Transcriptome Integrated with Metabolome Reveals the Molecular Mechanism of Phytoplasma Cherry Phyllody Disease on Stiff Fruit in Chinese Cherry (Cerasus pseudocerasus L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Total RNA Extraction, Expression Analysis of Genes with RNA-Seq
2.3. Metabolome Analysis
2.3.1. Metabolites Extraction
2.3.2. UPLC-MS Examination
2.3.3. Metabolomics Analysis
2.3.4. Quantification of Phytohormones
2.4. cDNA Synthesis, and Quantitative Real-Time PCR
3. Results
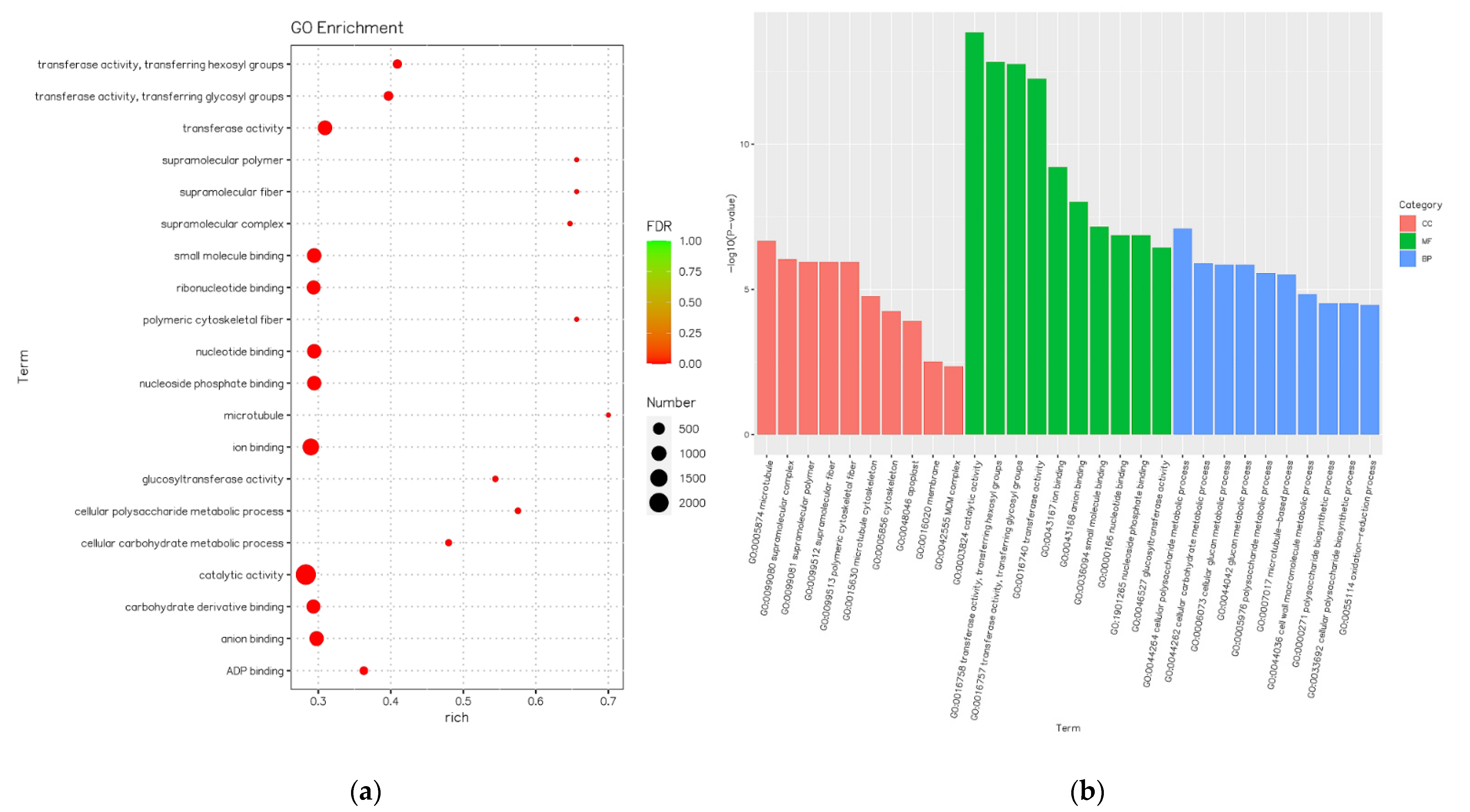
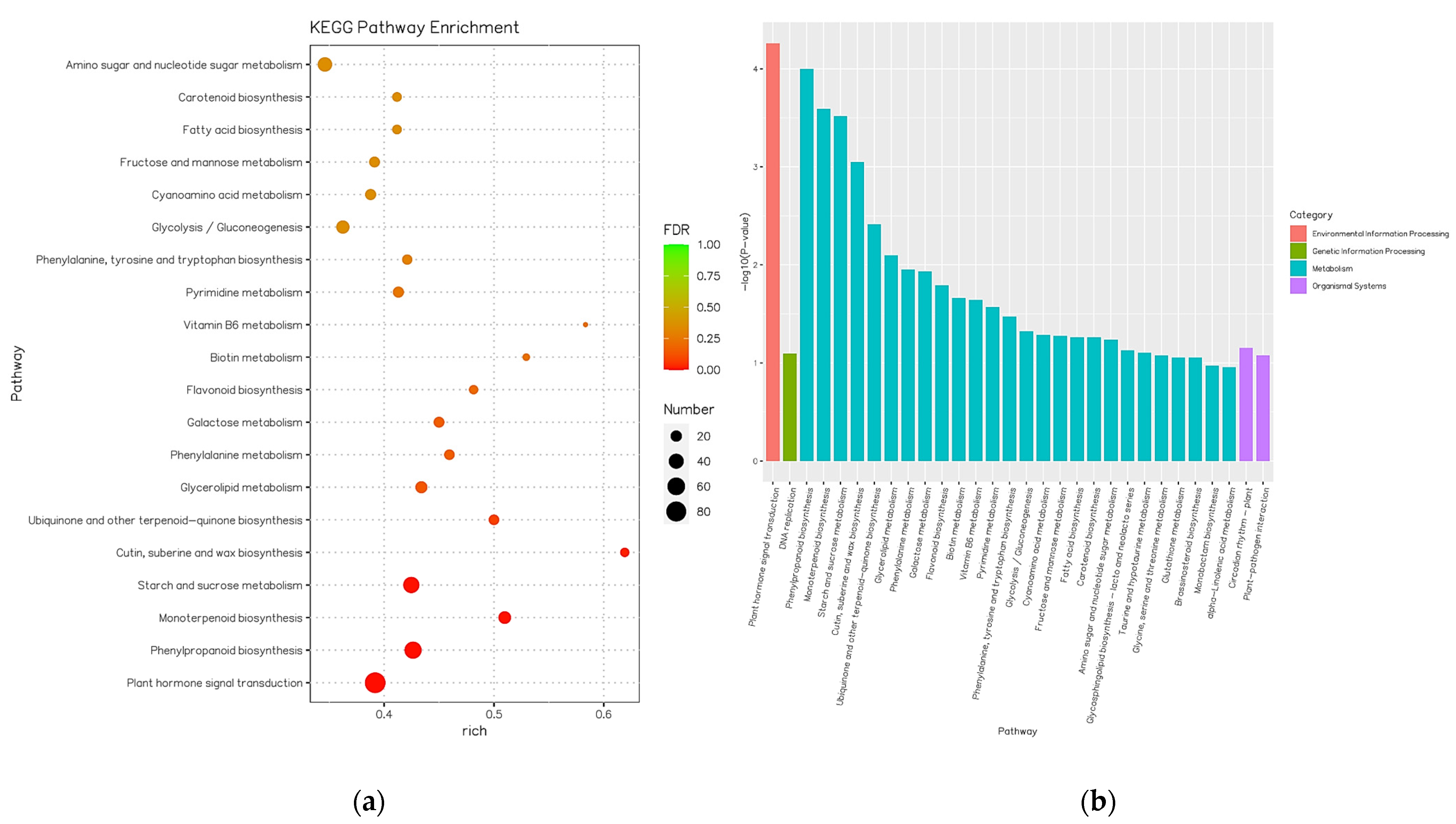
3.1. GO Enrichment and KEEG Pathway Enrichment Analysis of DEGs
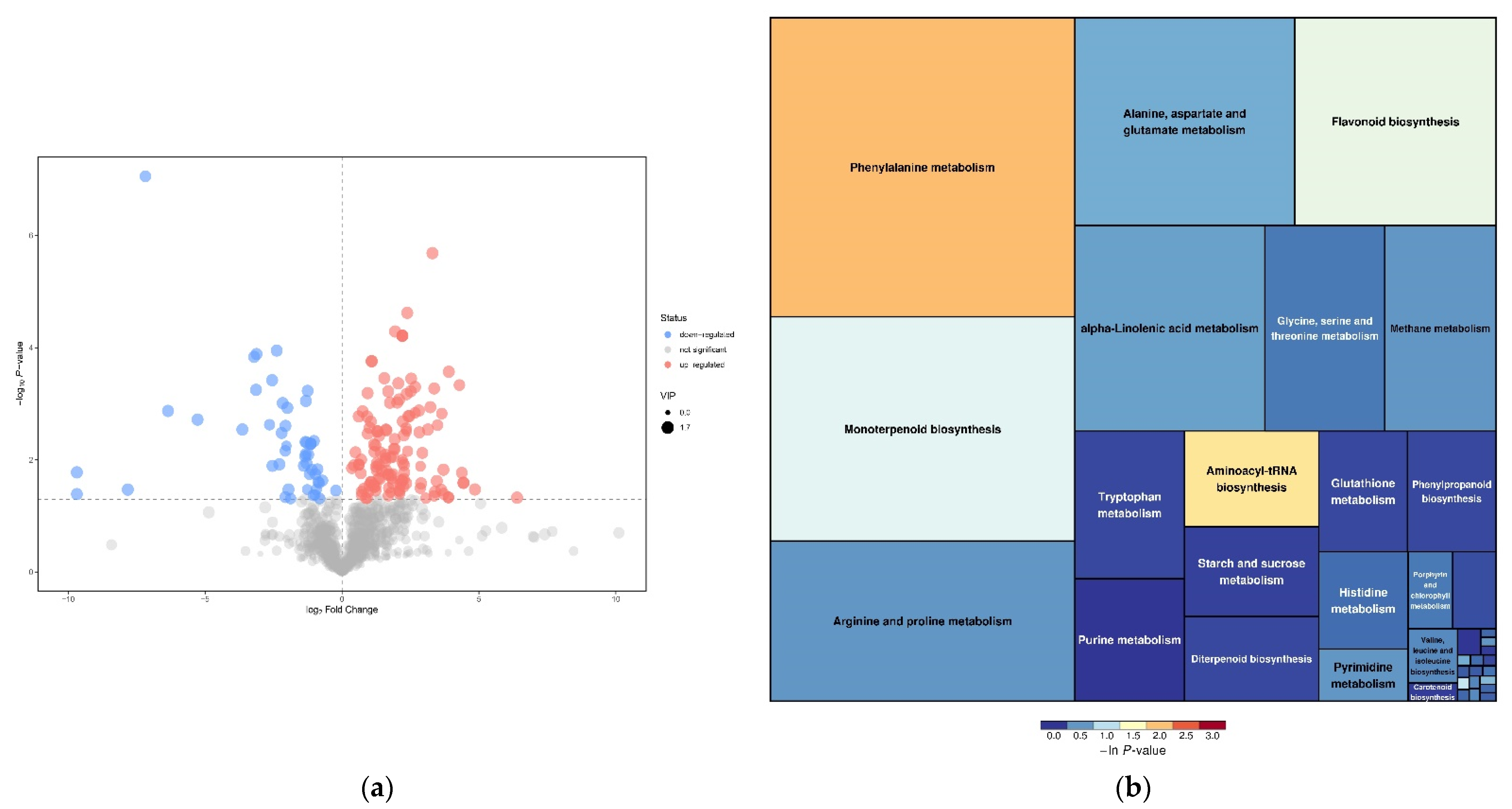
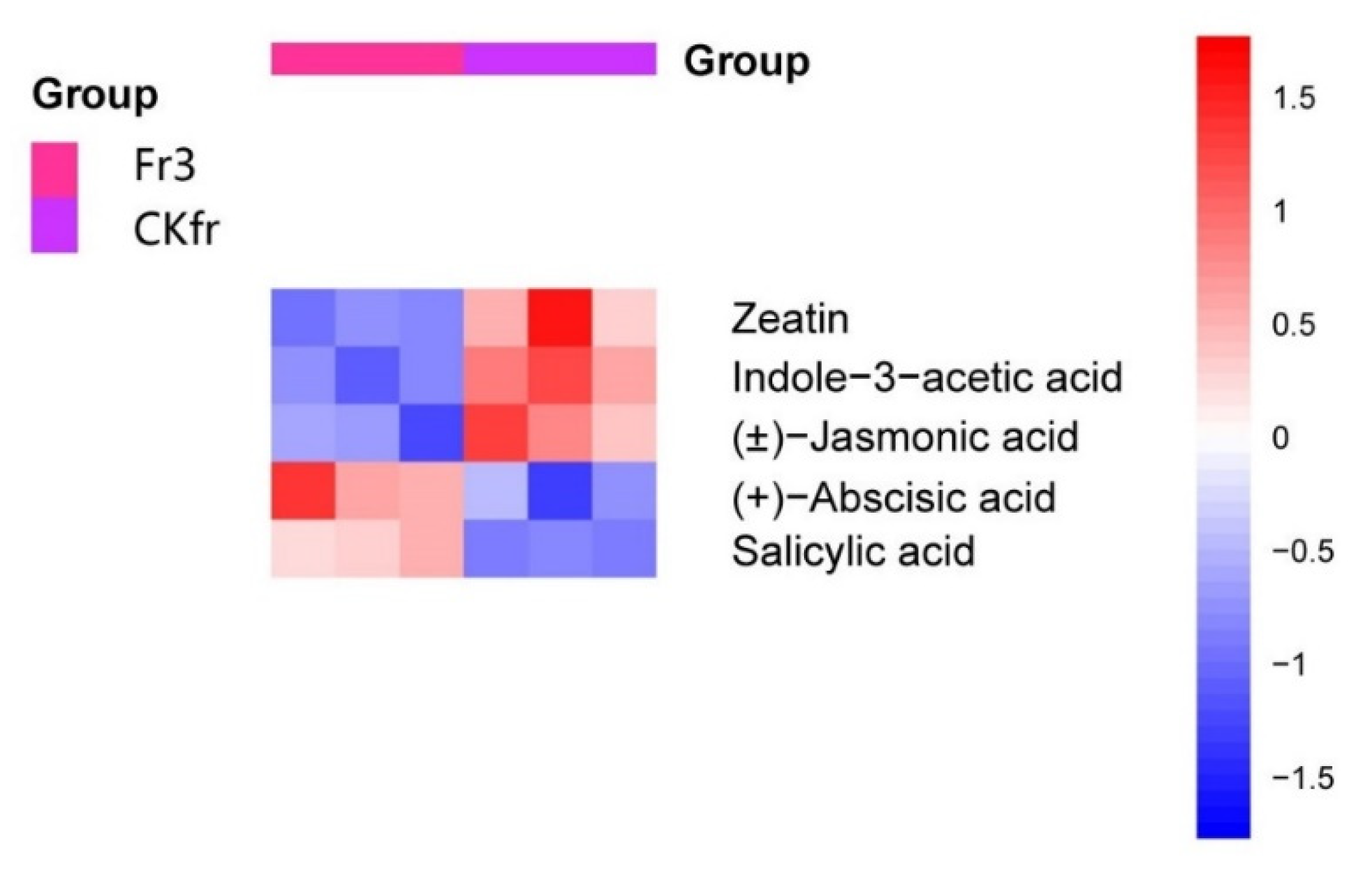
3.2. Differentially Accumulated Metabolites Analysis
3.3. Validation of RNA-Seq Data by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Granett, A.L.; Gilmer, R.M. Mycoplasmas associated with X- disease in various Prunus species. Phytopathology 1971, 61, 1036–1037. [Google Scholar] [CrossRef]
- Wang, J.W.; Zhu, S.F.; Liu, Q.; Davis, R.E.; Zhao, Y. First report of sweet cherry virescence disease in China and its association with infection by a ‘Candidatus Phytoplasma ziziphi’-related phytoplasma. Plant Dis. 2014, 98, 419. [Google Scholar] [CrossRef]
- Wang, J.; Ai, C.X.; Yu, X.M.; Zhang, K.P.; Gao, R.; Li, X.D. First report of ‘Candidatus phytoplasma asteris’ related strain associated with sweet cherry fasciation disease in China. Plant Dis. 2018, 102, 237. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Q.; Wei, W.; Davis, R.E.; Tan, Y.; Lee, I.M.; Zhu, D.; Wei, H.; Zhao, Y. Multilocus genotyping identifies a highly homogeneous phytoplasma lineage associated with sweet cherry virescence disease in China and its carriage by an erythroneurine leafhopper. Crop Prot. 2018, 106, 13–22. [Google Scholar] [CrossRef]
- Gao, R.; Yang, S.K.; Wang, J.; Lu, X.B.; Sun, Y.G.; Tian, Y.P.; Wang, W.X. Molecular detection and identification of subgroup 16SrV-B phytoplasma associated with Chinese cherry phyllody disease in China. Acta Sci. Hortic. 2019, 46, 1249–1256. (In Chinese) [Google Scholar] [CrossRef]
- Ermacora, P.; Ruggero, O. Symptoms of Phytoplasma Diseases. In Methods in Molecular Biology; Musetti, R., Pagliari, L., Eds.; Springer Nature, LLC.: Berlin/Heidelberg, Germany, 2019; Volume 1875, pp. 53–67. [Google Scholar] [CrossRef]
