.2. Analysis of Soil Chemical Properties
2.2.3. Metabarcoding Sequencing
2.2.4. Statistical Analysis
2.2.5. Isolation and Identification of Bacterial and Fungal Strains
3. Results
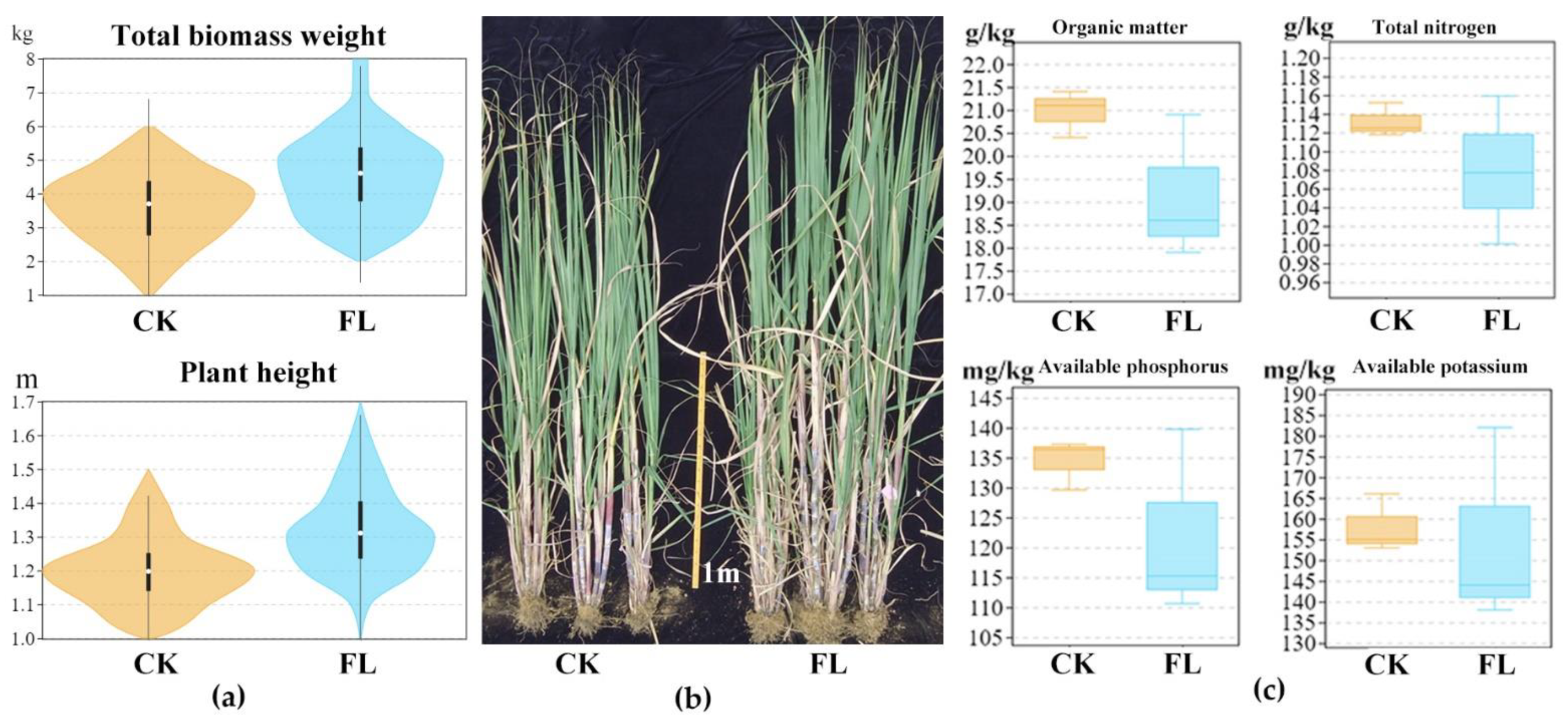
3.1. The Improved Agronomic Performace and the Altered Soil Properties Were Found in Fenlong Compared with the CK
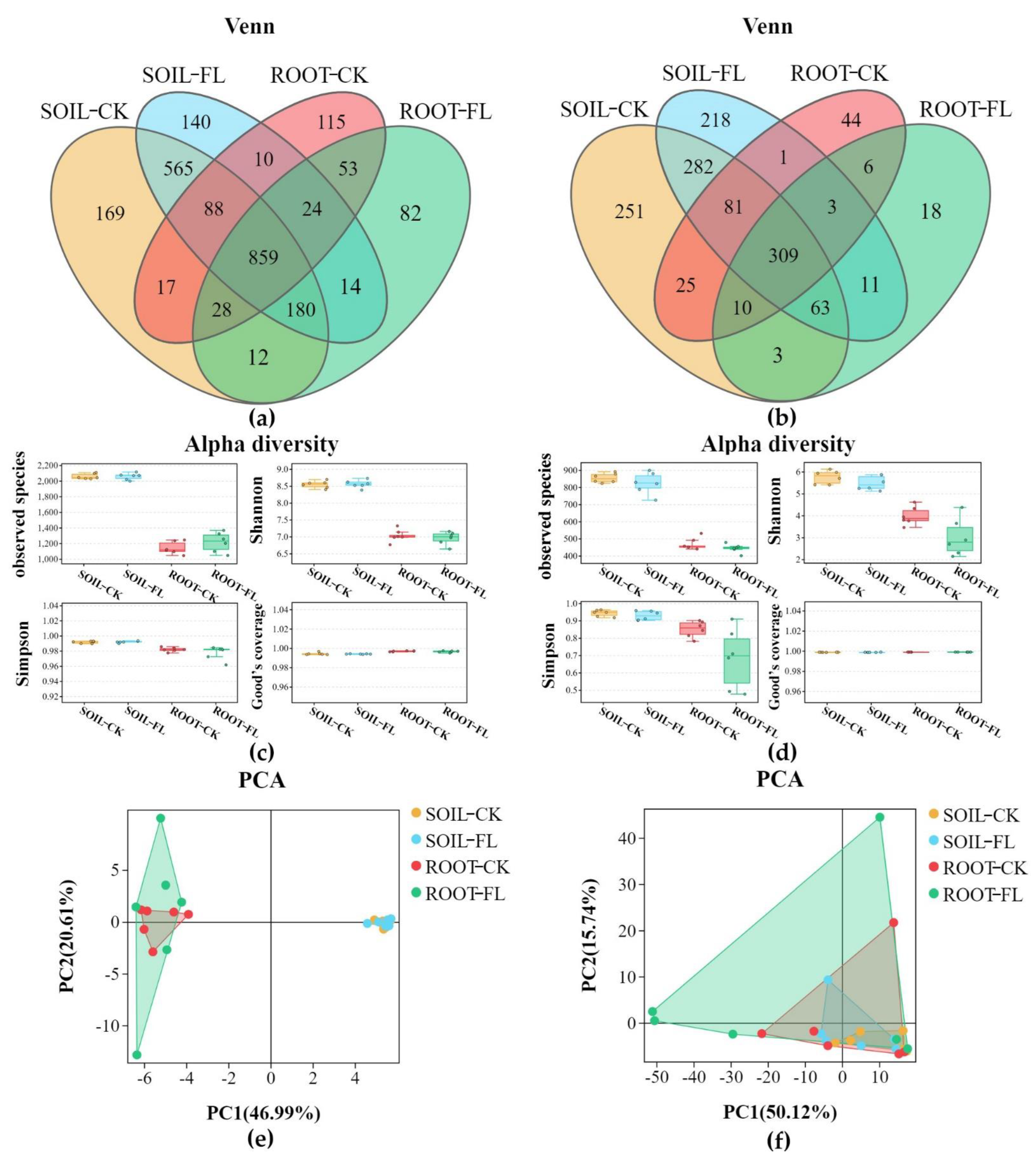
3.2. Metabarcoding Survey of Soil and Root Microbes
3.2.1. Sequencing Analysis Revealed the Greater Diversity in Fenlong Samples than in CK Samples for Fungi and/or Bacteria
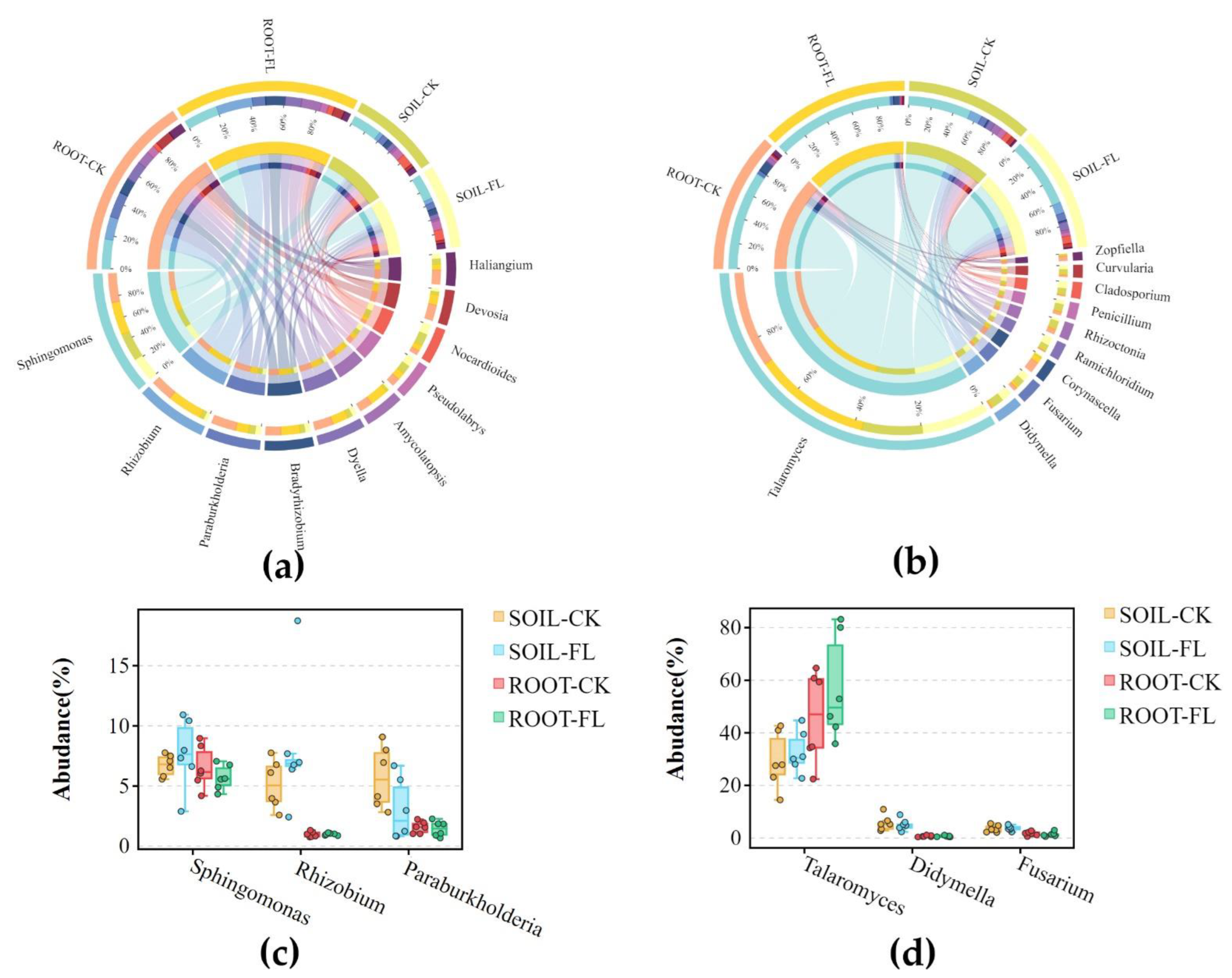
3.2.2. The Predominant Microbial Genera Identified in Fenlong Operation
3.3. Isolation and Classification of the Specific Endophytic Root Bacteria and Fungi from Sugarcane Rhizosphere
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, H.; Li, Y.; Zhu, J.K. Developing naturally stress-resistant crops for a sustainable agriculture. Nat. Plants 2018, 4, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Holland, J.M. The environmental consequences of adopting conservation tillage in Europe: Reviewing the evidence. Agric. Ecosyst. Environ. 2004, 103, 1–25. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Li, T.; Zhao, D.; Liao, Y. Conservation tillage decreases selection pressure on community assembly in the rhizosphere of arbuscular mycorrhizal fungi. Sci. Total Environ. 2020, 710, 136326. [Google Scholar] [CrossRef] [PubMed]
