Stepwise-Selected Bacillus amyloliquefaciens and B. subtilis Strains from Composted Aromatic Plant Waste Able to Control Soil-Borne Diseases
Abstract
1. Introduction
2. Materials and Methods
2.1. Composts
2.2. Isolation of Spore-Forming Bacteria
2.3. Characterization of Isolated Bacteria for In Vitro Antibiosis Activity
2.4. Characterization of the Potential Antagonistic Bacteria
2.5. Biocontrol Assays against Sclerotinia Minor and Rhizoctonia Solani on Rocket
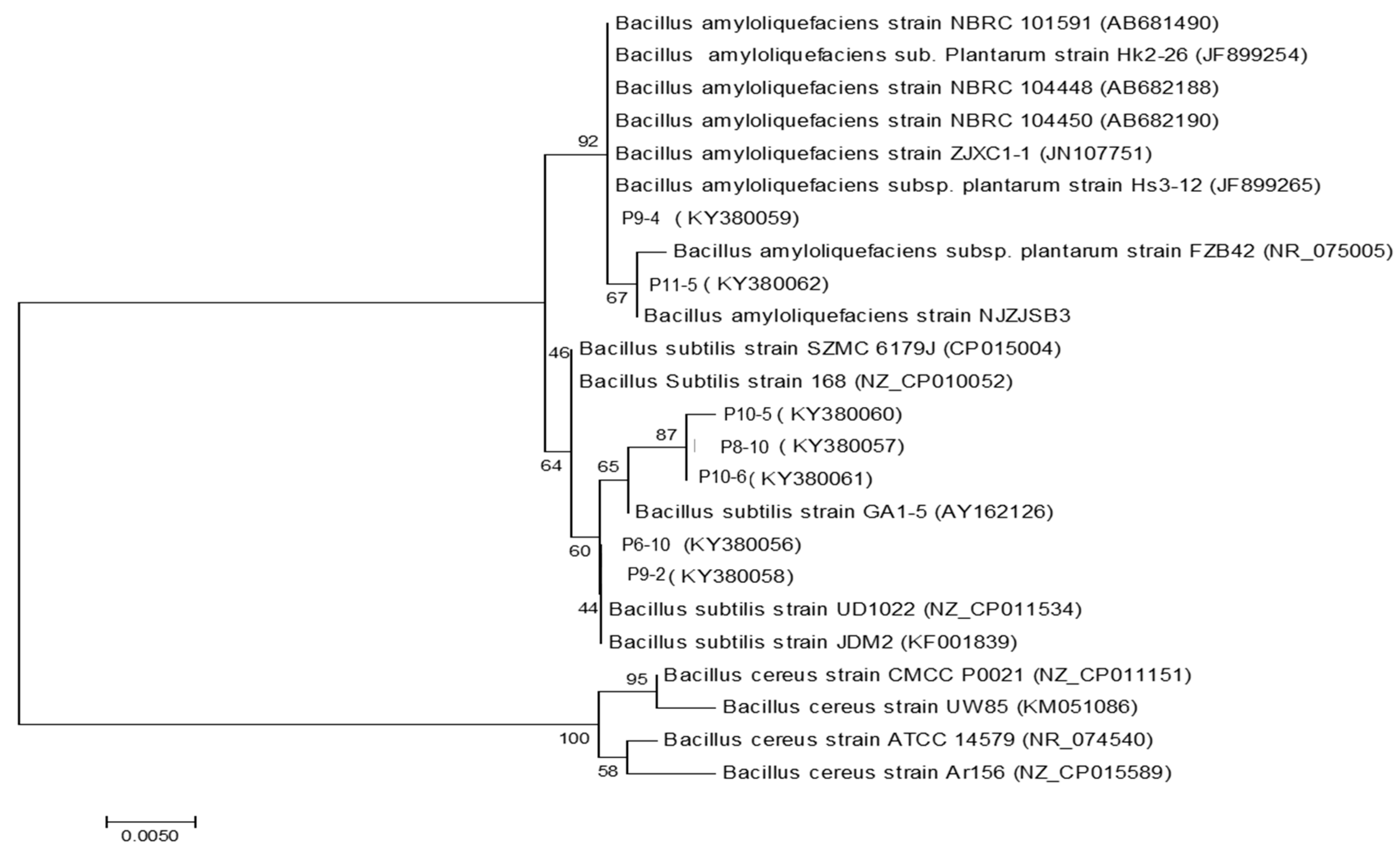
2.6. Identification of Antagonists/PGPR Isolates and Phylogentic Analisys
2.7. Detection of Antimicrobial Lipopeptides Genes
2.8. Statistical Analysis
3. Results
3.1. Isolation and Characterization for Antibiosis Activities of Spore-Forming Bacteria
3.2. Characterization of the Selected Bacterial Isolates
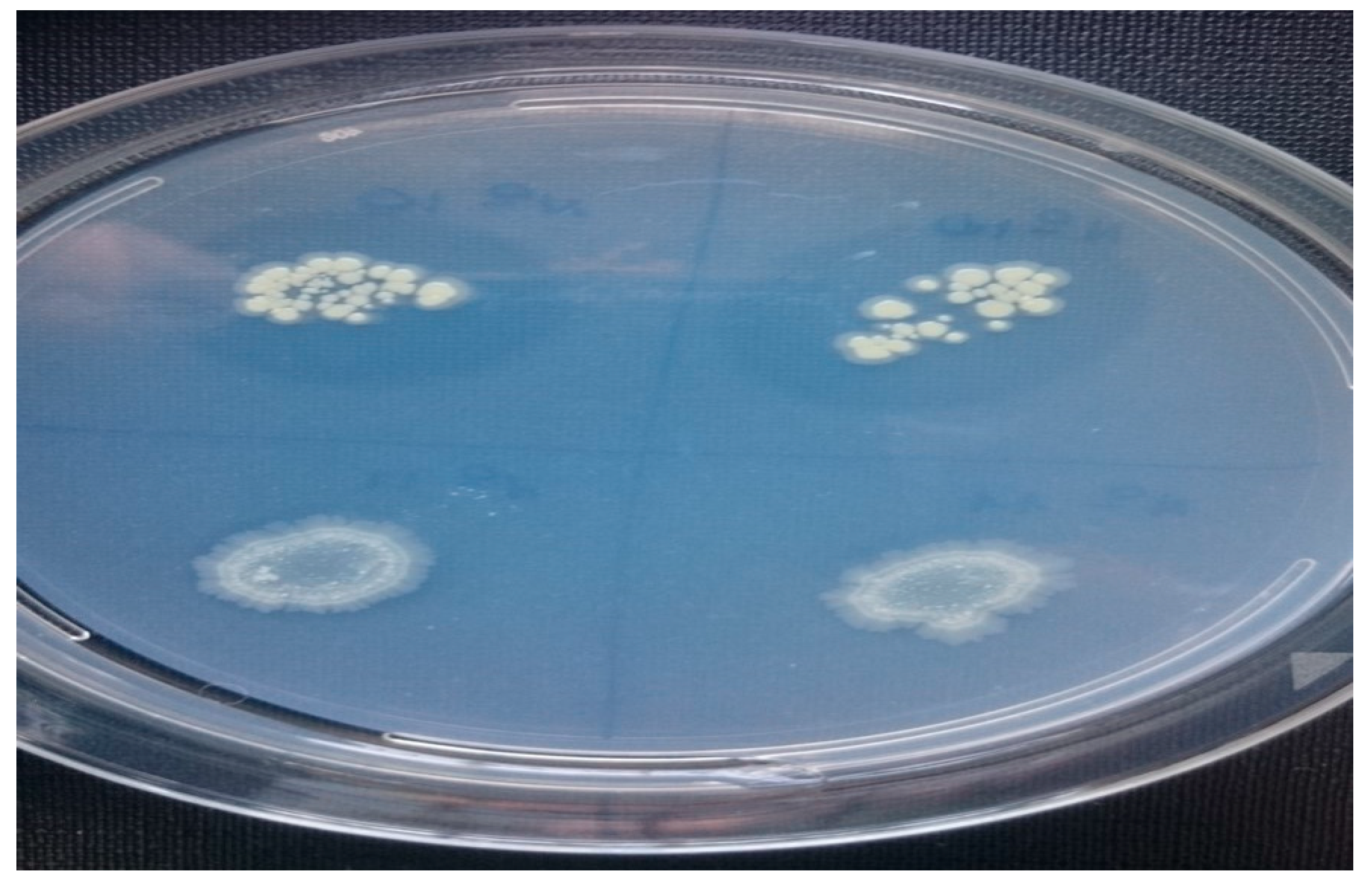
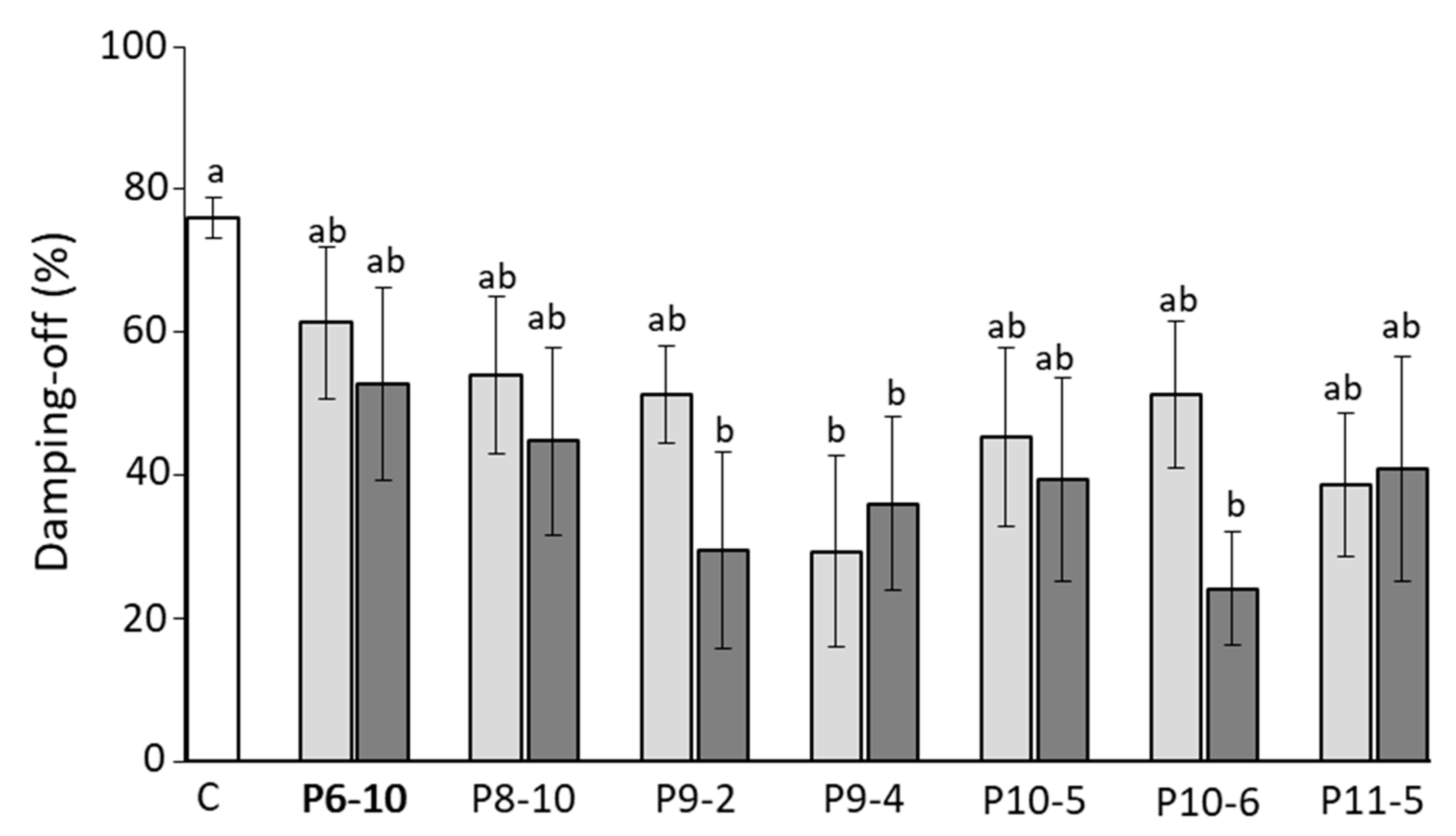
3.3. Biocontrol Assay, Identification of Bacterial Isolates and Detection of Lipopeptide Biosynthetic Gene Markers
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Strange, R.N.; Scott, P.R. Plant disease: A threat to global food security. Annu. Rev. Phytopathol. 2005, 43, 83–116. [Google Scholar] [CrossRef] [PubMed]
- Agrios, G.N. Plant Pathology; Academic Press: San Diego, CA, USA, 2005. [Google Scholar]
- Han, J.S.; Cheng, J.H.; Yoon, T.M.; Song, J.; Rajkarnikar, A.; Kim, W.G. Biological control agent of common scab disease by antagonistic strain Bacillus sp. sunhua. J. Appl. Microbiol. 2005, 99, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.H.; Park, J.L. Epidemiology of Pythium damping-off and Aphanomyces root-rot of peas after seed treatment with bacterial agents for biocontrol. Phytopathology 1993, 83, 1466–1473. [Google Scholar]
- Weller, M. Biological control of soil borne plant pathogens in the rhizosphere with bacteria. Ann. Rev. Phytopathol. 1998, 26, 379–407. [Google Scholar] [CrossRef]
- Touré, Y.; Ongena, M.; Jacques, P.; Guiro, A.; Thonart, P. Role of lipopeptides produced by Bacillus subtilis GA1 in the reduction of grey mould disease caused by Botrytis cinerea on apple. J. Appl. Microbiol. 2004, 96, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Darma, R.; Purnamasari, I.M.; Agustina, D.; Pramudito, T.E.; Sugiharti, M. A Strong antifungal-producing bacterium from bamboo powder for biocontrol of Sclerotium rolfsii in melon (Cucumis melo var. amanta). J. Plant Pathol. Microbiol. 2016, 7, 334. [Google Scholar]
- Gong, M.; Wang, J.D.; Zhang, J.; Yang, H.; Lu, X.F.; Pey, Y.; Cheng, J.Q. Study of the antifungal ability of Bacillus subtilis strain PY-1 in vitro and identification of its antifungal substance (Iturin A). Acta Biochim. Biophys. Sin. 2006, 38, 233–240. [Google Scholar] [CrossRef]