- MacLean, A.M.; Sugio, A.; Makarova, O.V.; Findlay, K.C.; Grieve, V.M.; To’th, R.; Nicolaisen, M.; Hogenhout, S.A. Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol. 2011, 157, 831–841. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.T.; Li, M.Y.; Cheng, K.T.; Tan, C.M.; Su, L.W.; Lin, W.Y.; Shih, H.T.; Chiou, T.J.; Yang, J.Y. Transgenic plants that express the phytoplasma effector SAP11 show altered phosphate starvation and defense responses. Plant Physiol. 2014, 164, 1456–1469. [Google Scholar] [CrossRef] [Green Version]
- Minato, N.; Himeno, M.; Hoshi, A.; Maejima, K.; Komatsu, K.; Takebayashi, Y.; Kasahara, H.; Yusa, A.; Yamaji, Y.; Oshima, K.; et al. Thephytoplasmal virulence factor TENGU causes plant sterility bydownregulating of the jasmonic acid and auxin pathways. Sci. Rep. 2014, 4, 7399. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.J.; MacLean, A.M.; Sugio, A.; Maqbool, A.; Busscher, M.; Cho, S.T.; Kanoun, S.; Kuo, C.H.; Immink, R.G.H.; Hogenhout, S.A. Parasitic modulation of host development by ubiquitin-independent protein degradation. Cell 2021, 184, 5201–5214.e1. [Google Scholar] [CrossRef]
- Janik, K.; Mithöfer, A.; Raffeiner, M.; Stellmach, H.; Hause, B.; Schlink, K. An effector of apple proliferation phytoplasma targets TCP transcription factors—a generalized virulence strategy of phytoplasma? Mol. Plant Pathol. 2017, 18, 435–442. [Google Scholar] [CrossRef]
- Wang, N.; Yang, H.; Yin, Z.; Liu, W.; Sun, L.; Wu, Y. Phytoplasma effector SWP1 induces witches’ broom symptom by destabilizing the TCP transcription factor BRANCHED1. Mol. Plant Pathol. 2018, 19, 2623–2634. [Google Scholar] [CrossRef]
- Zhou, J.; Ma, F.; Yao, Y.; Deng, M.; Chen, M.; Zhang, S.; Li, Y.; Yang, J.; Zhang, N.; Huang, J.; et al. JWB phytoplasma effectors SJP1 and SJP2 induce lateral bud outgrowth by repressing the ZjBRC1-controlled auxin efflux channel. Plant Cell Environ. 2021, 44, 3257–3272. [Google Scholar] [CrossRef]
- Sun, Y.L.; Detchemendy, T.W.; Pajerowska-Mukhtar, K.M.; Mukhtar, M.S. NPR1 in JazzSet with Pathogen Effectors. Trends Plant Sci. 2018, 23, 469–472. [Google Scholar] [CrossRef]
- Hagen, G.; Guilfoyle, T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef]
- Li, S.B.; Xie, Z.Z.; Hu, C.G.; Zhang, J.Z. A Review of Auxin Response Factors (ARFs) in Plants. Front. Plant Sci. 2016, 7, 47. [Google Scholar] [CrossRef] [Green Version]
