Strategy for Use of Genome-Wide Non-Invasive Prenatal Testing for Rare Autosomal Aneuploidies and Unbalanced Structural Chromosomal Anomalies
Abstract
1. Introduction
2. Experimental Section
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Huang, S.; Chang, C.; Cheng, P.; Hsiao, C.; Soong, Y.; Duan, T. First-trimester combined screening is effective for the detection of unbalanced chromosomal translocations at 11 to 12 weeks of gestation. Reprod. Sci. 2014, 21, 594–600. [Google Scholar] [CrossRef][Green Version]
- Torring, N.; Petersen, O.B.; Becher, N.; Vogel, I.; Uldbjerg, N. First trimester screening for other trisomies than trisomy 21, 18, and 13. Prenat. Diagn. 2015, 35, 612–619. [Google Scholar] [CrossRef]
- Lindquist, A.; Poulton, A.; Halliday, J.; Hui, L. Prenatal diagnostic testing and atypical chromosome abnormalities following combined first-trimester screening: Implications for contingent models of non-invasive prenatal testing. Ultrasound Obstet. Gynecol. 2018, 51, 487–492. [Google Scholar] [CrossRef]
- Bianchi, D.W.; Chiu, R.W.K. Sequencing of Circulating Cell-free DNA during Pregnancy. N. Engl. J. Med. 2018, 379, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, F.; Bono, S.; Pizzuti, F.; Duca, S.; Polverari, A.; Faieta, M.; Baldi, M.; Diano, L.; Spinella, F. The clinical utility of genome-wide non invasive prenatal screening. Prenat. Diagn. 2017, 37, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Lin, Y.; Qiao, F.; Li, H.; Wang, Y.; Zhang, J.; Liu, A.; Ji, X.; Ma, D.; Jiang, T.; et al. Perinatal outcomes following cell-free DNA screening in >32 000 women: Clinical follow-up data from a single tertiary center. Prenat. Diagn. 2018, 38, 755–764. [Google Scholar] [CrossRef]
- Akolekar, R.; Beta, J.; Picciarelli, G.; Ogilvie, C.; D’Antonio, F. Procedure-related risk of miscarriage following amniocentesis and chorionic villus sampling: A systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 2015, 45, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Salomon, L.J.; Sotiriadis, A.; Wulff, C.B.; Odibo, A.; Akolekar, R. Risk of miscarriage following amniocentesis or chorionic villus sampling: Systematic review of literature and updated meta-analysis. Ultrasound Obstet. Gynecol. 2019, 54, 442–451. [Google Scholar] [CrossRef]
- Michael, T.; Natalia, S.-L.; James, M.; Jörg, B.; Yuri, N.; Andreas, S.W.; Gregor, S. Mid-trimester preterm premature rupture of membranes (PPROM): Etiology, diagnosis, classification, international recommendations of treatment options and outcome. J. Perinat. Med. 2018, 46, 465–488. [Google Scholar] [CrossRef]
- Cederholm, M.; Haglund, B.; Axelsson, O. Maternal complications following amniocentesis and chorionic villus sampling for prenatal karyotyping. BJOG 2003, 110, 392–399. [Google Scholar] [CrossRef]
- Norton, M.E.; Baer, R.J.; Wapner, R.J.; Kuppermann, M.; Jelliffe-Pawlowski, L.L.; Currier, R.J. Cell-free DNA vs sequential screening for the detection of fetal chromosomal abnormalities. Am. J. Obstet. Gynecol. 2016, 214, e1–e6. [Google Scholar] [CrossRef] [PubMed]