- Kim, P.I.; Ryu, J.; Kim, Y.H.; Chl, Y.T. Production of biosurfactant lipopeptides iturin A, fengycin and surfactin A from Bacillus subtilis CMB32 for control of Colletotrichum gloeosporioides. J. Microbiol. Biotechnol. 2010, 20, 138–145. [Google Scholar] [CrossRef]
- Solanki, M.K.; Robert, A.S.; Singh, R.K.; Kumar, S. Characterization of mycolytic enzymes of Bacillus strains and their bio-protection role against Rhizoctonia solani in tomato. Curr. Microbiol. 2012, 65, 330–336. [Google Scholar] [CrossRef]
- Solanki, M.K.; Singh, R.K.; Srivastava, S.; Kumar, S.; Kashyap, P.L.; Srivastava, A.K. Characterization of antagonistic-potential of two Bacillus strains and their biocontrol activity against Rhizoctonia solani in tomato. J. Basic Microbiol. 2015, 55, 82–90. [Google Scholar] [CrossRef]
- Ouhaibi-Ben, A.N.; Vallance, J.; Gerbore, J.; Rey, P.; Daami-Remadi, M. Bio-suppression of Sclerotinia stem rot of tomato and biostimulation of plant growth using tomato-associated rhizobacteria. J. Plant Pathol. Microbiol. 2016, 7, 1000331. [Google Scholar]
- Bargabus, R.L.; Zidack, N.K.; Sherwood, J.E.; Jacobsen, B.J. Characterization of systemic resistance in sugar beet elicited by a non-pathogenic, phyllosphere-colonizing Bacillus mycoides, biological control agent. Physiol. Mol. Plant Pathol. 2002, 61, 289–298. [Google Scholar] [CrossRef]
- Zhang, S.; Moyne, A.L.; Reddy, M.S.; Kloepper, J.W. The role of salicylic acid induced systemic resistance elicited by plant growth-promoting rhizobacteria against blue mold of tobacco. Biol. Control 2002, 25, 288–296. [Google Scholar] [CrossRef]
- Romero, D.; Pérez-García, A.; Rivera, M.E.; Cazorla, F.M.; de Vicente, A. Isolation and evaluation of antagonistic bacteria towards the cucurbit powdery mildew fungus Podosphaera fusca. Appl. Microbiol. Biotechnol. 2004, 64, 263–269. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Ryu, C.M.; Zhang, S. Induce systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 2004, 94, 1259–1266. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Vlami, M.; de Souza, J.T. Antibiotic production by bacterial biocontrol agents. Antonie van Leeuwenhoek 2002, 81, 537–547. [Google Scholar] [CrossRef]
- Doncean, A.; Şumălan, R.; Şumălan, R. Comparative study of aerobic microorganisms in compost. J. Hortic. For. Biotechnol. 2011, 15, 29–34. [Google Scholar]