- Hong, R.; William, M.G. SAUR Proteins as Effectors of Hormonal and Environmental Signals in Plant Growth. Mol. Plant 2015, 8, 1153–1164. [Google Scholar] [CrossRef] [Green Version]
- Guilfoyle, T.J.; Ulmasov, T.; Hagen, G. The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cell. Mol. Life Sci. 1998, 54, 619–627. [Google Scholar] [CrossRef]
- Sarbottam, P.; Shrestha, S.K.; Brad, B.; Stewart, N.C.; Hewezi, T. Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front. Plant Sci. 2014, 5, 744. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 2003, 15, 533–543. [Google Scholar] [CrossRef] [Green Version]
- Kumar, N.; Tokas, J.; Raghavendra, M.; Singal, H.R. Impact of exogenous salicylic acid treatment on the cell wall metabolism and ripening process in postharvest tomato fruit stored at ambient temperature. Int. J. Food Sci. Technol. 2021, 56, 2961–2972. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, T.; Wang, Y.; Chen, Q.; Sun, B.; Luo, Y.; Zhang, Y.; Tang, H.R.; Wang, X.R. Genetic Diversity and Domestication Footprints of Chinese Cherry [Cerasus pseudocerasus (Lindl.) G.Don] as Revealed by Nuclear Microsatellites. Front. Plant Sci. 2018, 9, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcion, C.; Lohmann, A.; Lamodiere, E.; Catinot, J.; Buchala, A.; Doermann, P.; Metraux, J.-P. Characterization and Biological Function of the ISOCHORISMATE SYNTHASE2 Gene of Arabidopsis. Plant Physiol. 2008, 147, 1279–1287. [Google Scholar] [CrossRef] [Green Version]
- Shadle, G.L.; Wesley, S.V.; Korth, K.L.; Chen, F.; Lamb, C.; Dixon, R.-A. Phenylpropanoid compounds and disease resistance in transgenic tobacco with altered expression of l-phenylalanine ammonia-lyase. Phytochemistry 2003, 64, 153–161. [Google Scholar] [CrossRef] [Green Version]
- Napoleo, T.A.; Soares, G.; Vital, C.E.; Bastos, C.; Castro, R.; Loureiro, M.E.; Giordano, A. Methyl jasmonate and salicylic acid are able to modify cell wall but only salicylic acid alters biomass digestibility in the model grass brachypodium distachyon. Plant Sci. 2017, 263, 46. [Google Scholar] [CrossRef]
- Jeandet, P.; Clément, C.; Courot, E.; Cordelier, S. Modulation of Phytoalexin Biosynthesis in Engineered Plants for Disease Resistance. Int. J. Mol. Sci. 2013, 14, 14136–14170. [Google Scholar] [CrossRef] [Green Version]