- Wipf, H.M.; Xu, L.; Gao, C.; Spinner, H.B.; Taylor, J.; Lemaux, P.; Mitchell, J.; Coleman-Derr, D. Agricultural Soil Management Practices Differentially Shape the Bacterial and Fungal Microbiome of Sorghum bicolor. Appl. Environ. Microbiol. 2020, 87, e02345-20. [Google Scholar] [CrossRef]
- Li, Y.; Li, T.; Zhao, D.; Wang, Z.; Liao, Y. Different tillage practices change assembly, composition, and co-occurrence patterns of wheat rhizosphere diazotrophs. Sci. Total Environ. 2021, 767, 144252. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Zhang, J.; Li, F.; Liao, P.; Khan, A.; Hussain, I.; Iqbal, A.; Ali, I.; Wei, B.; Jiang, L. Smash ridge tillage strongly influence soil functionality, physiology and rice yield. Saudi J. Biol. Sci. 2021, 28, 1297–1307. [Google Scholar] [CrossRef]
- Suli, L.; Jinling, H.; Benhui, W.; Xiaoying, L.; Ruijie, L.; Lingqiang, W.; Zhigang, L. Effects of Fenlong Tillage on Photosynthetic and Physiological Characteristics, Yield and Quality of Sugarcane (Saccharumofficinarum). Chin. J. Trop. Crops 2021, 42, 726–731. [Google Scholar]
- Bin, W.; Yu-E, L.; An-Dong, C.; Shuo, L.; Tian-Jing, R.; Jia-Qi, Z. Global policies in agricultural greenhouse gas reduction and carbon sequestration and their enlightenment to China in the view of carbon neutrality. Clim. Chang. Res. 2022, 18, 1–13. [Google Scholar]
- Zhenke, Z.; Mouliang, X.; Liang, W.; Shuang, W.; Jina, D.; Jianping, C.; Tida, G. The key biogeochemical processes of carbon sequestration in paddy soil and its countermeasures for carbon neutralization. Chin. J. Eco-Agric. 2021, 1–11. Available online: http://kns.cnki.net/kcms/detail/13.1432.S.20211224.1121.001.html (accessed on 29 November 2021).