- Scotti, R.; Pane, C.; Spaccini, R.; Palese, A.M.; Piccolo, A.; Celano, G.; Zaccardelli, M. On-farm compost: A useful tool to improve soil quality under intensive farming systems. Appl. Soil Ecol. 2016, 107, 13–23. [Google Scholar] [CrossRef]
- Ronga, D.; Pane, C.; Zaccardelli, M.; Pecchioni, N. Use of spent coffee ground compost in peat-based growing media for the production of basil and tomato potting plants. Comm. Soil Sci. Plant Anal. 2016, 47, 356–368. [Google Scholar] [CrossRef]
- Pane, C.; Piccolo, A.; Spaccini, R.; Celano, G.; Villecco, D.; Zaccardelli, M. Agricultural waste-based composts exhibiting suppressivity to diseases caused by the phytopathogenic soil-borne fungi Rhizoctonia solani and Sclerotinia minor. Appl. Soil Ecol. 2013, 65, 43–51. [Google Scholar] [CrossRef]
- Chandna, P.; Mayilraj, S.; Kuhad, R.C. Bacillus pseudoflexus sp. nov., a moderately halophilic bacterium isolated from compost. Ann. Microbiol. 2016, 66, 895–905. [Google Scholar] [CrossRef]
- Pane, C.; Villecco, D.; Campanile, F.; Zaccardelli, M. Novel strains of Bacillus, isolated from compost and compost-amended soils, as biological control agents against soil-borne phytopathogenic fungi. Biocontrol Sci. Technol. 2012, 22, 1373–1388. [Google Scholar] [CrossRef]
- Pane, C.; Spaccini, R.; Piccolo, A.; Celano, G.; Zaccardelli, M. Disease suppressiveness of agricultural greenwaste composts as related to chemical and bio-based properties shaped by different on-farm composting methods. Biol. Control 2019, 137, 104026. [Google Scholar] [CrossRef]
- Sadfi, N.; Chérif, M.; Fliss, I.; Boudabbous, A.; Antoun, H. Evaluation of bacterial isolates from salty soils and Bacillus thuringiensis strains for the biocontrol of Fusarium dry rot of potato tubers. J. Plant Pathol. 2001, 83, 101–118. [Google Scholar]
- Boulter, J.I.; Trevors, J.T.; Boland, G.J. Microbial studies of compost: Bacterial identification, and their potential for turfgrass pathogen suppression. World J. Microbiol. Biotechnol. 2002, 18, 661–671. [Google Scholar] [CrossRef]
- Ryu, E. On the Gram-differentiation of bacteria by the simplest method. J. Jpn. Soc. Vet. Sci. 1938, 17, 31. [Google Scholar] [CrossRef]
- Ryu, E. A simple method of differentiation between Gram-positive and Gram- negative organisms without staining. Kitasato Arch. Exp. Med. 1940, 17, 58–63. [Google Scholar]
- Scher, F.M.; Baker, R. Effect of Pseudomonas putida and a synthetic iron chelator on induction of soil suppressiveness to Fusarium Wilt pathogen. Phytopathology 1982, 72, 1567–1573. [Google Scholar] [CrossRef]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 65–270. [Google Scholar] [CrossRef]