- Zanardo, D.I.L.; Lima, R.B.; Ferrarese, M.D.L.; Bubna, G.A.; Ferrarese, O. Soybean root growth inhibition and lignification induced by p-coumaric acid. Environ. Exp. Bot. 2009, 66, 25–30. [Google Scholar] [CrossRef]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D.G. A plant miRNA contributes to antibacterial resistance by repressing auxin signalling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef] [Green Version]
- Ferrari, S.; Galletti, R.; Pontiggia, D.; Manfredini, C.; Lionetti, V.; Bellincampi, D.; Cervone, F.; Lorenzo, G.D. Transgenic Expression of a Fungal endo-Polygalacturonase Increases Plant Resistance to Pathogens and Reduces Auxin Sensitivity. Plant Physiol. 2008, 146, 323–324. [Google Scholar] [CrossRef] [Green Version]
- Ludwig-Müller, J. Auxin conjugation: Growing out of the shade. Nat. Plants 2016, 2, 16044. [Google Scholar] [CrossRef]
- Zou, X.P.; Long, J.H.; Zhao, K.; Peng, A.H.; Chen, M.; Qin, L.; Chen, Y.R.; Chen, S.C.; Liu, J.H. Overexpressing GH3.1 and GH3.1L reduces susceptibility to xanthomonas citri subsp. citri by repressing auxin signalling in citrus (citrus sinensis osbeck). PLoS ONE 2019, 14, e0220017. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Jones, B.; Li, Z.G.; Frasse, P.; Delalande, C.; Regad, F.; Chaabouni, S.; Latche’, A.; Pech, J.C.; Bouzayen, M. The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 2005, 17, 2676–2692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Audran-Delalande, C.; Bassa, C.; Mila, I.; Regad, F.; Zouine, M.; Bouzayen, M. Genome-Wide Identification, Functional Analysis and Expression Profiling of the Aux/IAA Gene Family in Tomato. Plant Cell Physiol. 2012, 53, 659–672. [Google Scholar] [CrossRef] [Green Version]
- Bassa, C.; Mila, I.; Bouzayen, M.; Audran-Delalande, C. Phenotypes Associated with Down-Regulation of Sl-IAA27 Support Functional Diversity Among Aux/IAA Family Members in Tomato. Plant Cell Physiol. 2012, 53, 1583–1595. [Google Scholar] [CrossRef] [Green Version]
- Guillotin, B.; Etemadi, M.; Audran, C.; Bouzayen, M.; Becard, G.; Combier, J.P. Sl-IAA27 regulates strigolactone biosynthesis and mycorrhization in tomato (var. MicroTom). N. Phytol. 2017, 213, 1124–1132. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.M.; Wang, C.; He, L.G.; Wang, Z.J.; Tong, Z.; Song, F.; Tu, J.F.; Qiu, W.M.; Liu, J.H.; Jiang, Y.C.; et al. Transcriptome analysis unravels metabolic and molecular pathways related to fruit sac granulation in a late-ripening navel orange (citrus sinensis osbeck). Plants 2020, 9, 95. [Google Scholar] [CrossRef] [Green Version]