- Sehnert, A.J.; Rhees, B.; Comstock, D.; de Feo, E.; Heilek, G.; Burke, J.; Rava, R.P. Optimal detection of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell-free fetal DNA from maternal blood. Clin. Chem. 2011, 57, 1042–1049. [Google Scholar] [CrossRef] [PubMed]
- Ehrich, M.; Deciu, C.; Zwiefelhofer, T.; Tynan, J.A.; Cagasan, L.; Tim, R.; Lu, V.; McCullough, R.; McCarthy, E.; Nygren, A.O.; et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: A study in a clinical setting. Am. J. Obstet. Gynecol. 2011, 204, e1–e11. [Google Scholar] [CrossRef] [PubMed]
- Jensen, T.J.; Zwiefelhofer, T.; Tim, R.C.; Dzakula, Z.; Kim, S.K.; Mazloom, A.R.; Zhu, Z.; Tynan, J.; Lu, T.; McLennan, G.; et al. High-throughput massively parallel sequencing for fetal aneuploidy detection from maternal plasma. PLoS ONE 2013, 8, e57381. [Google Scholar] [CrossRef]
- Juneau, K.; Bogard, P.E.; Huang, S.; Mohseni, M.; Wang, E.T.; Ryvkin, P.; Kingsley, C.; Struble, C.A.; Oliphant, A.; Zahn, J.M. Microarray-based cell-free DNA analysis improves noninvasive prenatal testing. Fetal Diagn. Ther. 2014, 36, 282–286. [Google Scholar] [CrossRef]
- Zimmermann, B.; Hill, M.; Gemelos, G.; Demko, Z.; Banjevic, M.; Baner, J.; Ryan, A.; Sigurjonsson, S.; Chopra, N.; Dodd, M.; et al. Noninvasive prenatal aneuploidy testing of chromosomes 13, 18, 21, X, and Y, using targeted sequencing of polymorphic loci. Prenat. Diagn. 2012, 32, 1233–1241. [Google Scholar] [CrossRef]
- Pergament, E.; Cuckle, H.; Zimmermann, B.; Banjevic, M.; Sigurjonsson, S.; Ryan, A.; Hall, M.P.; Dodd, M.; Lacroute, P.; Stosic, M.; et al. Single-Nucleotide Polymorphism-Based Noninvasive Prenatal Screening in a High-Risk and Low-Risk Cohort. Obstet. Gynecol. 2014, 124, 210–218. [Google Scholar] [CrossRef]
- Fan, H.C.; Blumenfeld, Y.J.; Chitkara, U.; Hudgins, L.; Quake, S.R. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin. Chem. 2010, 56, 1279–1286. [Google Scholar] [CrossRef]
- Lo, Y.M.; Chan, K.C.; Sun, H.; Chen, E.Z.; Jiang, P.; Lun, F.M.; Zheng, Y.W.; Leung, T.Y.; Lau, T.K.; Cantor, C.R.; et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2010, 2, 61ra91. [Google Scholar] [CrossRef]
- Palomaki, G.E.; Kloza, E.M. Prenatal cell-free DNA screening test failures: A systematic review of failure rates, risks of Down syndrome, and impact of repeat testing. Genet. Med. 2018, 20, 1312–1323. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Ren, J.; Chen, F.; Zhou, Y.; Xie, J.; Dan, S.; Su, Y.; Xie, J.; Yin, B.; Su, W.; et al. Noninvasive Fetal Trisomy (NIFTY) test: An advanced noninvasive prenatal diagnosis methodology for fetal autosomal and sex chromosomal aneuploidies. BMC Med. Genom. 2012, 5, 57. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Yin, A.H.; Peng, C.F.; Fu, F.; Yang, J.X.; Li, R.; Chen, Y.Y.; Luo, D.H.; Zhang, Y.L.; Ou, Y.M.; et al. Noninvasive prenatal diagnosis of common aneuploidies by semiconductor sequencing. Proc. Natl. Acad. Sci. USA 2014, 111, 7415–7420. [Google Scholar] [CrossRef] [PubMed]
