CRISPR/Cas9 for Insect Pests Management: A Comprehensive Review of Advances and Applications
Abstract
1. Introduction
1.1. CRISPR/Cas-Mediated Genome Editing in Insects
1.2. Diptera
1.2.1. Drosophila
1.2.2. Anastrepha ludens
1.2.3. Bactrocera dorsalis
1.2.4. Ceratitis capitata
1.3. Lepidoptera
1.3.1. Helicoverpa armigera
1.3.2. Plutella xylostella, Spodoptera, Dendrolimus punctatus and Cydia pomonella
1.3.3. Dendrolimus punctatus
1.3.4. Cydia pomonella
1.3.5. Ostrinia furnacalis (Lepidoptera: Pyralidae)
1.3.6. Agrotis ipsilon
1.3.7. Hyphantria cunea
1.3.8. Mythimna separata
1.4. Hemiptera
1.4.1. Nilaparvata lugen
1.4.2. Diaphorina citri, Homalodisca vitripennis, Bemisia argentifolii and Bemisia tabaci
1.4.3. Euschistus heros
1.5. Coleoptera
1.5.1. Tribolium castaneum
1.5.2. Leptinotarsa decemlineata
1.6. Orthoptera
Locusta migratoria
1.7. Acarina
Tetranychus urticae
2. The CRISPR/Cas9 System in Pest Management: Challenges and Future Prospects
3. Discussion
| Insect Species | Target Gene | Accession Number | Genetic Trait | Mutation Type | Delivery of CRISPR Components | Findings | References |
|---|---|---|---|---|---|---|---|
| Drosophila melanogaster | yellow, rosy | NM_057444.3, NM_079613.3 | Pigmentation and Mating | Knockout, Knockin | Plasmid | This was the first report using the CRISPR/Cas9 system to mediate efficient genome engineering in Drosophila. | Gratz et al., 2013 |
| yellow, white | NM_057444.3, NM_079613.3 | Pigmentation and Mating | Knockout | Cas9 mRNA and sgRNA | sgRNA concentration-dependant knockout was shown for yellow gene, and highly efficient and varied genome editing efficiencies were shown by different sgRNAs. | Bassett et al., 2013 | |
| yellow | NM_057444.3 | Pigmentation and Mating | Knockout | Cas9 mRNA and gRNA | This report used the approach of targeting multiple genes with different sgRNAs, and it attained a remarkably effective targeted mutagenesis. | Yu et al., 2013 | |
| Ast, Eh, capa, Ccap, Crz, npf, Mip, mir-219, mir-315, white | NM_001300582.1, NM_079662.3, NM_079828.3, NM_001275917.2, NM_079626.3, NM_080493.3, NM_140714.4, NR_048289.1, NR_048297.1, X76202.1 | Knockout | Plasmid | To obtain a Cas9–sgRNA complex for achieving targeted mutagenesis, two transgene vectors harboring expression cassettes for Cas9 and sgRNA were delivered. | Kondo and Ueda, 2013 | ||
| rosy, DSH3PX1 | NM_079613.3, NM_140091.4 | Pigmentation and Mating | Knockout, Knockin | Plasmid | Executed efficient and complex genomic manipulations using CRISPR/Cas9-mediated HDR. | Gratz et al., 2014 | |
| ebony, yellow, wingless, wnt | NM_079707.4, NM_057444.3, NM_078778.5 | Segmentation | Knockout, Knockin | Plasmid | Different promoters were used to drive sgRNA expression, and based on promoter properties, different patterns of expression were observed. | Port et al., 2014 | |
| EGFP, mRFP | Chromogenic fluorophores | Knockout | Plasmid | Induction of mutations by injection of an sgRNA into Vasa-Cas9 transgenic fly embryos. | Sebo et al., 2014 | ||
| white, piwi | NM_057439.2, NM_001298896.1 | Pigmentation and Expression of group of small RNA | Knockout, Knockin | Plasmid | Used Cas9 nickase and sgRNA pairs to prevent off-target effects during the generation of indel mutants. | Ren et al., 2014 [110] | |
| ms(3)k81, white, yellow | NM_143253.2, NM_057439.2, NM_057444.3 | Pigmentation | Knockout, Knockin | Plasmid | CRISPR mediated genome editing was shown in Drosophila. | Xue et al., 2014a | |
| yellow, notch, bam, nos,ms(3)k81, cid | NM_057444.3, NM_001258581.2, NM_057452.4, NM_057310.4, NM_143253.2, NM_079006.4 | Physiology | Knockout | Plasmid | A CRISPR/Cas9-mediated conditional mutagenesis system combined with tissue-specific expression of Cas9 was used to temporally and spatially inhibit gene expression. | Xue et al., 2014b | |
| salm | NM_164966.3 | Zinc Finger Transcriptional Repressor | Knockin | mRNA, transgene | For flexible modification of fly genome, a two-step method was proposed. | Zhang X. et al., 2014 | |
| ebony, yellow, vermilion | NM_079707.4, NM_057444.3, NM_078558.3 | Pigmentation | Knockout, Knockin | Plasmid, transgene | Donor template and sgRNA plasmids were injected into Cas9 transgenic embryos in Drosophila. | Ren et al., 2014b | |
| ebony, yellow, white | NM_079707.4, NM_057444.3, NM_057439.2 | Pigmentation | Knockout, Knockin | Plasmid, transgene | A bicistronic Cas9/sgRNA vector was constructed which enhanced the efficiency of gene targeting. | Gokcezade et al., 2014 | |
| ebony, yellow, wg, wls, Lis1, Se | NM_079707.4, NM_057444.3, NM_078778.5, NM_140188.4, NM_057812.5, NM_139978.4, NM_057812.5 | Pigmentation and Physiology | Knockout, Knockin | Plasmid, transgene | Non-transgenic individuals exhibited lessefficient knockin than transgenic individuals did. | Port et al., 2015 [111] | |
| yellow | NM_057444.3 | Pigmentation | Knockin | Transgene | Heterozygous recessive mutation was converted to homozygous loss of function mutations utilizing mutagenic chain reaction (MCR) technology in Drosophila. | Gantz and Bier, 2015 | |
| Dα6 | NM_164874.3 | Insecticide resistance | Knockin | Plasmid | The G275E mutation of the nAChR Dα6 subunit is directly related to Spinosad resistance. | Zimmer et al., 2016 | |
| LUBEL | NM_001273232.2 | Growth and Development | Knockout | Plasmid | Flies with LUBEL mutations exhibited reduced survival and defective climbing in response to heat. | Asaoka et al., 2016 | |
| Scsα | NM_079181.4 | Growth and Development | Knockout | Plasmid | Mutant flies could not produce sufficient energy to promote normal growth. | Quan et al., 2017 | |