- De Jong, M.; Wolters-Arts, M.; Schimmel, B.-C.J.; Stultiens, C.L.M.; de Groot, P.-F.-M.; Powers, S.J.; Tikunov, Y.M.; Bovy, A.G.; Mariani, C.; Vriezen, W.H.; et al. Solanum lycopersicum AUXIN RESPONSE FACTOR 9 regulates cell division activity during early tomato fruit development. J. Exp. Bot. 2015, 66, 3405–3416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Dinh, T.T.; Li, D.; Shi, B.; Li, Y.; Cao, X.; Guo, L.; Pan, Y.; Jiao, Y.; Chen, X. AUXIN RESPONSEFACTOR 3 integrates the functions of AGAMOUS and APETALA2 in floral meristem determinacy. Plant J. 2014, 80, 629–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krogan, N.T.; Yin, X.; Ckurshumova, W.; Berleth, T. Distinct subclades of Aux/IAA genes are direct targets of ARF5/MP transcriptional regulation. N. Phytol. 2014, 204, 474–483. [Google Scholar] [CrossRef]
- Weijers, D.; Jürgens, G. Auxin and embryo axis formation: The ends in sight? Curr. Opin. Plant Biol. 2005, 8, 32–37. [Google Scholar] [CrossRef]
- Rinaldi, M.A.; Liu, J.; Enders, T.A.; Bartel, B.; Strader, L.C. A gain-of-function mutation in iaa16 confers reduced responses to auxin and abscisic acid and impedes plant growth and fertility. Plant Mol. Biol. 2012, 79, 359–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reed, J.W.; Wu, M.F.; Reeves, P.H.; Hodgens, C.; Yadav, V.; Hayes, S.; Pierik, R. Three auxin response factors promote hypocotyl elongation. Plant Physiol. 2018, 178, 864–875. [Google Scholar] [CrossRef] [Green Version]
- Sato, A.; Yamamoto, K.T. Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol. Plant. 2008, 133, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Domingo, C.; Andrés, F.; Tharreau, D.; Iglesias, D.J.; Talón, M. Constitutive expression of osgh3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol. Plant Microbe Interact. 2009, 22, 201–210. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Stone, J.M. Arabidopsis thaliana GH3.9 influences primary root growth. Planta 2007, 226, 21–34. [Google Scholar] [CrossRef] [Green Version]