- Ben-hui, W.; Xiu-qin, G.; Zhang-you, S.; Xiu-cheng, N.; Liu-ying, L.; Guang-po, W.; Yan-ying, L.; Po, H.; Bin, L.; Yan-yong, W. Yield Increase of Smash-Ridging Cultivation of Sugarcane. Sci. Agric. Sin. 2011, 44, 4544–4550. [Google Scholar] [CrossRef]
- Benhui, W.; Zhangyou, S.; Jia, Z.; Lingzhi, Z.; Po, H.; Xian, Z. Study on Effect and Mechanism of Improving Saline-alkali Soil by Fenlong Tillage. Soils 2020, 52, 699–703. [Google Scholar] [CrossRef]
- Jianghan, L.; Wenshou, H. Effects of Smash-ridging Technology on Soil Properties and Potato Yield. J. Northeast Agric. Sci. 2020, 45, 20–25. [Google Scholar] [CrossRef]
- Shijia, W.; Daihua, J.; Wenguo, Z.; Rongrong, Z.; Junwei, L.; Benhui, W. Effect of Deep Vertical Rotary Tillage on Aggregate Structure in Farmland of Lateritic Red Soil. Acta Pedol. Sin. 2020, 57, 326–335. [Google Scholar] [CrossRef]
- Xiaohua, D.; Xinyue, W.; Hongwu, Y.; Yongjun, L.; Yongsheng, D.; Miliang, Z.; Mingfa, Z.; Jiongping, Z.; Qi, L.; Weimin, W.; et al. Effects of Smashing Ridge Tillage on Growth, Dry Matter Accumulation, Output and Quality of Flue-cured Tobacco. Chin. Tob. Sci. 2020, 41, 28–35. [Google Scholar] [CrossRef]
- Hao, L.; Jin-ling, H.; Zhi-gang, L.; Ben-hui, W.; Xiao-ru, C.; Shi-jian, H.; Xiao-ying, L.; Su-li, L. Fenlong tillage increase soil nutrient availability, and benefit vascular tissue structure and nutrient absorption of sugarcane. J. Plant Nutr. Fertil. 2021, 27, 204–214. [Google Scholar] [CrossRef]