- Stumm, M.; Entezami, M.; Haug, K.; Blank, C.; Wustemann, M.; Schulze, B.; Raabe-Meyer, G.; Hempel, M.; Schelling, M.; Ostermayer, E.; et al. Diagnostic accuracy of random massively parallel sequencing for non-invasive prenatal detection of common autosomal aneuploidies: A collaborative study in Europe. Prenat. Diagn. 2014, 34, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Samango-Sprouse, C.; Banjevic, M.; Ryan, A.; Sigurjonsson, S.; Zimmermann, B.; Hill, M.; Hall, M.P.; Westemeyer, M.; Saucier, J.; Demko, Z.; et al. SNP-based non-invasive prenatal testing detects sex chromosome aneuploidies with high accuracy. Prenat. Diagn. 2013, 33, 643–649. [Google Scholar] [CrossRef]
- Mazloom, A.R.; Dzakula, Z.; Oeth, P.; Wang, H.; Jensen, T.; Tynan, J.; McCullough, R.; Saldivar, J.S.; Ehrich, M.; van den Boom, D.; et al. Noninvasive prenatal detection of sex chromosomal aneuploidies by sequencing circulating cell-free DNA from maternal plasma. Prenat. Diagn. 2013, 33, 591–597. [Google Scholar] [CrossRef]
- Helgeson, J.; Wardrop, J.; Boomer, T.; Almasri, E.; Paxton, W.B.; Saldivar, J.S.; Dharajiya, N.; Monroe, T.J.; Farkas, D.H.; Grosu, D.S.; et al. Clinical outcome of subchromosomal events detected by whole-genome noninvasive prenatal testing. Prenat. Diagn. 2015, 35, 999–1004. [Google Scholar] [CrossRef]
- Martin, K.; Iyengar, S.; Kalyan, A.; Lan, C.; Simon, A.L.; Stosic, M.; Kobara, K.; Ravi, H.; Truong, T.; Ryan, A.; et al. Clinical experience with a single-nucleotide polymorphism-based non-invasive prenatal test for five clinically significant microdeletions. Clin. Genet. 2018, 93, 293–300. [Google Scholar] [CrossRef]
- van der Meij, K.R.M.; Sistermans, E.A.; Macville, M.V.E.; Stevens, S.J.C.; Bax, C.J.; Bekker, M.N.; Bilardo, C.M.; Boon, E.M.J.; Boter, M.; Diderich, K.E.M.; et al. TRIDENT-2: National Implementation of Genome-wide Non-invasive Prenatal Testing as a First-Tier Screening Test in the Netherlands. Am. J. Hum. Genet. 2019, 105, 1091–1101. [Google Scholar] [CrossRef]
- Pertile, M.D.; Halks-Miller, M.; Flowers, N.; Barbacioru, C.; Kinnings, S.L.; Vavrek, D.; Seltzer, W.K.; Bianchi, D.W. Rare autosomal trisomies, revealed by maternal plasma DNA sequencing, suggest increased risk of feto-placental disease. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef]
- Pescia, G.; Guex, N.; Iseli, C.; Brennan, L.; Osteras, M.; Xenarios, I.; Farinelli, L.; Conrad, B. Cell-free DNA testing of an extended range of chromosomal anomalies: Clinical experience with 6388 consecutive cases. Genet. Med. 2017, 19, 169–175. [Google Scholar] [CrossRef]
- Agence de la BioMédecine. Rapport médical et scientifique de l’assistance médicale à la procréation et de la génétique humaines en France. In Prenatal Diagnosis and Preimplantation Genetic Diagnosis; 2017. Available online: https://rams.agence-biomedecine.fr/diagnostic-sur-lembryon-et-le-foetus (accessed on 23 September 2019).