- Takase, T.; Nakazawa, M.; Ishikawa, A.; Manabe, K.; Matsui, M. DFL2, a New Member of the Arabidopsis GH3 Gene Family, is Involved in Red Light-Specific Hypocotyl Elongation. Plant Cell Physiol. 2003, 44, 1071–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Sun, N.; Zhang, F.; Yu, R.; Wei, N. Differential regulation of Arabidopsis PP2C-D1 by SAUR17 and SAUR50 in apical hook development and cotyledon opening. Plant Cell 2020, 32, 3792–3811. [Google Scholar] [CrossRef]
- Zeng, Y.; Zhao, T.; Kermode, A.R. A conifer ABI3-interacting protein plays important roles during key transitions of the plant life cycle. Plant Physiol. 2012, 161, 179–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, T.; Qi, M.; Ding, X.; Zheng, Y.; Wang, J. The SAUR41 subfamily of SMALL AUXIN UP RNA genes is abscisic acid-inducible to modulate cell expansion and salt tolerance in Arabidopsis thaliana seedlings. Ann. Bot. 2019, 125, 805–819. [Google Scholar] [CrossRef] [Green Version]
- He, Y.J.; Liu, Y.; Li, M.Z.; Lamin-Samu, A.T.; Yang, D.-D.; Yu, X.L.; Izhar, M.; Jan, I.; Ali, M.; Lu, G. The Arabidopsis SMALL AUXIN UP RNA32 Protein Regulates ABA-Mediated Responses to Drought Stress. Front. Plant Sci. 2021, 12, 259. [Google Scholar] [CrossRef]
- Mambro, D.R.; Svolacchia, N.; Dello Ioio, R.; Pierdonati, E.; Salvi, E.; Pedrazzini, E.; Vitale, A.; Perilli, S.; Sozzani, R.; Benfey, P.-N.; et al. The Lateral Root Cap Acts as an Auxin Sink that Controls Meristem Size. Curr. Biol. 2019, 29, 1199–1205.e4. [Google Scholar] [CrossRef] [Green Version]
- Spartz, A.K.; Sang, H.L.; Wenger, J.P.; Gonzalez, N.; Itoh, H.; Inzé, D.; Peer, W.A.; Murphy, A.S.; Overvoorde, P.J.; Gray, W.M. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012, 70, 978–990. [Google Scholar] [CrossRef] [Green Version]
- Overvoorde, P.J.; Okushima, Y.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Liu, A.; Onodera, C.; Quach, H.; et al. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 2005, 17, 3282–3300. [Google Scholar] [CrossRef] [Green Version]
- Cao, F.Y.; Yoshioka, K.; Desveaux, D. The roles of ABA in plant–pathogen interactions. J. Plant. Res. 2011, 124, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; Kobayashi, M.; et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009, 50, 1345–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, M.-J.; Zhang, Y.L.; Liu, X.; Huang, H.; Zhou, X.E.; Wang, W.L.; Zeng, A.; Zhao, C.Z.; Si, T.; Du, J.M.; et al. Combining chemical and genetic approaches to increase drought resistance in plants. Nat. Commun. 2017, 8, 1183. [Google Scholar] [CrossRef]