- Smibert, R.M.; Krieg, N.R. Phenotypic characterization. In Methods for General and Molecular Bacteriology; Gerhardt, P., Murray, G.E., Wood, W.A., Krieg, N.R., Eds.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 607–654. [Google Scholar]
- Yang, D.; Wang, B.; Wang, J.; Chen, Y.; Zhou, M. Activity and efficacy of Bacillus subtilis strain NJ-18 against Rice sheath blight and Sclerotinia stem rot of rape. Biol. Control 2009, 51, 61–65. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Mora, I.; Cabrefiga, J.; Montesinos, E. Antimicrobial peptide genes in Bacillus strains from plant environments. Int. Microbiol. 2011, 14, 213–223. [Google Scholar] [PubMed]
- Heydari, A.; Pessarakli, M. A review on biological control of fungal plant pathogens using microbial antagonist. J. Biol. Sci. 2010, 10, 273–290. [Google Scholar]
- Huang, X.; Zhang, N.; Yong, X.; Yang, X.; Shen, Q. Biocontrol of Rhizoctonia Solani damping-off disease in Cucumber with Bacillus pumilus SQR-N43. Microbiol. Res. 2012, 167, 135–143. [Google Scholar] [CrossRef]
- Khedher, S.B.; Kilani-Feki, O.; Dammak, M.; Jabnoun-Khiareddine, H.; Daami-Remadi, M.; Slim Tounsi, S. Efficacy of Bacillus subtilis V26 as a biological control agent against Rhizoctonia solani on potato. Comptes Rendus Biol. 2015, 338, 784–792. [Google Scholar] [CrossRef]
- Pane, C.; Zaccardelli, M. Evaluation of Bacillus strains isolated from Solanaceous phylloplane for biocontrol of Alternaria early blight of tomato. Biol. Control 2015, 84, 11–18. [Google Scholar] [CrossRef]
- Kamal, M.M.; Lindbeck, K.D.; Savocchia, S.; Ash, G.J. Biological control of Sclerotinia stem rot of canola using antagonistic bacteria. Plant Pathol. 2015, 64, 1375–1384. [Google Scholar] [CrossRef]
- Phae, C.G.; Sasaki, M.; Shoda, M.; Kubota, H. Characteristics of Bacillus subtilis isolated from composts suppressing phytopathogenic microorganisms. Soil Sci. Plant Nutr. 1990, 36, 575–586. [Google Scholar] [CrossRef]
- Bonanomi, G.; Antignani, V.; Capodilupo, M.; Scala, F. Identifying the characteristics of organic soil amendments that suppress soilborne plant diseases. Soil Biol. Biochem. 2010, 42, 136–144. [Google Scholar] [CrossRef]
- Fuchs, J.G. Practical use of quality compost for plant health and vitality improvement. In Microbiology of Composting; Insam, H., Riddech, N., Klammer, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2002; pp. 435–444. [Google Scholar]
- Zaccardelli, M.; De Nicola, F.; Villecco, D.; Scotti, R. The development and suppressive activity of soil microbial communities under compost amendment. J. Sci. Nutr. 2013, 13, 730–742. [Google Scholar] [CrossRef]