| clamp | NM_136293.4 | Sex Specific | Knockout | Plasmid | The expression of a sex-specific gene was regulated by an essential transcription factor. | Urban et al., 2016 | |
| chameau, CG4221, CG5961 | NM_135273.5,NM_141949.4 | Knockin | mRNA | HDR-mediated genome modifications efficiency was tested, and a problem associated with “ends-in” recombination was resolved. | Yu et al., 2014 | ||
| fdl | NM_165908.2 | Knockout | Plasmid | Capability of CRISPR/Cas9 system for analysing or manipulating protein glycosylation pathways. | Mabashi-Asazuma et al., 2015 [112] | ||
| mod(mdg4) | NM_163878.2 | Knockout | Plasmid | Validation of a functional gene involved in trans-splicing that influenced the development in flies. | Gao et al., 2015 [113] | ||
| act5C, lig4, mus308 | NM_167053.2, NM_132679.3, L76559.1 | Knockout, Knockin | Plasmid, transgene | Offered a comprehensive technique for genome editing in Drosophila S2 cells. | Kunzelmann et al., 2016 [114] | ||
| yellow, white, tan | NM_057444.3, NM_057439.2, NM_132315.1 | Pigmentation | Knockin | Plasmid, transgene | Proposed a new process of attaining single or multiple allelic substitutions. | Lamb et al., 2016 | |
| wntless | NM_140188.4 | Growth and development | Knockout | Plasmid | A complex of tRNA–sgRNA was proposed to amplify the cleavage efficiency of the Cpf1 and Cas9 nucleases. | Port and Bullock, 2016 | |
| TpnC | NM_078895.4 | Growth and development | Knockout | Plasmid | Confirmed that the myofibril assembly is related to TpnC gene. | Chechenova et al., 2017 | |
| Alk | NM_144343.3 | Growth and development | Knockout | Plasmid | Revealed that transcription factors can affect Alk gene expression by establishing mutations in Alk enhancer regions. | Mendoza-Garcia et al., 2017 | |
| Drosophila suzukii | white (w-) | NM_057439.2 | Pigmentation | Knockout | Plasmid | Absence of mating and copulation failure was reported. The mutation also caused pigmentation deficiency in testis sheath, which could be a probable reason for copulation failure. | Yan et al., 2020 |
| white, Sxl | NM_057439.2, XM_017083263.2 | Sex determination | Knockout | Plasmid | Sxl gene was proved as excellent gene to suppress the population growth of this destructive pest. | Li and Scott, 2016 | |
| DsRed (red fluorescence protein) | knockin | Plasmid, transgene | The enhancer/promoter of the spermatogenesis-specific beta-2-tubulin (β2t) gene was used for expression of fluorescent proteins or effector molecules in testes of pests, and this providing basis for reproductive biology studies sexing and monitoring. | Ahmed, H. M. et al., 2019 | |||
| Drosophila subobscura | yellow, white | XM_034814491.1, XM_034808177.1 | Pigmentation | Knockout | mRNA | Gene functions were analyzed in a non-model Drosophila species. | Tanaka et al., 2016 [115] |
| Anastrepha ludens | Astra-2 | EU024509.1 | Sex determination | Knockout | RNP complex | The mutation caused sterility, thus, the target gene was proposed for helping in pest control. | Li et al., 2019 |
| Bactrocera dorsalis | White and transformer | AY055817.1, KP342062.1 | Sex determination and reproduction | Knockout | RNP complex | CRISPR/Cas9 mediated mutation of white and transformer genes caused various phenotypic effects. | Zhao et al., 2018 |
| Ceratitis capitata |
| Homology directed repair | knockin | RNP complex and a single-stranded oligo donor | Conversion of eGFP-to-BFP was demonstrated for establishing an efficient HDR through CRISPR-based genome editing. | Aumann, R. A. et al., 2018 | |
| X89933.1, XM_020858622.2 | Segmentation | Knockout | RNP complex | A simple and highly efficient RNP complex-based genome editing approach was reported with the details of designing and preparation. | Meccariello, A. et al., 2017 | |
| Helicoverpa armigera | NPC1b | MK555324.1 | Growth and dietary uptake of Cholesterol | Knockout | RNP complex | NPC1b is vital for the growth and dietary cholesterol uptake. Thus, a novel pest-management technique can be developed using NPC1b as an insecticidal target. | Zheng, J. C. et al., 2020 |
| HaCad | JX23382.1 | cell–cell adhesion | Knockout | sgRNAs and Cas9 mRNA | sgRNAs and Cas9 mRNA were injected into the fresh eggs, and a high editing efficiency of the HaCad locus was achieved. | Wang, J. et al., 2016 | |
| HaABCA2 | KP259911.1 | Regulation of enzymes | Knockout | Cas9 mRNA and sgRNA | The knockout of HaABCA2 confirmed the role of HaABCA2 in mediating toxicity of both Cry2Aa and Cry2Ab against H. armigera. | Wang, J. et al., 2017 | |
| odorant receptor 16 (OR16) | KF768670.1 | Olfaction | Knockout | Cas9 mRNA + sgRNA and RNP complex | The results represent the basis for novel olfactory-based strategies of pest population control. | Chang, H. et al., 2017 | |
| white, ok, brown, and scarlet | XM_021344759.2, KU754490.1, KU754480.1, KU754478.1 | Pigmentation | Knockout | Cas9 mRNA | The report represented differential distribution of eye pigments in the mutants; this finding may be helpful in elucidation of biosynthetic pathway. | Khan, S. A.,et al., 2017 | |
| cluster of nine P450 genes | KM016735.1, R095600.1, KM016739.1, KM016740.1, DQ256407.1, KM016743.1, KM016741 | Regulation of detoxifying enzymes | Knockout | Cas9 protein and multiple sgRNAs | The report identified the key players in the insecticide metabolism. | Wang, H. et al., 2018 | |
| Plutella xylostella | Pxabd-A | XM_011570968.3 | Body segmentation | Knockout | Cas9 mRNA | CRISPR/Cas9 was used to target genes in P. xylostella for the first time which provided new ideas for pest control. | Huang et al., 2016 |