- Ji, H.L.; Kyndt, T.; He, W.; Vanholme, B.; Gheysen, G. β-Aminobutyric Acid–Induced Resistance Against Root-Knot Nematodes in Rice Is Based on Increased Basal Defense. Mol. Plant Microbe Interact. 2015, 28, 519–533. [Google Scholar] [CrossRef] [Green Version]
- Tran, L.S.P.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of ahk1/athk1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 20623–20628. [Google Scholar] [CrossRef] [Green Version]
- De Torres-Zabala, M.; Truman, W.; Bennett, M.; Lafforgue, G.; Mansfield, J.; Egea, P.; Bögre, L.; Grant, M. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 2007, 26, 1434–1443. [Google Scholar] [CrossRef] [PubMed]
- Galstyan, A.; Nemhauser, J.L. Auxin promotion of seedling growth via ARF5 is dependent on the brassinosteroid-regulated transcription factors BES1 and BEH4. Plant Direct 2019, 3, e0016. [Google Scholar] [CrossRef] [Green Version]


| Gene | log2FC(FR3/CKfr) | −∆∆Ct | Relative Expression |
|---|---|---|---|
| Pav_sc0000998.1_g200.1.mk (IAA4) | 2.31 | 3.73 | 13.29 |
| Pav_sc0002327.1_g560.1.mk (IAA9) | 1.53 | 3.36 | 10.25 |
| Pav_sc0000716.1_g230.1.mk (IAA17) | 1.13 | 2.26 | 4.78 |
| Pav_sc0000478.1_g250.1.mk (IAA31) | 2.49 | 3.66 | 12.62 |
| Pav_sc0000752.1_g190.1.br (SAUR20) | / | 3.82 | 14.17 |
| Pav_sc0000586.1_g800.1.mk (SAUR32) | 2.66 | 4.34 | 20.21 |
| Pav_sc0001583.1_g270.1.br (SAUR72) | 1.52 | 2.92 | 7.55 |
| Pav_sc0004359.1_g050.1.br (ARF5) | 4.92 | 3.06 | 8.31 |
| Pav_sc0002250.1_g170.1.mk (ARF9) | 1.44 | 4.82 | 28.27 |
| Pav_sc0000049.1_g070.1.mk (GH3.1) | 1.50 | 2.29 | 4.90 |
| Pav_sc0001540.1_g090.1.mk (GH3.17) | 4.81 | / | / |
| Pav_sc0000554.1_g390.1.mk (PP2CA) | 1.46 | 2.22 | 4.66 |
| Pav_sc0000848.1_g040.1.mk (AHP2) | 2.60 | 2.54 | 5.80 |
| Pav_sc0000157.1_g130.1.mk (AHK2) | 1.49 | 5.15 | 35.45 |
| Pav_sc0001335.1_g500.1.mk (PYL8) | 1.26 | 1.90 | 3.73 |
| Pav_sc0001242.1_g120.1.mk (SnRK2.3) | 1.29 | 3.09 | 8.53 |
| Pav_sc0008321.1_g030.1.mk (TGA10) | 4.19 | 4.94 | 30.72 |
| Pav_sc0004469.1_g010.1.br (ARG7) | 3.79 | 4.20 | 18.36 |
| Pav_sc0000567.1_g900.1.mk (PAN) | 2.29 | 3.72 | 13.19 |
| Pav_sc0000568.1_g810.1.br (PR1a) | 4.94 | / | / |
| Pav_co4011651.1_g010.1.br (PRB1) | 3.28 | 3.44 | 10.85 |
| Pav_sc0006499.1_g050.1.mk (MYC2) | 1.96 | 3.20 | 9.21 |