- Yu, G.Y.; Sinclair, J.B.; Hartman, G.L.; Bertagnolli, B.L. Production of iturin A by Bacillus amyloliquefaciens suppressing Rhizoctonia solani. Soil Biol. Biochem. 2002, 34, 955–963. [Google Scholar] [CrossRef]
- Choudhary, D.K.; Johri, B.N. Interactions of Bacillus spp. and plants with special reference to induced systemic resistance (ISR). Microbiol. Res. 2009, 164, 493–513. [Google Scholar] [CrossRef] [PubMed]
- Ryu, C.M.; Farag, M.A.; Hu, C.H.; Reddy, M.S.; Wei, H.X.; Kloepper, J.W. Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 2004, 134, 1017–1026. [Google Scholar] [CrossRef]
- Whipps, J.M. Effect of media on growth and interactions between a range of soil-borne glasshouse pathogens and antagonistic fungi. New Phytol. 1987, 107, 127–142. [Google Scholar] [CrossRef]
- Asaka, O.; Shoda, M. Biocontrol of Rhizoctonia solani damping-off of tomato with Bacillus subtilis RB14. Appl. Environ. Microbiol. 1996, 62, 4081–4085. [Google Scholar] [CrossRef]
- Moyne, A.L.; Shelby, R.; Cleveland, T.E.; Tuzun, S. Bacillomycin D: An iturin with antifungal activity against Aspergillus flavus. J. Appl. Microbiol. 2001, 90, 622–629. [Google Scholar] [CrossRef]
- Abriouel, H.; Franz, C.M.A.P.; Omar, N.B.; Galvez, A. Diversity and applications of bacillus bacteriocins. FEMS Microbiol. Rev. 2011, 35, 201–232. [Google Scholar] [CrossRef]
- Dimkic, I.; Zivkovic, S.; Beric, T.; Ivanovic, Z.; Gavrilovic, V.; Stankovic, S.; Fira, D. Characterization and evaluation of two Bacillus strains, SS-12.6 and SS-13.1, as potential agents for the control of phytopathogenic bacteria and fungi. Biol. Control 2013, 65, 312–321. [Google Scholar] [CrossRef]
- Cao, Y.; Pi, H.; Chandrangsu, P.; Li, Y.; Wang, Y.; Zhou, H.; Xiong, H.; Helmann, J.D.; Cai, Y. Antagonism of two plant-growth promoting Bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum. Sci. Rep. 2018, 8, 4360. [Google Scholar] [CrossRef] [PubMed]
- Kakar, K.U.; Duan, Y.P.; Nawaz, Z.; Sun, G.; Almoneafy, A.A.; Hassan, M.A.; Elshakh, A.; Li, B.; Xie, G.L. A novel rhizobacterium Bk7 for biological control of brown sheath rot of rice caused by Pseudomonas fuscovaginae and its mode of action. Eur. J. Plant Pathol. 2014, 138, 819–834. [Google Scholar] [CrossRef]
- Arima, K.; Kakinuma, A.; Tamura, G. Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: Isolation, characterization and its inhibition of fibrin clot formation. Biochem. Biophys. Res. Commun. 1968, 31, 488–494. [Google Scholar] [CrossRef]
- Peypoux, F.; Bonmatin, J.M.; Wallach, J. Recent trends in the biochemistry of surfactin. Appl. Micobiol Biotechnol. 1999, 51, 553–563. [Google Scholar] [CrossRef]