- Bayindir, B.; Dehaspe, L.; Brison, N.; Brady, P.; Ardui, S.; Kammoun, M.; Van der Veken, L.; Lichtenbelt, K.; Van den Bogaert, K.; Van Houdt, J.; et al. Noninvasive prenatal testing using a novel analysis pipeline to screen for all autosomal fetal aneuploidies improves pregnancy management. Eur. J. Hum. Genet. 2015, 23, 1286–1293. [Google Scholar] [CrossRef] [PubMed]
- Van Opstal, D.; van Maarle, M.C.; Lichtenbelt, K.; Weiss, M.M.; Schuring-Blom, H.; Bhola, S.L.; Hoffer, M.J.V.; Huijsdens-van Amsterdam, K.; Macville, M.V.; Kooper, A.J.A.; et al. Origin and clinical relevance of chromosomal aberrations other than the common trisomies detected by genome-wide NIPS: Results of the TRIDENT study. Genet. Med. 2018, 20, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Scott, F.; Bonifacio, M.; Sandow, R.; Ellis, K.; Smet, M.E.; McLennan, A. Rare autosomal trisomies: Important and not so rare. Prenat. Diagn. 2018, 38, 765–771. [Google Scholar] [CrossRef] [PubMed]
- Chatron, N.; Till, M.; Abel, C.; Bardel, C.; Ramond, F.; Sanlaville, D.; Schluth-Bolard, C. Detection of rare autosomal trisomies through non-invasive prenatal testing: Benefits for pregnancy management. Ultrasound Obstet. Gynecol. 2019, 53, 129–130. [Google Scholar] [CrossRef] [PubMed]
- Holland, A.J.; Cleveland, D.W. Chromoanagenesis and cancer: Mechanisms and consequences of localized, complex chromosomal rearrangements. Nat. Med. 2012, 18, 1630–1638. [Google Scholar] [CrossRef] [PubMed]
- Grati, F.R. Chromosomal Mosaicism in Human Feto-Placental Development: Implications for Prenatal Diagnosis. J. Clin. Med. 2014, 3, 809–837. [Google Scholar] [CrossRef]