| Pxdsx | XM_048630440.1 | Sex determination | Knockout | Microinjection of RNP complex | The results showed CRISPR/Cas9 system led to altered expression of sex biased genes. | Wang et al., 2019 | |
| PxCHS1 | AB271784.1 | Development | Knockout | Plasmid | Description of the resistance management strategies for insect pests, it and explained the MoA behind the resistance using CRISPR/Cas9 system. | Douris et al., 2016 | |
| LW | Locomotion | Knockout | RNP complex | The results showed weaker phototaxis and reduced locomotion, thus making it a helpful method for pest control | Chen et al., 2021 [116] | ||
| Spodoptera frugiperda | Sfabd-A | MH541836.1 | Body segmentation | Knockout | RNP complex | The results showed that gene function validation and the understanding of resistance mechanism can be performed using CRISPR/Cas9 system which can lead to the development of novel pest management approaches. | Wu et al., 2018 |
| XM_035582273.2 XM_050696092.1 XM_050696079.1 | Growth and development | Knockout | Cas9 protein and multiple sgRNAs | The developed mutants were helpful to understand the crucial pathways of S. frugiperda and the strategy can also applied for other invasive pests. | Zhu, G. H. et al., 2020 [117] | |
| Spodoptera litura | Slabd-A | GCA_002706865.1 | Body segmentation | Knockout | Cas9 mRNA and sgRNA | The direct injection of Cas9-coding mRNA and Slabd-A-specific sgRNA into the embryos of the S. litura led to the induction of the typical abd-A deficient phenotypes showing irregular segmentation and unusual pigmentation at the larval stage. | Bi, H. L. et al., 2016 |
| SlitBLOS2 | XM_022977403.1 | Molecular marker | Knockout | Cas9 mRNA and sgRNA | The study demonstrated that SlitBLOS2 has a role in the coloration of the integuments, and thus, it provided a marker gene for functional studies and pest control strategies. | Zhu, G. H. et al., 2017 | |
| Spodoptera littoralis | SlitOrco | Olfaction | Knockout | mRNA | The Orco gene was investigated in the insect Spodoptera littoralis. The results were helpful in making a pest control strategy and in gene function analysis. | Koutroumpa et al., 2016 | |
| Spodoptera exigua | Seα6 | MN714701.1 | Knockout | RNP complex | The study demonstrated that knocked-out Seα6 was highly resistant to insecticides. | Zuo et al., 2020 | |
| Dendrolimus punctatus | DpWnt-1 | KU640201.1 | Development and segmentation | Knockout | mRNA | Proved the necessity of DpWnt-1 signaling in appendage development and anterior segmentation. | Liu H. et al., 2017 |
| Cydia pomonella | CpomOR1 | FJ385021.1 | Olfaction | Knockout | Cas9 mRNA and sgRNA | The report demonstrated mutation in the CpomOR1 gene via CRISPR/Cas9 affected the egg production and viability in the insect. | Garczynski, S. F. et al., 2017 |
| Ostrinia furnacalis | OfAgo1 | Growth and development | knockout | sgRNA and Cas9 mRNA | Mutation in OfAgo1 gene through CRISPR/Cas9 technology caused cuticle disruption. | You et al., 2019 | |
| Agrotis ipsilon | AiTH | Growth and development | Knockout | sgRNA and Cas9 mRNA | The AiTH gene knockout by CRISPR/Cas9 caused narrowing in the egg shell. | Yang et al., 2018 | |
| Hyphantria cunea | Hcdsx | Reproduction | Knockout | sgRNA and Cas9 mRNA | Knocked-out Hcdsx gene by CRISPR/Cas9 caused sex-specific sterility, thus making it a pest control method. | Li et al., 2020 | |
| Mythimna separata | NPC1b | MZ209049.1 | Intestinal absorption and sterol trafficking | Knockout | RNP complex | Knockout of NPC1b can hamper nutrient absorption. | Tang et al. 2022 |
| Nilaparvata Lugens | Nl-cn and Nl-w | MH105806.1 | Pigmentation | Knockout | Cas9 mRNA and sgRNA | Two genes for eye pigmentation were targeted using CRISPR/Cas9, and the results paved path for gene-function interrogation. | Xue. et al., 2018 |
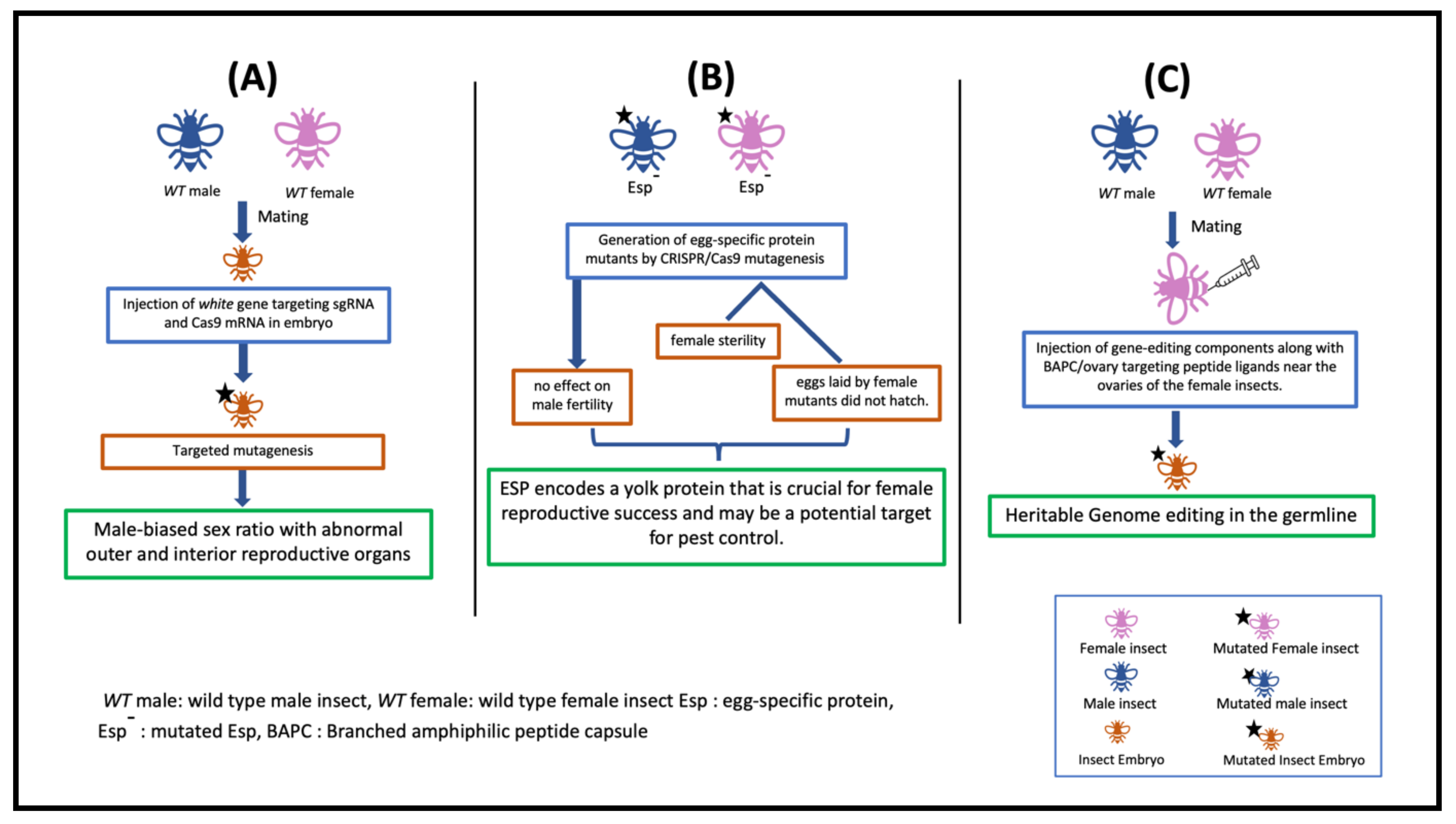
| Diaphorina citri | ACP-TRX-2 | XM_026831570.1 | Physiology | Knockout | BAPC-assisted delivery of CRISPR components | The method incorporated BAPC-assisted delivery of CRISPR/Cas9 into nymphs and adults, thus resulting an innovative breakthrough in gene editing, it and has shown a significant improvement over efforts using injection of eggs. | Hunter et al., 2018 |
| Diaphorina citriHomalodisca vitripennis, Bemisia argentifolii | Thioredoxin and Vermillion | XM_046819472.1 | Physiology andEye color | Knockout | BAPC, plasmid, dsRNA | The BAPC-assisted delivery system developed gene editing methods across the all hemipteran pests by permitting the use of nymphs and adults. BAPC-assisted CRISPR delivery transformed the approaches to protect food crops from different pathogens and insect vectors. | Hunteret al., 2019 |
| Bemisia tabaci | white | XM_019053144.1 | Pigmentation | Knockout | SgRNA + Cas9 protein fused with overy targeting peptide ligand (BtKV) | The method has significantly expanded the capability of CRISPR techniques for whitefly research. | Heu et al., 2020 |
| Euschistus heros | abnormal wing disc (awd), tyrosine hydroxylase (th) and yellow (yel) | NP_001119625.1, XP_008182999.1, XP_001948479.1 | Body segmentation and pattern | Knockdown and knockout | dsRNA, RNP complex | Use of RNAi and CRISPR/Cas9 techniques for managing insect pests. | Cagliari et al., 2020 |
| Tribolium castaneum | Tribolium E-cadherin | XM_961215.3 | Dorsal closure defect | Knockout | Plasmid | Tribolium E-cadherin gene was targeted for knockout study. | Gilles et al., 2015 |