- Rolfe, S.A.; Griffiths, J.; Ton, J. Crying out for help with root exudates: Adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes. Curr. Opin. Microbiol. 2019, 49, 73–82. [Google Scholar] [CrossRef]
- Nesme, J.; Achouak, W.; Agathos, S.N.; Bailey, M.; Baldrian, P.; Brunel, D.; Frostegård, Å.; Heulin, T.; Jansson, J.K.; Jurkevitch, E.; et al. Back to the Future of Soil Metagenomics. Front. Microbiol. 2016, 7, 73. [Google Scholar] [CrossRef]
- Liu, Q.; Zhao, X.; Liu, Y.; Xie, S.; Xing, Y.; Dao, J.; Wei, B.; Peng, Y.; Duan, W.; Wang, Z. Response of Sugarcane Rhizosphere Bacterial Community to Drought Stress. Front. Microbiol. 2021, 12, 716196. [Google Scholar] [CrossRef]
- Achouak, W.; Abrouk, D.; Guyonnet, J.; Barakat, M.; Ortet, P.; Simon, L.; Lerondelle, C.; Heulin, T.; Haichar, F.E.Z. Plant hosts control microbial denitrification activity. FEMS Microbiol. Ecol. 2019, 95, fiz021. [Google Scholar] [CrossRef] [PubMed]
- Guyonnet, J.P.; Guillemet, M.; Dubost, A.; Simon, L.; Ortet, P.; Barakat, M.; Heulin, T.; Achouak, W.; Haichar, F.E.Z. Plant Nutrient Resource Use Strategies Shape Active Rhizosphere Microbiota Through Root Exudation. Front. Plant Sci. 2018, 9, 1662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.K.; Singh, P.; Li, H.B.; Song, Q.Q.; Guo, D.J.; Solanki, M.K.; Verma, K.K.; Malviya, M.K.; Song, X.P.; Lakshmanan, P.; et al. Diversity of nitrogen-fixing rhizobacteria associated with sugarcane: A comprehensive study of plant-microbe interactions for growth enhancement in Saccharum spp. BMC Plant Biol. 2020, 20, 220. [Google Scholar] [CrossRef] [PubMed]