- Gil, M.M.; Accurti, V.; Santacruz, B.; Plana, M.N.; Nicolaides, K.H. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: Updated meta-analysis. Ultrasound Obstet. Gynecol. 2017, 50, 302–314. [Google Scholar] [CrossRef]
- de Wergifosse, S.; Bevilacqua, E.; Mezela, I.; El Haddad, S.; Gounongbe, C.; de Marchin, J.; Maggi, V.; Conotte, S.; Badr, D.A.; Fils, J.F.; et al. Cell-free DNA analysis in maternal blood: Comparing genome-wide versus targeted approach as a first-line screening test. J. Matern. Fetal Neonatal Med. 2019. [Google Scholar] [CrossRef]
- Grati, F.R.; Ferreira, J.; Benn, P.; Izzi, C.; Verdi, F.; Vercellotti, E.; Dalpiaz, C.; D’Ajello, P.; Filippi, E.; Volpe, N.; et al. Outcomes in pregnancies with a confined placental mosaicism and implications for prenatal screening using cell-free DNA. Genet. Med. 2020, 22, 309–316. [Google Scholar] [CrossRef]
- Gadsbøll, K.; Petersen, O.B.; Gatinois, V.; Strange, H.; Jacobsson, B.; Wapner, R.; Vermeesch, J.R.; Group, T.N.-m.S.; Vogel, I. Current use of noninvasive prenatal testing in Europe, Australia and the USA: A graphical presentation. Acta Obstet. Gynecol. Scand. 2020, 99, 722–730. [Google Scholar] [CrossRef]
- Benachi, A.; Caffrey, J.; Calda, P.; Carreras, E.; Jani, J.C.; Kilby, M.D.; Klein, H.G.; Rizzo, G.; Yaron, Y. Understanding attitudes and behaviors towards cell-free DNA-based noninvasive prenatal testing (NIPT): A survey of European health-care providers. Eur. J. Med. Genet. 2020, 63, 103616. [Google Scholar] [CrossRef] [PubMed]
- Ehrich, M.; Tynan, J.; Mazloom, A.; Almasri, E.; McCullough, R.; Boomer, T.; Grosu, D.; Chibuk, J. Genome-wide cfDNA screening: Clinical laboratory experience with the first 10,000 cases. Genet. Med. 2017, 19, 1332–1337. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Cram, D.S.; Tan, H.; Linpeng, S.; Liu, Y.; Sun, H.; Zhang, Y.; Tian, F.; Zhu, H.; Xu, M.; et al. Clinical utility of noninvasive prenatal screening for expanded chromosome disease syndromes. Genet. Med. 2019. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhao, G.; Li, H.; Zhang, Q.; Lu, J.; Yu, B.; Wang, T. Non-invasive prenatal testing to detect chromosome aneuploidies in 57,204 pregnancies. Mol. Cytogenet. 2019, 12, 29. [Google Scholar] [CrossRef] [PubMed]
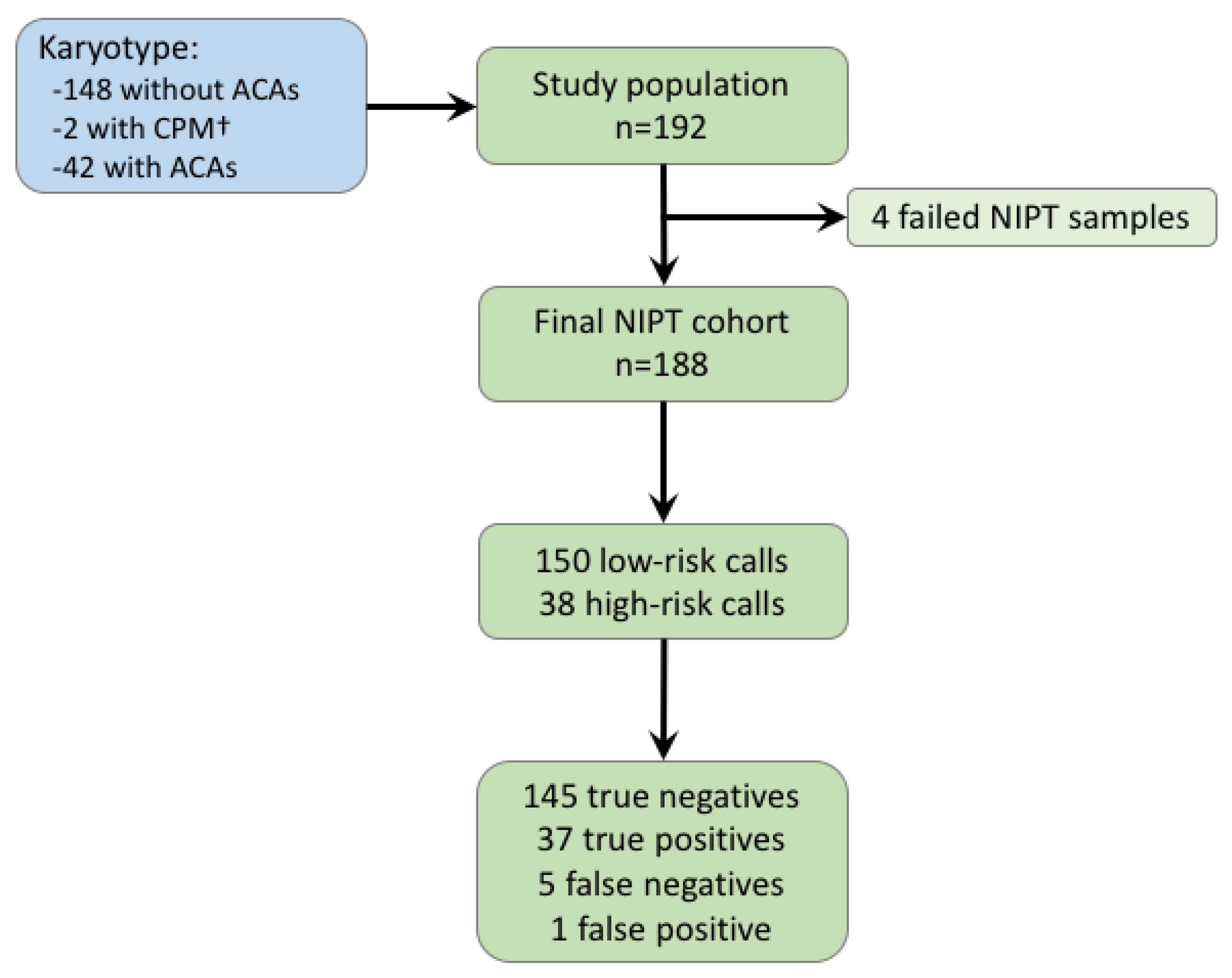
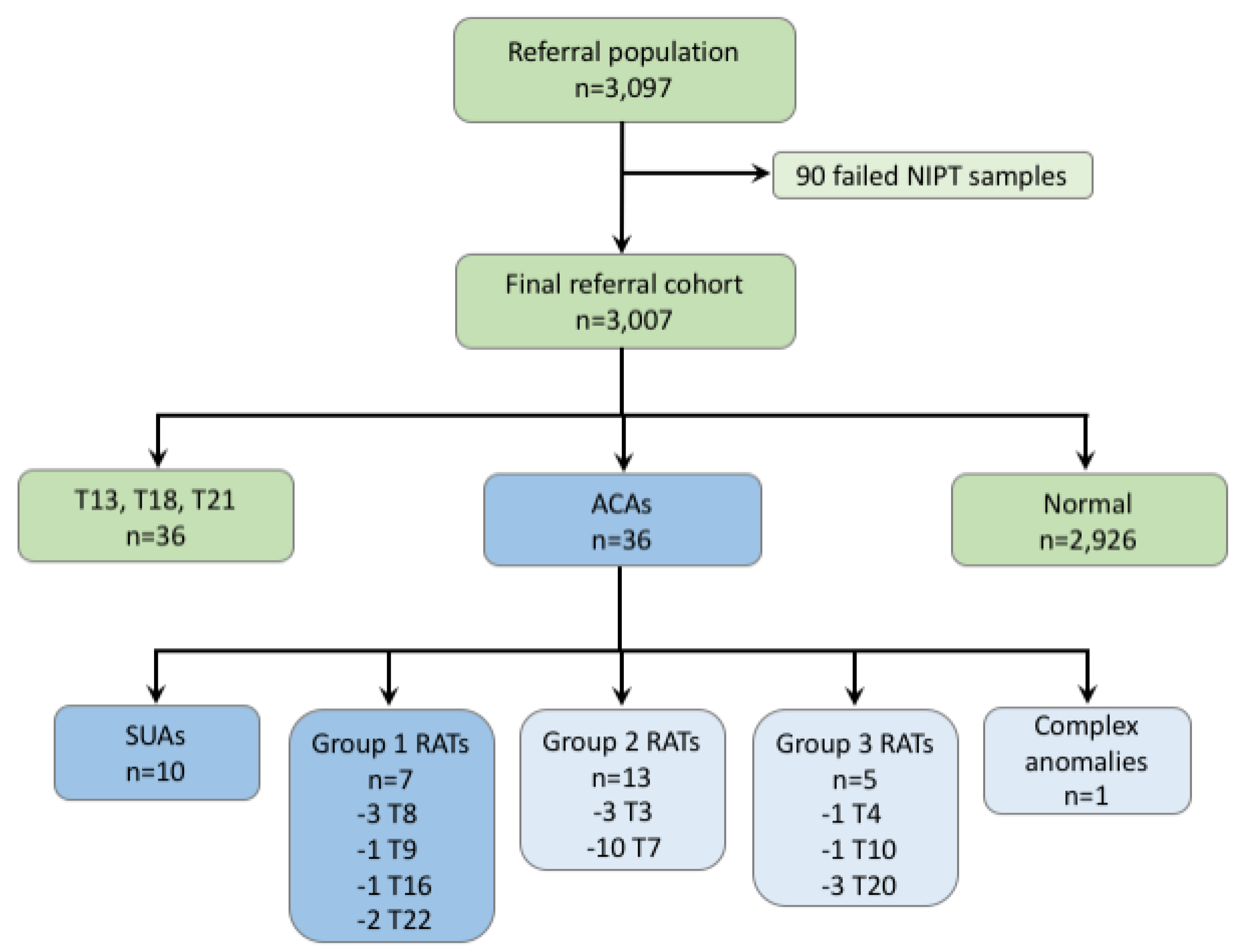
| Sample | Fetal Fraction | Karyotype | Size (Mb) | Comments |
|---|---|---|---|---|
| 1 | 10% | 46,XX,del(4)(p16.3).ish del(4)(WHS-,D4S3359-) | 5–8, based on the karyotype | Possible size < 7 Mb |
| 2 | 15% | arr[GRCh37] 12p13.33q11(173786_37876500)x3 | 37.7 | Suspected mosaicism |
| 3 | 9% | 46,XX,der(8)?add(8)(p?)?dup(8)(q22q23)dn.ish der(8)(qter->?::?->qter)(D8S504, VIJyRM2053+,wcp8+,VIJyRM2053+).arr[GRCh38] 8p23.3p23.1(158048_6935930)x1,8p23.1p11.23(12585435_38267493)x3, 8p11.22(38314367_39246760)x3, 8p11.22(39247087_39386852)x1,8p11.22(39389765_40264413)x3, 8q22.3q23.2(104688373_111952230)x3, 8q24.3(144972747_146295771)x3 | Total deletion = 6.9 Total duplication = 36.2 | Chromoanasynthesis |
| 4 | 3% | 46,XX,add(4)(qter).ish add(4)(wcp4-).arr[GRCh37] 5q31.2q35.3(138522878_180715096)x3 | 41.2 | |
| 5 | 4% | 46,XY,i(18)(q10) |
| Sample | NIPT | Array |
|---|---|---|
| 1 | 18.2 | 12.8 |
| 2 | 9.7 | 9.8 |
| 3 | 11.5 | 11.1 |
| 4 | 28.8 | 29.9 |
| 5 | Trisomy 11 | 2.7 |
| 6 | 8.1 | 6.7 |
| 7 | 10.7 | 11.5 |
| 8 | 11.3 | 17.4 |
| 9 | 26 | 26.6 |
| 10 | 17.2 | 7.9 |
| 11 | 18.3 | 18.3 |
| 12 | 9 | 8.8 |
| 13 | 12.5 | 12.5 |
| 14 | 13.7 | 11.8 |
| 15 | 60.1 | 59.9 |