| Leptinotarsa decemlineata | vestigial gene (vest) | XM_023168389.1 | Growth and development | Knockout | RNP complex | Functionally characterized vest gene and CRISPR/Cas9 protocol was established for mutagenesis. | Gui, S. et al., 2020 |
| Locusta migratoria | Orco | JN989549.1 | Olfaction | Knockout | mRNA | Functional genetic studies of locusts by generation of loss-of-function mutation for managing insect pests. | Li Y. et al., 2016 |
| Tetranychus urticae | PSST | KX806605.1 | Knockout | Plasmid | Substitution in the H92R amino acid of the PSST homolog was related to pyridaben resistance and the mutation into the Drosophila PSST homolog using CRISPR/Cas9 genome-editing tools. | Bajda et al., 2017 | |
| phytoene desaturase | MF167355.1 | Knockout | RNP complex | Induction of two mutagenetic events using CRISPR/Cas9 providing basis for functional studies. | Dermauw, W. et al., 2020 |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. The State of Food Security and Nutrition in the World; FAO: Rome, Italy, 2017. [Google Scholar]
- Sun, D.; Guo, Z.; Liu, Y.; Zhang, Y. Progress and prospects of CRISPR/Cas systems in insects and other arthropods. Front. Physiol. 2017, 8, 608. [Google Scholar] [CrossRef]
- Scheben, A.; Wolter, F.; Batley, J.; Puchta, H.; Edwards, D. Towards CRISPR/Cas crops–bringing together genomics and genome editing. New Phytol. 2017, 216, 682–698. [Google Scholar] [CrossRef] [PubMed]
- Broekgaarden, C.; Snoeren, T.A.; Dicke, M.; Vosman, B. Exploiting natural variation to identify insect-resistance genes. Plant Biotechnol. J. 2011, 9, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Chaerani, R.; Voorrips, R.E. Tomato early blight (Alternaria solani): The pathogen, genetics, and breeding for resistance. J. Gen. Plant. Pathol. 2006, 72, 335–347. [Google Scholar] [CrossRef]
- Flowers, T.J.; Yeo, A.R. Breeding for salinity resistance in crop plants: Where next? Funct. Plant. Biol. 1995, 22, 875–884. [Google Scholar] [CrossRef]
- Paszkowski, J.; Baur, M.; Bogucki, A.; Potrykus, I. Gene targeting in plants. EMBO J. 1988, 7, 4021–4026. [Google Scholar] [CrossRef] [PubMed]
- Puchta, H.; Dujon, B.; Hohn, B. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 1993, 21, 5034–5040. [Google Scholar] [CrossRef]
- Upadhyay, S.K. Genome Engineering for Crop Improvement; John Wiley & Sons: Hoboken, NJ, USA, 2021; pp. 1–394. [Google Scholar] [CrossRef]
- Sushmita; Kaur, G.; Upadhyay, S.K.; Verma, P.C. An Overview of Genome-Engineering Methods. In Genome Engineering for Crop Improvement; Upadhyay, S.K., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2021; pp. 1–21. [Google Scholar] [CrossRef]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Ezura, H. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef]
- Tang, X.; Lowder, L.G.; Zhang, T.; Malzahn, A.A.; Zheng, X.; Voytas, D.F.; Kirkland, E.R. A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plant. 2017, 3, 1–5. [Google Scholar]
- Zong, Y.; Wang, Y.; Li, C.; Zhang, R.; Chen, K.; Ran, Y.; Gao, C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 438. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Rath, D.; Amlinger, L.; Rath, A.; Lundgren, M. The CRISPR-Cas immune system: Biology, mechanisms and applications. Biochimie 2015, 117, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Botella, J.R.; Zhu, J.K. Heritability of targeted gene modifications induced by plant-optimized CRISPR systems. Cell. Mol. Life Sci. 2017, 74, 1075–1093. [Google Scholar] [CrossRef] [PubMed]
- Char, S.N.; Neelakandan, A.K.; Nahampun, H.; Frame, B.; Main, M.; Spalding, M.H.; Yang, B. An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant. Biotechnol. J. 2017, 15, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Mir, A.; Alterman, J.F.; Hassler, M.R.; Debacker, A.J.; Hudgens, E.; Echeverria, D.; Brodsky, M.H.; Khvorova, A.; Watts, J.K.; Sontheimer, E.J. Heavily and fully modified RNAs guide efficient SpyCas9-mediated genome editing. Nat. Commun. 2018, 9, 2641. [Google Scholar] [CrossRef] [PubMed]
- Handler, A.M. Prospects for using genetic transformation for improved SIT and new biocontrol methods. Genetica 2002, 116, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, J.C.; Li, X.; Henry, R.A.; Haack, N.; Stringfellow, L.; Heath, A.C.G.; Scott, M.J. Germ-line transformation of the Australian sheep blowfly Luciliacuprina. Insect Mol. Biol. 2002, 11, 1–10. [Google Scholar] [CrossRef]
- Schetelig, M.F.; Caceres, C.; Zacharopoulou, A.; Franz, G.; Wimmer, E.A. Conditional embryonic lethality to improve the sterile insect technique in Ceratitis capitata (Diptera: Tephritidae). BMC Biol. 2009, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Schetelig, M.F.; Wimmer, E.A. Insect transgenesis and the sterile insect technique. In Insect Biotechnology; Springer: Dordrecht, The Netherlands, 2011; pp. 169–194. [Google Scholar]
- Schetelig, M.F.; Handler, A.M. Strategy for enhanced transgenic strain development for embryonic conditional lethality in Anastrephasuspensa. Proc. Natl. Acad. Sci. USA 2012, 109, 9348–9353. [Google Scholar] [CrossRef]
- Schetelig, M.F.; Targovska, A.; Meza, J.S.; Bourtzis, K.; Handler, A.M. Tetracycline-suppressible female lethality and sterility in the Mexican fruit fly, Anastrephaludens. Insect Mol. Biol. 2016, 5, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Ogaugwu, C.E.; Schetelig, M.F.; Wimmer, E.A. Transgenic sexing system for Ceratitis capitata (Diptera: Tephritidae) based on female-specific embryonic lethality. Insect Biochem. Mol. Biol. 2013, 43, 1–8. [Google Scholar] [CrossRef]