- Beckers, B.; De Beeck, M.O.; Thijs, S.; Truyens, S.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Performance of 16s rDNA Primer Pairs in the Study of Rhizosphere and Endosphere Bacterial Microbiomes in Metabarcoding Studies. Front. Microbiol. 2016, 7, 650. [Google Scholar] [CrossRef] [Green Version]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. 38—Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Avşar, C.; Aras, E.S. Community structures and comparison of nosZ and 16S rRNA genes from culturable denitrifying bacteria. Folia Microbiol. 2020, 65, 497–510. [Google Scholar] [CrossRef]
- Busari, M.A.; Kukal, S.S.; Kaur, A.; Bhatt, R.; Dulazi, A.A. Conservation tillage impacts on soil, crop and the environment. Int. Soil Water Conserv. Res. 2015, 3, 119–129. [Google Scholar] [CrossRef] [Green Version]
- Selman, P. Learning to Love the Landscapes of Carbon-Neutrality. Landsc. Res. 2010, 35, 157–171. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.-K.; Guo, Z.; Li, J.-B.; Tian, C.; Hua, D.-W.; Wang, H.-Y.; Han, J.-C.; Xu, Y. Effects of conservation tillage on soil physicochemical properties and crop yield in an arid Loess Plateau, China. Sci. Rep. 2020, 10, 4716. [Google Scholar] [CrossRef] [PubMed]
- Kwong, K. The effects of potassium on growth, development, yield and quality of sugarcane. In Pasricha and Bansal. Potassium for Sustainable Crop Production: Potash Research Institute of India and International Potash Institute; Springer: Horgen, Switzerland, 2002; pp. 430–440. Available online: https://www.ipipotash.org/uploads/udocs/Potassium%20in%20sugarcane.pdf (accessed on 29 November 2021).