| Type of Rearrangement | Observed on Karyotype, n | Detected by NIPT, n | Detection Rate, % (95% CI) |
|---|---|---|---|
| Deletion | 13 | 11 | 84.6 (54.6–98.1) |
| Duplication | 28 | 24 | 85.7 (67.3–96.0) |
| Interstitial | 5 | 5 | 100 (47.8–100) |
| Terminal | 36 | 28 | 77.8 (60.9–89.9) |
| Measurement | General Population | MSS Score 1/51–1/1000 | MSS Score >1/1000 | MSS Score 1/51–1/300 | MSS Score >1/300 |
|---|---|---|---|---|---|
| Prevalence 1 | 0.10% | 0.37% | 0.61% | 1.01% | 1.40% |
| PPV | 11% | 32% | 44% | 56% | 64% |
| Population Type | Failed | No Anomalies | Common Trisomies | ACAs | Total, n (%) |
|---|---|---|---|---|---|
| MSS ≥ 1/1000 | 71 | 2596 | 35 | 29 | 2731 (88) |
| MSS < 1/1000 | 9 | 139 | 0 | 1 | 149 (5) |
| Parental Robertsonian translocation | 0 | 2 | 0 | 0 | 2 (0) |
| Previous history of fetal trisomy | 2 | 51 | 0 | 1 | 54 (2) |
| First-tier screening | 8 | 147 | 1 | 5 | 161 (5) |
| Total | 90 | 2935 | 36 | 36 | 3097 |
| Measurement | Biobank Samples (Cohort A) (n = 189) | Normal Biobank Samples from Cohort A (n = 147) | Abnormal Biobank Samples from Cohort A (n = 42) | Referral Samples (Cohort B) (n = 3007) |
|---|---|---|---|---|
| Average | 12.27% | 12.40% | 11.76% | 11% |
| Median | 11% | 11% | 10.5% | 10% |
| Range | 3–35% | 4–35% | 3–24% | 2–35% |
| Population Type | Total, Excluding Failures | Group 1 + SUAs | Group 2 | Group 3 | Prevalence of All ACAs | Prevalence of Group 1 + SUAs |
|---|---|---|---|---|---|---|
| MSS ≥ 1/1000 | 2660 | 14 | 10 | 5 | 1.09% | 0.53% |
| MSS < 1/1000 | 140 | 1 | 0 | 0 | 0.71% | 0.71% |
| First-tier screening | 153 | 1 | 3 | 1 | 3.27% | 0.65% |