- Gantz, V.M.; Akbari, O.S. Gene editing technologies and applications for insects. Curr. Opin. Insect Sci. 2018, 28, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Bassett, A.R.; Liu, J.L. CRISPR/Cas9 and genome editing in Drosophila. J. Genet. Genom. 2014, 41, 7–19. [Google Scholar] [CrossRef]
- Gratz, S.J.; Cummings, A.M.; Nguyen, J.N.; Hamm, D.C.; Donohue, L.K.; Harrison, M.M.; Wildonger, J.; O’Connor-Giles, K.M. Genome engineering of Drosophila with the CRISPR RNA-Guided Cas9 nuclease. Genetics 2013, 194, 1029–1035. [Google Scholar] [CrossRef]
- Gratz, S.J.; Wildonger, J.; Harrison, M.M.; O’connor-Giles, K.M. CRISPR/Cas9-mediated genome engineering and the promise of designer flies on demand. Fly 2013, 7, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Baena-Lopez, L.A.; Alexandre, C.; Mitchell, A.; Pasakarnis, L.; Vincent, J.P. Accelerated hmologous recombination and subsequent genome modification in Drosophila. Development 2013, 140, 4818–4825. [Google Scholar] [PubMed]
- Yu, Z.; Chen, H.; Liu, J.; Zhang, H.; Yan, Y.; Zhu, N.; Guo, Y.; Yang, B.; Chang, Y.; Dai, F.; et al. Various applications of TALEN- and CRISPR/Cas9-mediated homologous recombination to modify the Drosophila genome. Biol. Open 2014, 3, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Kondo, S.; Ueda, R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 2013, 195, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Port, F.; Chen, H.M.; Lee, T.; Bullock, S.L. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 2014, 111, E2967–E2976. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Yang, Z.; Mao, D.; Chang, Z.; Qiao, H.H.; Wang, X.; Sun, J.; Hu, Q.; Cui, Y.; Liu, L.P.; et al. Performance of the Cas9 nickase system in Drosophila melanogaster. G3 2014, 4, 1955–1962. [Google Scholar] [CrossRef] [PubMed]
- Sebo, Z.L.; Lee, H.B.; Peng, Y.; Guo, Y. A simplified and efficient germline-specific CRISPR/Cas9 system for Drosophila genomic engineering. Fly 2014, 8, 52–57. [Google Scholar] [CrossRef]
- Xue, Z.; Ren, M.D.; Wu, M.H.; Dai, J.B.; Rong, Y.K.S.; Gao, G.J. Efficient gene knock-out and knock-in with transgenic Cas9 in Drosophila. G3 2014, 4, 925–929. [Google Scholar] [CrossRef]
- Xue, Z.; Wu, M.; Wen, K.; Ren, M.; Long, L.; Zhang, X.; Gao, G. CRISPR/Cas9 mediates efficient conditional mutagenesis in Drosophila. G3 2014, 4, 2167–2173. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Koolhaas, W.H.; Schnorrer, F. A versatile two-step CRISPR-and RMCE-based strategy for efficient genome engineering in Drosophila. G3 Genes Genomes Genet. 2014, 4, 2409–2418. [Google Scholar] [CrossRef] [PubMed]
- Gokcezade, J.; Sienski, G.; Duchek, P. Efficient CRISPR/Cas9 plasmids for rapid and versatile genome editing in Drosophila. G3 2014, 4, 2279–2282. [Google Scholar] [CrossRef] [PubMed]
- Port, F.; Muschalik, N.; Bullock, S.L. Systematic evaluation of Drosophila CRISPR tools reveals safe and robust alternatives to autonomous gene drives in basic research. G3 2015, 5, 1493–1502. [Google Scholar] [CrossRef]
- Gantz, V.M.; Bier, E. The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science 2015, 348, 442–444. [Google Scholar] [CrossRef]
- Zimmer, C.T.; Garrood, W.T.; Puinean, A.M.; Eckel-Zimmer, M.; Williamson, M.S.; Davies, T.G.; Bass, C. A CRISPR/Cas9 mediated point mutation in the alpha 6 subunit of the nicotinic acetylcholine receptor confers resistance to spinosad in Drosophila melanogaster. Insect Biochem. Mol. Biol. 2016, 73, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Douris, V.; Denecke, S.; Van Leeuwen, T.; Bass, C.; Nauen, R.; Vontas, J. Using CRISPR/Cas9 genome modification to understand the genetic basis of insecticide resistance: Drosophila and beyond. Pestic. Biochem. Physiol. 2020, 167, 104595. [Google Scholar] [CrossRef] [PubMed]
- Bassett, A.R.; Tibbit, C.; Ponting, C.P.; Liu, J.L. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell. Rep. 2013, 4, 220–228. [Google Scholar] [CrossRef]
- Yu, Z.; Ren, M.; Wang, Z.; Zhang, B.; Rong, Y.S.; Jiao, R.; Gao, G. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics 2013, 195, 289–291. [Google Scholar] [CrossRef]
- Li, F.; Scott, M.J. CRISPR/Cas9-mediated mutagenesis of the white and Sex lethal loci in the invasive pest, Drosophila suzukii. Biochem. Biophys. Res. Commun. 2016, 469, 911–916. [Google Scholar] [CrossRef] [PubMed]
- Quan, X.; Sato-Miyata, Y.; Tsuda, M.; Muramatsu, K.; Asano, T.; Takeo, S.; Aigaki, T. Deficiency of succinyl-CoA synthetase alpha subunit delays development, impairs locomotor activity and reduces survival under starvation in Drosophila. Biochem. Biophys. Res. Commun. 2017, 483, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Asaoka, T.; Almagro, J.; Ehrhardt, C.; Tsai, I.; Schleiffer, A.; Deszcz, L. Linear ubiquitination by LUBEL has a role in Drosophila heat stress response. EMBO Rep. 2016, 17, 1624–1640. [Google Scholar] [CrossRef] [PubMed]
- Lamb, A.M.; Walker, E.A.; Wittkopp, P.J. Tools and strategies for scarless allele replacement in Drosophila using CRISPR/Cas9. Fly 2016, 11, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Urban, J.A.; Doherty, C.A.; Jordan, W.T., III; Bliss, J.E.; Feng, J.; Soruco, M.M.; Rieder, L.E.; Tsiarli, M.A.; Larschan, E.N. The essential Drosophila CLAMP protein differentially regulates non-coding roX RNAs in male and females. Chromosome Res. 2016, 25, 101–113. [Google Scholar] [CrossRef]
- Chechenova, M.B.; Maes, S.; Oas, S.T.; Nelson, C.; Kiani, K.G.; Bryantsev, A.L. Functional redundancy and nonredundancy between two Troponin C isoforms in Drosophila adult muscles. Mol. Biol. Cell. 2017, 28, 760–770. [Google Scholar] [CrossRef]