- Almoneafy, A.A.; Kakar, K.U.; Nawaz, Z.; Li, B.; Saand, M.A.; Chun-Lan, Y.; Xie, G. Tomato plant growth promotion and antibacterial related-mechanisms of four rhizobacterial Bacillus strains against Ralstonia solanacearum. Symbiosis 2014, 63, 59–70. [Google Scholar] [CrossRef]
- Baysal, O.; Çalişkan, M.; Yeşilova, O. An inhibitory effect of a new Bacillus subtilis strain (EU07) against Fusarium oxysporum f. sp. radicis-lycopersici. Physiol. Mol. Plant Pathol. 2008, 73, 25–32. [Google Scholar] [CrossRef]
- Müller, H.; Westendorf, C.; Leitner, E.; Chernin, L.; Riedel, K.; Schmidt, S.; Eberl, L.; Berg, G. Quorum-sensing effects in the antagonistic rhizosphere bacterium Serratia plymuthica HRO-C48. FEMS Microbiol. Ecol. 2009, 67, 468–478. [Google Scholar] [CrossRef]
- Chet, I.; Chernin, L. Biocontrol microbial agents in soil. In Encyclopedia of Environmental Microbiology; Bitton, G., Ed.; Willey: New York, NY, USA, 2002; pp. 450–465. [Google Scholar]
- Huang, A. Control of soil borne crop diseases by soil amendments. Plant Prot. Bull. 1991, 33, 113–123. [Google Scholar]
| Gene | Primer | Sequence | Annealing T (°C) | Amplicon Length (bp) |
|---|---|---|---|---|
| srfAA | SRFAF | TCGGGACAGGAAGACATCAT | 60 | 201 |
| SRFAR | CCACTCAAACGGATAATCCTGA | |||
| bacA | BACF | CAGCTCATGGGAATGCTTTT | 60 | 498 |
| BACR | CTCGGTCCTGAAGGGACAAG | |||
| ituC | ITUCF | GGCTGCTGCAGATGCTTTAT | 60 | 423 |
| ITUCR | TCGCAGATAATCGCAGTGAG | |||
| bmyB | BMYBF | GAATCCCGTTGTTCTCCAAA | 60 | 370 |
| BMYBR | GCGGGTATTGAATGCTTGTT | |||
| fenD | FENDF | GGCCCGTTCTCTAAATCCAT | 60 | 269 |
| FENDR | GTCATGCTGACGAGAGCAAA | |||
| 16S | fD1 | CCGAATTCGTCGACAACAGAGTTTGATCCTGGCTCAG | 63 | 1600 |
| rD1 | CCCGGGATCCAAGCTTAAGGAGGTGATCCAGCC |
| Compost | N° of Isolates | ||||
|---|---|---|---|---|---|
| All | without in-Vitro Antibiosis | with in-Vitro Antibiosis Against Sclerotinia Minor | with in-Vitro Antibiosis Against Rhizoctonia Solani | with in-Vitro Antibiosis Against both Pathogens | |
| P1 | 10 | 2 | 3 | 0 | 5 |
| P2 | 10 | 3 | 2 | 0 | 5 |
| P3 | 11 | 1 | 0 | 6 | 4 |
| P4 | 13 | 0 | 0 | 0 | 13 |
| P5 | 16 | 4 | 0 | 2 | 10 |
| P6 | 11 | 0 | 0 | 0 | 11 |
| P7 | 9 | 3 | 3 | 0 | 3 |
| P8 | 17 | 0 | 0 | 0 | 17 |
| P9 | 9 | 0 | 0 | 0 | 9 |
| P10 | 7 | 0 | 0 | 0 | 7 |
| P11 | 20 | 0 | 0 | 0 | 20 |
| Total | 133 | 13 | 8 | 8 | 104 |
| Compost | Isolate | Inhibition (%) | Compost | Isolate | Inhibition (%) | ||
|---|---|---|---|---|---|---|---|
| RS | SM | RS | SM | ||||
| P1 | 1 | 26.1 | 35.6 | 3 | 32.2 | 35.0 | |
| 2 | 33.3 | 35.0 | 4 | 32.8 | 36.1 | ||
| 3 | 27.8 | 40.6 | 5 | 39.4 | 39.4 | ||