| Pav_sc0002181.1_g160.1.mk (IAA11) | −2.78 | / | / |
| Pav_sc0000545.1_g110.1.mk (IAA12) | −5.76 | −3.99 | 0.06 |
| Pav_sc0000800.1_g500.1.mk (IAA26) | −2.43 | −1.42 | 0.37 |
| Pav_sc0000212.1_g380.1.mk (IAA27) | −2.28 | −0.81 | 0.57 |
| Pav_sc0000568.1_g290.1.br (SAUR24) | −4.29 | −1.49 | 0.35 |
| Pav_sc0000395.1_g310.1.mk (SAUR40) | / | −5.24 | 0.03 |
| Pav_sc0001248.1_g030.1.mk (SAUR50) | −3.85 | −1.09 | 0.47 |
| Pav_sc0000752.1_g020.1.mk (SAUR71) | −1.17 | 0.39 | 1.31 |
| Pav_sc0000373.1_g220.1.mk (LAX2) | −3.82 | −3.05 | 0.12 |
| Pav_sc0001900.1_g150.1.mk (ARF3) | −2.58 | −3.04 | 0.12 |
| Pav_sc0000852.1_g400.1.mk (CYCD3-1) | −1.57 | −0.50 | 0.71 |
| Pav_sc0001807.1_g180.1.mk (CYCD3-2) | −2.50 | −1.10 | 0.47 |
| Pav_sc0009346.1_g010.1.mk (AREB3) | −1.56 | 0.08 | 1.05 |
| Pav_sc0000893.1_g690.1.mk (WOL) | −4.27 | −4.79 | 0.036 |
| Pav_sc0000700.1_g840.1.mk (AHP1) | −1.99 | −1.02 | 0.49 |
| Pav_sc0001801.1_g110.1.mk (TGA9) | −2.31 | −1.16 | 0.45 |
| Pav_sc0000848.1_g820.1.mk (GH3.9) | / | / | / |
| Pav_sc0002360.1_g300.1.mk (GH3.10) | −3.14 | −1.19 | 0.44 |
| Pav_sc0001341.1_g250.1.mk (PYL4) | −1.80 | −0.46 | 0.73 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Chen, S.; Wang, W.; Li, C. Transcriptome Integrated with Metabolome Reveals the Molecular Mechanism of Phytoplasma Cherry Phyllody Disease on Stiff Fruit in Chinese Cherry (Cerasus pseudocerasus L.). Agriculture 2022, 12, 12. https://doi.org/10.3390/agriculture12010012
Li J, Chen S, Wang W, Li C. Transcriptome Integrated with Metabolome Reveals the Molecular Mechanism of Phytoplasma Cherry Phyllody Disease on Stiff Fruit in Chinese Cherry (Cerasus pseudocerasus L.). Agriculture. 2022; 12(1):12. https://doi.org/10.3390/agriculture12010012
Chicago/Turabian StyleLi, Jihan, Silei Chen, Weixing Wang, and Chunyan Li. 2022. "Transcriptome Integrated with Metabolome Reveals the Molecular Mechanism of Phytoplasma Cherry Phyllody Disease on Stiff Fruit in Chinese Cherry (Cerasus pseudocerasus L.)" Agriculture 12, no. 1: 12. https://doi.org/10.3390/agriculture12010012
APA StyleLi, J., Chen, S., Wang, W., & Li, C. (2022). Transcriptome Integrated with Metabolome Reveals the Molecular Mechanism of Phytoplasma Cherry Phyllody Disease on Stiff Fruit in Chinese Cherry (Cerasus pseudocerasus L.). Agriculture, 12(1), 12. https://doi.org/10.3390/agriculture12010012