- Mendoza-Garcia, P.; Hugosson, F.; Fallah, M.; Higgins, M.L.; Iwasaki, Y.; Pfeifer, K.; Wolfstetter, G.; Varshney, G.; Popichenko, D.; Gergen, J.P.; et al. The Zic family homologueOdd-paired regulates Alk expression in Drosophila. PLoS Genet. 2017, 13, e1006617. [Google Scholar] [CrossRef]
- Knipling, E.F. Possibilities of insect control or eradication through the use of sexually sterile males. J. Econ. Entomol. 1955, 48, 459–462. [Google Scholar] [CrossRef]
- Dyck, V.A.; Hendrichs, J.; Robinson, A.S. (Eds.) Sterile Insect Technique: Principles and Practice in Area-Wide Integrated Pest Management; Springer Science & Business Media: Berlin, Germany, 2006. [Google Scholar]
- Kalajdzic, P.; Schetelig, M.F. CRISPR/Cas-mediated gene editing using purified protein in D rosophilasuzukii. Entomol. Exp. Appl. 2017, 164, 350–362. [Google Scholar] [CrossRef]
- Choo, A.; Crisp, P.; Saint, R.; O’Keefe, L.V.; Baxter, S.W. CRISPR/Cas9-mediated mutagenesis of the white gene in the tephritid pest Bactroceratryoni. J. Appl. Entomol. 2018, 142, 52–58. [Google Scholar] [CrossRef]
- Sim, S.B.; Kauwe, A.N.; Ruano, R.E.; Rendon, P.; Geib, S.M. The ABCs of CRISPR in Tephritidae: Developing methods for inducing heritable mutations in the genera Anastrepha, Bactrocera and Ceratitis. Insect Mol. Biol. 2019, 28, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.M.; Hildebrand, L.; Wimmer, E.A. Improvement and use of CRISPR/Cas9 to engineer a sperm-marking strain for the invasive fruit pest Drosophila suzukii. BMC Biotechnol. 2019, 19, 85. [Google Scholar] [CrossRef]
- Yan, Y.; Ziemek, J.; Schetelig, M.F. CRISPR/Cas9 mediated disruption of the white gene leads to pigmentation deficiency and copulation failure in Drosophila suzukii. J. Insect Physiol. 2020, 126, 104091. [Google Scholar] [CrossRef] [PubMed]
- Stone, A. The Fruitflies of the Genus Anastrepha. USDA Miscellaneous Publications, 1942; p. 439. [Google Scholar]
- Aluja, M. Bionomics and Management of Anastrepha. Annu. Rev. Entomol. 1994, 39, 155–178. [Google Scholar] [CrossRef]
- Weens Jr., H.V.; Heppner, J.B.; Gary, J. Steck. Mexican Fruit Fly (Anastrephaludens); Featured Creatures Entomology & Nematology Department. University of Florida: Gainesville, FL, USA, 2019. [Google Scholar]
- Li, J.; Handler, A.M. CRISPR/Cas9-mediated gene editing in an exogenous transgene and an endogenous sex determination gene in the Caribbean fruit fly, Anastrephasuspensa. Gene 2019, 691, 160–166. [Google Scholar] [CrossRef]
- United States Department of Agriculture. Animal and Plant Health Inspection Service Fruit Fly Exclusion and Detection Program. In Fruit Fly Exclusion and Detection Strategic Plan FY 2019–2023; Washington, DC, USA, 2019; p. 1. [Google Scholar]
- Zhao, S.; Xing, Z.; Liu, Z.; Liu, Y.; Liu, X.; Chen, Z.; Yan, R. Efficient somatic and germline genome engineering of Bactrocera dorsalis by the CRISPR/Cas9 system. Pest Manag. Sci. 2019, 75, 1921–1932. [Google Scholar] [CrossRef] [PubMed]
- Aumann, R.A.; Schetelig, M.F.; Häcker, I. Highly efficient genome editing by homology-directed repair using Cas9 protein in Ceratitis capitata. Insect Mol. Biol. 2018, 101, 85–93. [Google Scholar] [CrossRef]
- Meccariello, A.; Monti, S.M.; Romanelli, A.; Colonna, R.; Primo, P.; Inghilterra, M.G.; Patti, F. Highly efficient DNA-free gene disruption in the agricultural pest Ceratitis capitata by CRISPR-Cas9 ribonucleoprotein complexes. Sci. Rep. 2017, 7, 10061. [Google Scholar] [CrossRef] [PubMed]
- Terenius, O.; Papanicolaou, A.; Garbutt, J.S.; Eleftherianos, I.; Huvenne, H.; Kanginakudru, S.; Bebas, P. RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 2011, 57, 231–245. [Google Scholar] [CrossRef]
- Kolliopoulou, A.; Swevers, L. Recent progress in RNAi research in Lepidoptera: Intracellular machinery, antiviral immune response and prospects for insect pest control. Curr. Opin. Insect Sci. 2014, 6, 28–34. [Google Scholar] [CrossRef]
- Li, J.J.; Shi, Y.; Wu, J.N.; Li, H.; Smagghe, G.; Liu, T.X. CRISPR/Cas9 in lepidopteran insects: Progress, application and prospects. J. Insect Physiol. 2021, 135, 104325. [Google Scholar] [CrossRef] [PubMed]
- Voght, S.P.; Fluegel, M.L.; Andrews, L.A.; Pallanck, L.J. Drosophila NPC1b promotes an early step in sterol absorption from the midgut epithelium. Cell. Metab. 2007, 5, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.C.; Yue, X.R.; Kuang, W.Q.; Li, S.L.; Tang, R.; Zhang, Z.F.; Jing, X. NPC1b as a novel target in controlling the cotton bollworm, Helicoverpa armigera. Pest Manag. Sci. 2020, 76, 2233–2242. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, H.; Wang, H.; Zhao, S.; Zuo, Y.; Yang, Y.; Wu, Y. Functional validation of cadherin as a receptor of Bt toxin Cry1Ac in Helicoverpa armigera utilizing the CRISPR/Cas9 system. Insect Biochem. Mol. Biol. 2016, 76, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Liu, Y.; Ai, D.; Jiang, X.; Dong, S.; Wang, G. A pheromone antagonist regulates optimal mating time in the moth Helicoverpa armigera. Curr. Biol. 2017, 27, 1610–1615. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, H.; Liu, S.; Liu, L.; Tay, W.T.; Walsh, T.K.; Wu, Y. CRISPR/Cas9 mediated genome editing of Helicoverpa armigera with mutations of an ABC transporter gene HaABCA2 confers resistance to Bacillus thuringiensis Cry2A toxins. Insect Biochem. Mol. Biol. 2017, 87, 147–153. [Google Scholar] [CrossRef]