| Previous history of fetal trisomy or parental Robertsonian translocation | 54 | 1 | 0 | 0 | 1.85% | 1.85% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kleinfinger, P.; Lohmann, L.; Luscan, A.; Trost, D.; Bidat, L.; Debarge, V.; Castaigne, V.; Senat, M.-V.; Brechard, M.-P.; Guilbaud, L.; et al. Strategy for Use of Genome-Wide Non-Invasive Prenatal Testing for Rare Autosomal Aneuploidies and Unbalanced Structural Chromosomal Anomalies. J. Clin. Med. 2020, 9, 2466. https://doi.org/10.3390/jcm9082466
Kleinfinger P, Lohmann L, Luscan A, Trost D, Bidat L, Debarge V, Castaigne V, Senat M-V, Brechard M-P, Guilbaud L, et al. Strategy for Use of Genome-Wide Non-Invasive Prenatal Testing for Rare Autosomal Aneuploidies and Unbalanced Structural Chromosomal Anomalies. Journal of Clinical Medicine. 2020; 9(8):2466. https://doi.org/10.3390/jcm9082466
Chicago/Turabian StyleKleinfinger, Pascale, Laurence Lohmann, Armelle Luscan, Detlef Trost, Laurent Bidat, Véronique Debarge, Vanina Castaigne, Marie-Victoire Senat, Marie-Pierre Brechard, Lucie Guilbaud, and et al. 2020. "Strategy for Use of Genome-Wide Non-Invasive Prenatal Testing for Rare Autosomal Aneuploidies and Unbalanced Structural Chromosomal Anomalies" Journal of Clinical Medicine 9, no. 8: 2466. https://doi.org/10.3390/jcm9082466
APA StyleKleinfinger, P., Lohmann, L., Luscan, A., Trost, D., Bidat, L., Debarge, V., Castaigne, V., Senat, M.-V., Brechard, M.-P., Guilbaud, L., Le Guyader, G., Satre, V., Laurichesse Delmas, H., Lallaoui, H., Manca-Pellissier, M.-C., Boughalem, A., Valduga, M., Hodeib, F., Benachi, A., & Costa, J. M. (2020). Strategy for Use of Genome-Wide Non-Invasive Prenatal Testing for Rare Autosomal Aneuploidies and Unbalanced Structural Chromosomal Anomalies. Journal of Clinical Medicine, 9(8), 2466. https://doi.org/10.3390/jcm9082466