- Poole, P.; Ramachandran, V.; Terpolilli, J. Rhizobia: From saprophytes to endosymbionts. Nat. Rev. Microbiol. 2018, 16, 291–303. [Google Scholar] [CrossRef]
- Júnior, I.D.A.M.; de Matos, G.F.; de Freitas, K.M.; da Conceição Jesus, E.; Rouws, L.F.M. Occurrence of diverse Bradyrhizobium spp. in roots and rhizospheres of two commercial Brazilian sugarcane cultivars. Braz. J. Microbiol. 2019, 50, 759–767. [Google Scholar] [CrossRef]
- An, D.S.; Liu, Q.M.; Lee, H.G.; Jung, M.S.; Kim, S.C.; Lee, S.T.; Im, W.T. Sphingomonas ginsengisoli sp. nov. and Sphingomonas sediminicola sp. nov. Int. J. Syst. Evol. Microbiol. 2013, 63, 496–501. [Google Scholar] [CrossRef] [Green Version]
- Cyle, K.T.; Klein, A.R.; Aristilde, L.; Martínez, C.E. Ecophysiological Study of Paraburkholderia sp. Strain 1N under Soil Solution Conditions: Dynamic Substrate Preferences and Characterization of Carbon Use Efficiency. Appl. Environ. Microbiol. 2020, 86, e01851-20. [Google Scholar] [CrossRef]
- Miljaković, D.; Marinković, J.; Balešević-Tubić, S. The Significance of Bacillus spp. in Disease Suppression and Growth Promotion of Field and Vegetable Crops. Microorganisms 2020, 8, 1037. [Google Scholar] [CrossRef]
- Inthama, P.; Pumas, P.; Pekkoh, J.; Pathom-Aree, W.; Pumas, C. Plant Growth and Drought Tolerance-Promoting Bacterium for Bioremediation of Paraquat Pesticide Residues in Agriculture Soils. Front. Microbiol. 2021, 12, 604662. [Google Scholar] [CrossRef] [PubMed]
- Tahir, H.A.S.; Gu, Q.; Wu, H.; Raza, W.; Safdar, A.; Huang, Z.; Rajer, F.U.; Gao, X. Effect of volatile compounds produced by Ralstonia solanacearum on plant growth promoting and systemic resistance inducing potential of Bacillus volatiles. BMC Plant Biol. 2017, 17, 133. [Google Scholar] [CrossRef] [PubMed]
- Rubio, P.J.S.; Godoy, M.S.; Mónica, I.F.D.; Pettinari, M.J.; Godeas, A.M.; Scervino, J.M. Carbon and Nitrogen Sources Influence Tricalcium Phosphate Solubilization and Extracellular Phosphatase Activity by Talaromyces flavus. Curr. Microbiol. 2016, 72, 41–47. [Google Scholar] [CrossRef] [PubMed]


| Sample ID | 16S rRNA | ITS | ||||
|---|---|---|---|---|---|---|
| Tags | N90 (bp) | OTUs | Tags | N90 (bp) | OTUs | |
| SOIL-CK-1 | 118718 | 409 | 2098 | 123409 | 301 | 823 |
| SOIL-CK-2 | 103827 | 408 | 2041 | 115703 | 297 | 878 |
| SOIL-CK-3 | 113775 | 409 | 2031 | 119342 | 301 | 833 |
| SOIL-CK-4 | 105755 | 409 | 2027 | 127228 | 301 | 891 |
| SOIL-CK-5 | 112379 | 408 | 2044 | 127989 | 300 | 865 |
| SOIL-CK-6 | 112899 | 409 | 2107 | 128203 | 300 | 835 |
| SOIL-FL-1 | 116253 | 409 | 2069 | 119658 | 302 | 725 |
| SOIL-FL-2 | 103995 | 406 | 2019 | 131919 | 302 | 899 |
| SOIL-FL-3 | 112407 | 409 | 2075 | 120641 | 302 | 788 |
| SOIL-FL-4 | 110659 | 403 | 2070 | 117866 | 301 | 821 |
| SOIL-FL-5 | 110378 | 409 | 1998 | 114546 | 301 | 829 |
| SOIL-FL-6 | 111498 | 408 | 2114 | 125921 | 301 | 880 |
| ROOT-CK-1 | 119801 | 409 | 1234 | 127742 | 310 | 491 |
| ROOT-CK-2 | 103773 | 409 | 1095 | 113294 | 285 | 457 |
| ROOT-CK-3 | 109902 | 409 | 1244 | 120016 | 301 | 451 |
| ROOT-CK-4 | 112555 | 409 | 1111 | 116902 | 320 | 451 |
| ROOT-CK-5 | 114501 | 409 | 1045 | 127469 | 302 | 531 |