- Khan, S.A.; Reichelt, M.; Heckel, D.G. Functional analysis of the ABCs of eye color in Helicoverpa armigera with CRISPR/Cas9-induced mutations. Sci. Rep. 2017, 7, 40025. [Google Scholar] [CrossRef]
- Wang, H.; Shi, Y.; Wang, L.; Liu, S.; Wu, S.; Yang, Y.; Wu, Y. CYP6AE gene cluster knockout in Helicoverpa armigera reveals role in detoxification of phytochemicals and insecticides. Nat. Commun. 2018, 9, 4820. [Google Scholar] [CrossRef]
- Van Leeuwen, T.; Demaeght, P.; Osborne, E.J.; Dermauw, W.; Gohlke, S.; Nauen, R.; Clark, R.M. Population bulk segregant mapping uncovers resistance mutations and the mode of action of a chitin synthesis inhibitor in arthropods. Proc. Natl. Acad. Sci. USA 2012, 109, 4407–4412. [Google Scholar] [CrossRef]
- Douris, V.; Steinbach, D.; Panteleri, R.; Livadaras, I.; Pickett, J.A.; Van Leeuwen, T.; Vontas, J. Resistance mutation conserved between insects and mites unravels the benzoylurea insecticide mode of action on chitin biosynthesis. Proc. Natl. Acad. Sci. USA 2016, 113, 14692–14697. [Google Scholar]
- Huang, Y.; Chen, Y.; Zeng, B.; Wang, Y.; James, A.A.; Gurr, G.M.; You, M. CRISPR/Cas9 mediated knockout of the abdominal-A homeotic gene in the global pest, diamondback moth (Plutellaxylostella). Insect Biochem. Mol. Biol. 2016, 75, 98–106. [Google Scholar] [CrossRef]
- Wu, K.; Shirk, P.D.; Taylor, C.E.; Furlong, R.B.; Shirk, B.D.; Pinheiro, D.H.; Siegfried, B.D. CRISPR/Cas9 mediated knockout of the abdominal-A homeotic gene in fall armyworm moth (Spodoptera frugiperda). PLoS ONE 2018, 13, e0208647. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Z.; Xu, J.; Li, X.; Bi, H.; Andongma, A.A.; Huang, Y. Mutation of doublesex induces sex-specific sterility of the diamondback moth Plutellaxylostella. Insect Biochem. Mol. Biol. 2019, 112, 103180. [Google Scholar] [CrossRef]
- Bi, H.L.; Xu, J.; Tan, A.J.; Huang, Y.P. CRISPR/Cas9-mediated targeted gene mutagenesis in Spodoptera litura. Insect Sci. 2016, 23, 469–477. [Google Scholar] [CrossRef]
- Zhu, G.H.; Peng, Y.C.; Zheng, M.Y.; Zhang, X.Q.; Sun, J.B.; Huang, Y.; Dong, S.L. CRISPR/Cas9 mediated BLOS2 knockout resulting in disappearance of yellow strips and white spots on the larval integument in Spodoptera litura. J. Insect Physiol. 2017, 103, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Vogt, R.G.; Riddiford, L.M. Pheromone binding and inactivation by moth antennae. Nature 1981, 293, 161–163. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.H.; Xu, J.; Cui, Z.; Dong, X.T.; Ye, Z.F.; Niu, D.J.; Dong, S.L. Functional characterization of SlitPBP3 in Spodoptera litura by CRISPR/Cas9 mediated genome editing. Insect Biochem. Mol. Biol. 2016, 75, 1–9. [Google Scholar] [CrossRef]
- Koutroumpa, F.A.; Monsempes, C.; François, M.C.; De Cian, A.; Royer, C.; Concordet, J.P.; Jacquin-Joly, E. Heritable genome editing with CRISPR/Cas9 induces anosmia in a crop pest moth. Sci. Rep. 2016, 6, 29620. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Xue, Y.; Lu, W.; Ma, H.; Chen, M.; Wu, Y.; Hu, Z. Functional validation of nicotinic acetylcholine receptor (nAChR) α6 as a target of spinosyns in Spodoptera exiguautilizing the CRISPR/Cas9 system. Pest Manag. Sci. 2020, 76, 2415–2422. [Google Scholar] [CrossRef]
- Liu, H.; Liu, Q.; Zhou, X.; Huang, Y.; Zhang, Z. Genome editing of Wnt-1, a gene associated with segmentation, via CRISPR/Cas9 in the pine caterpillar moth, Dendrolimus punctatus Front. Physiol. 2017, 7, 666. [Google Scholar] [CrossRef] [PubMed]
- Garczynski, S.F.; Martin, J.A.; Griset, M.; Willett, L.S.; Cooper, W.R.; Swisher, K.D.; Unruh, T.R. CRISPR/Cas9 editing of the codling moth (Lepidoptera: Tortricidae) CpomOR1 gene affects egg production and viability. J. Econ. Entomol. 2017, 110, 1847–1855. [Google Scholar] [CrossRef] [PubMed]
- You, L.; Bi, H.L.; Wang, Y.H.; Li, X.W.; Chen, X.E.; Li, Z.Q. CRISPR/Cas9-based mutation reveals Argonaute 1 is essential for pigmentation in Ostriniafurnacalis. Insect Sci. 2019, 26, 1020–1028. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, Y.H.; Chen, X.E.; Tian, D.; Xu, X.; Li, K.; He, L. CRISPR/Cas9-mediated Tyrosine hydroxylase knockout resulting in larval lethality in Agrotisipsilon. Insect Sci. 2018, 25, 1017–1024. [Google Scholar] [CrossRef]
- Li, X.; Liu, Q.; Liu, H.; Bi, H.; Wang, Y.; Chen, X.; Chen, H. Mutation of doublesex in Hyphantria cunea results in sex-specific sterility. Pest Manag. Sci. 2020, 76, 1673–1682. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Li, S.; Liang, J.; Yi, H.; Jing, X.; Liu, T.X. Optimization of the application of the CRISPR/Cas9 system in Mythimna separata. Entomol. Exp. Appl. 2022. [Google Scholar] [CrossRef]
- Xue, W.H.; Xu, N.; Yuan, X.B.; Chen, H.H.; Zhang, J.L.; Fu, S.J.; Xu, H.J. CRISPR/Cas9-mediated knockout of two eye pigmentation genes in the brown planthopper, Nilaparvatalugens (Hemiptera: Delphacidae). Insect Biochem. Mol. Biol. 2018, 93, 19–26. [Google Scholar] [CrossRef]
- Hunter, W.B.; Gonzalez, M.T.; Tomich, J. BAPC-assisted CRISPR/Cas9 System: Targeted Delivery into Adult Ovaries for Heritable Germline Gene Editing (Arthropoda: Hemiptera). BioRxiv 2018, 478743. [Google Scholar] [CrossRef]
- Hunter, W.B.; Gonzalez, M.T.; Tomich, J. BAPC-assisted-CRISPR-Cas9 Delivery into Nymphs and Adults for Heritable Gene Editing (Hemiptera). FASEB J. 2019, 33 (Suppl. 1), 626.2. [Google Scholar] [CrossRef]
- Heu, C.C.; McCullough, F.M.; Luan, J.; Rasgon, J.L. CRISPR-Cas9-Based Genome Editing in the Silverleaf Whitefly (Bemisiatabaci). CRISPR J. 2020, 3, 89–96. [Google Scholar] [CrossRef]