| 4 | 28.9 | 23.3 | P8 | 1 | 32.8 | 32.8 | |
| P2 | 1 | 23.3 | 20.6 | 2 | 13.3 | 33.3 | |
| 2 | 21.7 | 21.1 | 3 | 12.8 | 35.6 | ||
| 3 | 29.4 | 22.8 | 4 | 27.8 | 38.3 | ||
| 4 | 28.3 | 38.9 | 5 | 39.4 | 43.3 | ||
| 5 | 25.6 | 13.9 | 6 | 41.7 | 45.0 | ||
| 6 | 25.6 | 13.9 | 7 | 44.4 | 47.2 | ||
| 7 | 28.9 | 21.7 | 8 | 10.0 | 40.6 | ||
| 8 | 12.8 | 38.9 | 9 | 29.4 | 38.9 | ||
| P3 | 1 | 10.3 | 21.7 | 10 | 41.1 | 50.6 | |
| 2 | 31.7 | 40.6 | 11 | 16.7 | 39.4 | ||
| P4 | 1 | 20.6 | 21.7 | 12 | 25.0 | 40.6 | |
| 2 | 26.1 | 34.4 | 13 | 11.7 | 39.4 | ||
| 3 | 30.0 | 28.3 | 14 | 38.9 | 35.0 | ||
| 4 | 28.9 | 28.9 | 15 | 11.1 | 41.1 | ||
| 5 | 31.7 | 36.1 | P9 | 1 | 43.3 | 44.4 | |
| 6 | 34.4 | 37.2 | 2 | 50.0 | 55.0 | ||
| 7 | 25.6 | 31.1 | 3 | 22.2 | 38.9 | ||
| 8 | 33.3 | 25.0 | 4 | 45.0 | 57.2 | ||
| 9 | 28.3 | 26.7 | 5 | 41.1 | 36.1 | ||
| 10 | 25.0 | 21.7 | 6 | 40.6 | 43.3 | ||
| 11 | 30.6 | 26.7 | 7 | 45.6 | 31.7 | ||
| 12 | 30.0 | 24.4 | 8 | 38.3 | 35.0 | ||
| P5 | 1 | 32.8 | 32.2 | 9 | 38.3 | 35.0 | |
| 2 | 30.6 | 27.8 | P10 | 1 | 11.1 | 35.6 | |
| 3 | 32.2 | 23.9 | 2 | 29.4 | 38.3 | ||
| 4 | 35.0 | 27.8 | 3 | 13.3 | 36.7 | ||
| 5 | 32.2 | 30.8 | 4 | 16.7 | 37.2 | ||
| 6 | 29.4 | 27.8 | 5 | 41.7 | 52.8 | ||
| 7 | 28.3 | 36.7 | 6 | 45.0 | 51.7 | ||
| 8 | 32.2 | 25.8 | 7 | 31.7 | 36.7 | ||
| 9 | 31.1 | 26.7 | P11 | 1 | 32.2 | 31.1 | |
| 10 | 32.2 | 30.0 | 2 | 43.3 | 46.7 | ||
| 11 | 19.4 | 26.7 | 3 | 29.4 | 48.9 | ||
| 12 | 30.6 | 21.9 | 4 | 33.9 | 39.4 | ||
| 13 | 34.4 | 34.7 | 5 | 56.1 | 59.4 | ||
| P6 | 1 | 41.7 | 41.7 | 6 | 45.0 | 34.4 | |
| 2 | 38.3 | 32.8 | 7 | 40.6 | 31.7 | ||
| 3 | 37.8 | 30.6 | 8 | 36.7 | 42.8 | ||
| 4 | 40.6 | 37.8 | 9 | 28.9 | 32.2 | ||
| 5 | 36.1 | 33.1 | 10 | 38.3 | 36.1 | ||
| 6 | 36.1 | 31.1 | 11 | 30.6 | 41.1 | ||
| 7 | 37.8 | 32.8 | 12 | 32.2 | 48.3 | ||
| 8 | 34.4 | 32.2 | 13 | 33.3 | 37.2 | ||
| 9 | 34.4 | 37.2 | 14 | 14.4 | 38.9 | ||
| 10 | 39.2 | 50.0 | 15 | 36.7 | 29.4 | ||
| 11 | 38.3 | 41.1 | 16 | 38.9 | 34.4 | ||
| P7 | 1 | 25.3 | 26.7 | 17 | 21.1 | 26.7 | |
| 2 | 30.8 | 28.6 | 18 | 32.2 | 33.3 | ||
| Class of Morphology | Shape | Thickness | Edge | Pigmentation |
|---|---|---|---|---|
| A | Circular/Fried-egg | Umbonate | Regular | Glossy-white |
| B | Circular | Convex | Regular | Glossy-white |
| C | Circular | - | Regular | Matt-white |
| D | Circular | Cupuliform | Regular | Yellowish |
| E | Circular | Flat | Regular-full | Matt-white |
| F | Fried egg | Umbonate | Ondulate | White |
| G | Flat | Flat | Ondulate | Unpigmented |
| Isolate | Class of Morphology | Gram Reaction | Siderophore | P-Solubilization | |
|---|---|---|---|---|---|