| ROOT-FL-1 | 109488 | 409 | 1122 | 128731 | 297 | 440 |
| ROOT-FL-1 | 109488 | 409 | 1257 | 126253 | 302 | 401 |
| ROOT-FL-2 | 111747 | 410 | 1098 | 124065 | 339 | 438 |
| ROOT-FL-3 | 107628 | 409 | 1202 | 128470 | 320 | 444 |
| ROOT-FL-4 | 111854 | 410 | 1047 | 113670 | 318 | 449 |
| ROOT-FL-5 | 116919 | 409 | 1367 | 124252 | 296 | 478 |
| ROOT-FL-6 | 108549 | 409 | 1323 | 121863 | 301 | 455 |
| Bacteria | Fungi | ||
|---|---|---|---|
| Genbank ID | Taxon | Genbank ID | Taxon |
| NR026067 | Rhizobium tropici | NR138339 | Penicillium ludwigii |
| NR115466 | Rhizobium tropici | NR121230 | Penicillium raperi |
| NR118084 | Rhizobium tropici | NR138263 | Penicillium brefeldi |
| NR113739 | Rhizobium tropici | NR158825 | Penicillium panissan |
| NR044063 | Rhizobium miluonense | NR131276 | Aspergillus terreus |
| NR109703 | Rhizobium mayense | NR077153 | Penicillium crustosu |
| EF061096 | Rhizobium miluonense | NR135337 | Aspergillus glaucus |
| AY738130 | Rhizobium lusitanum | NR103665 | Talaromyces calidica |
| NR118139 | Rhizobium mesoameric | NR147413 | Talaromyces flavus |
| FN908229 | Rhizobium mesoamerica | NR170732 | Talaromyces annesoph |
| NR117203 | Rhizobium nepotum | NR172395 | Talaromyces coprophi |
| NR115953 | Bacillus aryabhattai | NR165525 | Talaromyces argentin |
| NR118439 | Bacillus aerius | NR138223 | Curvularia lunata |
| NR041794 | Bacillus safensis | NR158448 | Curvularia petersoni |
| NR113945 | Bacillus safensis | ||
| NR114126 | Ralstonia sp. | ||
| Bacteria | Fungi | ||
|---|---|---|---|
| Strain ID | Clustering of Specie | Strain ID | Clustered Species |
| R1 | Rhizobium sp. | T16 | Penicillium ludwigii |
| R3 | Bacillus aryabhattai | T13 | |
| Lx2.2 | RT8 | Penicillium raperi | |
| Rx11 | T5 | Penicillium brefeldi | |
| R2 | T24 | Penicillium sp. | |
| Rx4 | Bacillus aerius | T3 | Aspergillus terreus |
| Lx2.1 | T19 | Talaromyces sp. | |
| Rx12 | Bacillus safensis | RT4 | Talaromyces argentin |
| Rx18 | T8 | ||
| Rx1 | Ralstonia sp. | R3 | |
| Rx16 | T18 | Curvularia petersoni | |
| Rx5 | |||
| Rx13 | |||
| R5 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, M.; Long, Y.; Fan, H.; Ma, L.; Han, S.; Li, S.; Wei, B.; Wang, L. Fenlong-Ridging Promotes Microbial Activity in Sugarcane: A Soil and Root Metabarcoding Survey. Agriculture 2022, 12, 244. https://doi.org/10.3390/agriculture12020244
Duan M, Long Y, Fan H, Ma L, Han S, Li S, Wei B, Wang L. Fenlong-Ridging Promotes Microbial Activity in Sugarcane: A Soil and Root Metabarcoding Survey. Agriculture. 2022; 12(2):244. https://doi.org/10.3390/agriculture12020244
Chicago/Turabian StyleDuan, Mingzheng, Yanyan Long, Hongzeng Fan, Li Ma, Shijian Han, Suli Li, Benhui Wei, and Lingqiang Wang. 2022. "Fenlong-Ridging Promotes Microbial Activity in Sugarcane: A Soil and Root Metabarcoding Survey" Agriculture 12, no. 2: 244. https://doi.org/10.3390/agriculture12020244
APA StyleDuan, M., Long, Y., Fan, H., Ma, L., Han, S., Li, S., Wei, B., & Wang, L. (2022). Fenlong-Ridging Promotes Microbial Activity in Sugarcane: A Soil and Root Metabarcoding Survey. Agriculture, 12(2), 244. https://doi.org/10.3390/agriculture12020244