- Cagliari, D.; Smagghe, G.; Zotti, M.; Taning, C.N.T. RNAi and CRISPR/Cas9 as Functional Genomics Tools in the Neotropical Stink Bug, Euschistusheros. Insects 2020, 11, 838. [Google Scholar] [CrossRef]
- Richards, S.; Gibbs, R.A.; Weinstock, G.M.; Brown, S.J.; Denell, R.; Beeman, R.W.; Sattelle, D.B. The genome of the model beetle and pest Tribolium castaneum. Nature 2008, 452, 949–955. [Google Scholar]
- Kim, K.H.; Jeong, J.Y.; Surh, Y.J.; Kim, K.W. Expression of stress-response ATF3 is mediated by Nrf2 in astrocytes. Nucleic Acids Res. 2010, 38, 48–59. [Google Scholar] [CrossRef]
- Pavlopoulos, A.; Berghammer, A.J.; Averof, M.; Klingler, M. Efficient transformation of the beetle Triboliumcastaneum using the Minos transposable element: Quantitative and qualitative analysis of genomic integration events. Genetic 2004, 167, 737–746. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gilles, A.F.; Schinko, J.B.; Averof, M. Efficient CRISPR-mediated gene targeting and transgene replacement in the beetle Triboliumcastaneum. Development 2015, 142, 2832–2839. [Google Scholar]
- Berghammer, A.J.; Weber, M.; Trauner, J.; Klingler, M. Red flour beetle (Tribolium) germline transformation and insertional mutagenesis. Cold Spring Harb. Protoc. 2009, 2009, pdb-prot5259. [Google Scholar] [CrossRef] [PubMed]
- Gui, S.; Taning, C.N.T.; Wei, D.; Smagghe, G. First report on CRISPR/Cas9-targeted mutagenesis in the Colorado potato beetle, Leptinotarsa decemlineata. J. Insect Physiol. 2020, 121, 104013. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Chen, Q.; Wei, Y.; Jiang, F.; Yang, M.; Hao, S.; Kang, L. MicroRNA-276 promotes egg-hatching synchrony by up-regulating brm in locusts. Proc. Natl. Acad. Sci. USA 2016, 113, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.; Chen, D.; Yang, P.; Jiang, F.; Wang, X.; Kang, L. CRISPR/Cas9 in locusts: Successful establishment of an olfactory deficiency line by targeting the mutagenesis of an odorant receptor co-receptor (Orco). Insect Biochem. Mol. Biol. 2016, 79, 27–35. [Google Scholar]
- Bajda, S.; Dermauw, W.; Panteleri, R.; Sugimoto, N.; Douris, V.; Tirry, L. A mutation in the PSST homologue of complex I (NADH: Ubiquinone oxidoreductase) from Tetranychusurticae is associated with resistance to METI acaricides. Insect Biochem. Mol. Biol. 2017, 80, 79–90. [Google Scholar] [CrossRef]
- Dermauw, W.; Jonckheere, W.; Riga, M.; Livadaras, I.; Vontas, J.; Van Leeuwen, T. Targeted mutagenesis using CRISPR-Cas9 in the chelicerate herbivore Tetranychusurticae. Insect Biochem. Mol. Bio. 2020, 120, 103347. [Google Scholar] [CrossRef]
- Ren, X.; Yang, Z.; Xu, J.; Sun, J.; Mao, D.; Hu, Y.; Yang, S.J.; Qiao, H.H.; Wang, X.; Hu, Q.; et al. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell. Rep. 2014, 9, 1151–1162. [Google Scholar] [CrossRef]
- Port, F.; Bullock, S.L. Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat. Methods 2016, 13, 852–854. [Google Scholar] [CrossRef]
- Mabashi-Asazuma, H.; Sohn, B.H.; Kim, Y.S.; Kuo, C.W.; Khoo, K.H.; Kucharski, C.A.; Jarvis, D.L. Targeted glycoengineering extends the protein N-glycosylation pathway in the silkworm silk gland. Insect Biochem. Mol. Biol. 2015, 65, 20–27. [Google Scholar] [CrossRef][Green Version]
- Gao, J.L.; Fan, Y.J.; Wang, X.Y.; Zhang, Y.; Pu, J.; Li, L.; Xu, Y.Z. A conserved intronic U1 snRNP-binding sequence promotes trans-splicing in Drosophila. Genes Dev. 2015, 29, 760–771. [Google Scholar] [CrossRef] [PubMed]
- Kunzelmann, S.; Böttcher, R.; Schmidts, I.; Förstemann, K. A comprehensive toolbox for genome editing in cultured Drosophila melanogaster cells. G3 2016, 6, 1777–1785. [Google Scholar] [CrossRef]
- Tanaka, R.; Murakami, H.; Ote, M.; Yamamoto, D. Clustered regulatory interspaced short palindromic repeats (CRISPR)-mediated mutagenesis and phenotype rescue by piggyBac transgenesis in a nonmodel Drosophila species. Insect Mol. Biol. 2016, 25, 355–361. [Google Scholar] [CrossRef]
- Chen, E.H.; Hou, Q.L. Identification and expression analysis of cuticular protein genes in the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Pestic. Biochem. Physiol. 2021, 178, 104943. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.H.; Chereddy, S.C.; Howell, J.L.; Palli, S.R. Genome editing in the fall armyworm, Spodoptera frugiperda: Multiple sgRNA/Cas9 method for identification of knockouts in one generation. Insect Biochem. Mol. Biol. 2020, 122, 103373. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, S.; Rahangdale, S.; Pandita, S.; Saxena, G.; Upadhyay, S.K.; Mishra, G.; Verma, P.C. CRISPR/Cas9 for Insect Pests Management: A Comprehensive Review of Advances and Applications. Agriculture 2022, 12, 1896. https://doi.org/10.3390/agriculture12111896
Singh S, Rahangdale S, Pandita S, Saxena G, Upadhyay SK, Mishra G, Verma PC. CRISPR/Cas9 for Insect Pests Management: A Comprehensive Review of Advances and Applications. Agriculture. 2022; 12(11):1896. https://doi.org/10.3390/agriculture12111896
Chicago/Turabian StyleSingh, Sanchita, Somnath Rahangdale, Shivali Pandita, Gauri Saxena, Santosh Kumar Upadhyay, Geetanjali Mishra, and Praveen C. Verma. 2022. "CRISPR/Cas9 for Insect Pests Management: A Comprehensive Review of Advances and Applications" Agriculture 12, no. 11: 1896. https://doi.org/10.3390/agriculture12111896
APA StyleSingh, S., Rahangdale, S., Pandita, S., Saxena, G., Upadhyay, S. K., Mishra, G., & Verma, P. C. (2022). CRISPR/Cas9 for Insect Pests Management: A Comprehensive Review of Advances and Applications. Agriculture, 12(11), 1896. https://doi.org/10.3390/agriculture12111896