| P1-2 | B | + | - | - | |
| P3-2 | F | + | - | + | |
| P4-5 | E | + | - | - | |
| P4-6 | C | + | - | + | |
| P6-1 | A | + | - | - | |
| P6-4 | F | + | - | - | |
| P6-9 | B | + | - | + | |
| P6-10 | B | + | - | + | |
| P6-11 | B | + | - | - | |
| P7-3 | B | + | - | - | |
| P7-4 | C | + | - | + | |
| P7-5 | E | + | - | - | |
| P8-5 | E | + | - | - | |
| P8-6 | E | + | - | - | |
| P8-7 | B | + | - | - | |
| P8-10 | B | + | - | - | |
| P8-14 | B | + | - | - | |
| P9-1 | B | + | - | - | |
| P9-2 | B | + | - | - | |
| P9-4 | C | + | - | - | |
| P9-5 | C | + | - | - | |
| P9-6 | A | + | - | - | |
| P9-8 | B | + | - | - | |
| P9-9 | A | + | - | - | |
| P10-5 | G | + | - | + | |
| P10-6 | B | + | - | + | |
| P10-7 | C | - | - | + | |
| P11-2 | C | + | - | + | |
| P11-4 | D | - | - | + | |
| P11-5 | B | + | - | - | |
| P11-6 | C | + | - | - | |
| P11-8 | C | + | - | - | |
| P11-10 | B | + | - | + | |
| P11-12 | B | - | - | + | |
| P11-13 | D | + | - | - |
| BCA strain | Identified as | bacA | bmyB | fenD | ituC | srfAA |
|---|---|---|---|---|---|---|
| P6-10 | BS | + | - | + | + | + |
| P8-10 | BS | + | + | + | - | + |
| P9-2 | BS | + | - | - | - | + |
| P9-4 | BA | + | + | + | - | + |
| P10-5 | BS | + | + | + | - | + |
| P10-6 | BS | + | + | + | - | + |
| P11-5 | BA | + | + | + | + | + |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaccardelli, M.; Sorrentino, R.; Caputo, M.; Scotti, R.; De Falco, E.; Pane, C. Stepwise-Selected Bacillus amyloliquefaciens and B. subtilis Strains from Composted Aromatic Plant Waste Able to Control Soil-Borne Diseases. Agriculture 2020, 10, 30. https://doi.org/10.3390/agriculture10020030
Zaccardelli M, Sorrentino R, Caputo M, Scotti R, De Falco E, Pane C. Stepwise-Selected Bacillus amyloliquefaciens and B. subtilis Strains from Composted Aromatic Plant Waste Able to Control Soil-Borne Diseases. Agriculture. 2020; 10(2):30. https://doi.org/10.3390/agriculture10020030
Chicago/Turabian StyleZaccardelli, Massimo, Roberto Sorrentino, Michele Caputo, Riccardo Scotti, Enrica De Falco, and Catello Pane. 2020. "Stepwise-Selected Bacillus amyloliquefaciens and B. subtilis Strains from Composted Aromatic Plant Waste Able to Control Soil-Borne Diseases" Agriculture 10, no. 2: 30. https://doi.org/10.3390/agriculture10020030
APA StyleZaccardelli, M., Sorrentino, R., Caputo, M., Scotti, R., De Falco, E., & Pane, C. (2020). Stepwise-Selected Bacillus amyloliquefaciens and B. subtilis Strains from Composted Aromatic Plant Waste Able to Control Soil-Borne Diseases. Agriculture, 10(2), 30. https://doi.org/10.3390/agriculture10020030








