Epigenetic Regulation of Alternative mRNA Splicing in Dilated Cardiomyopathy
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Enrollment and Phenotyping
2.2. Library Preparation and Next Generation Sequencing
2.3. Quality Control of Sequencing Data
2.4. Data Normalization and Batch Effect Correction
2.5. Bioinformatic Computation and Analysis
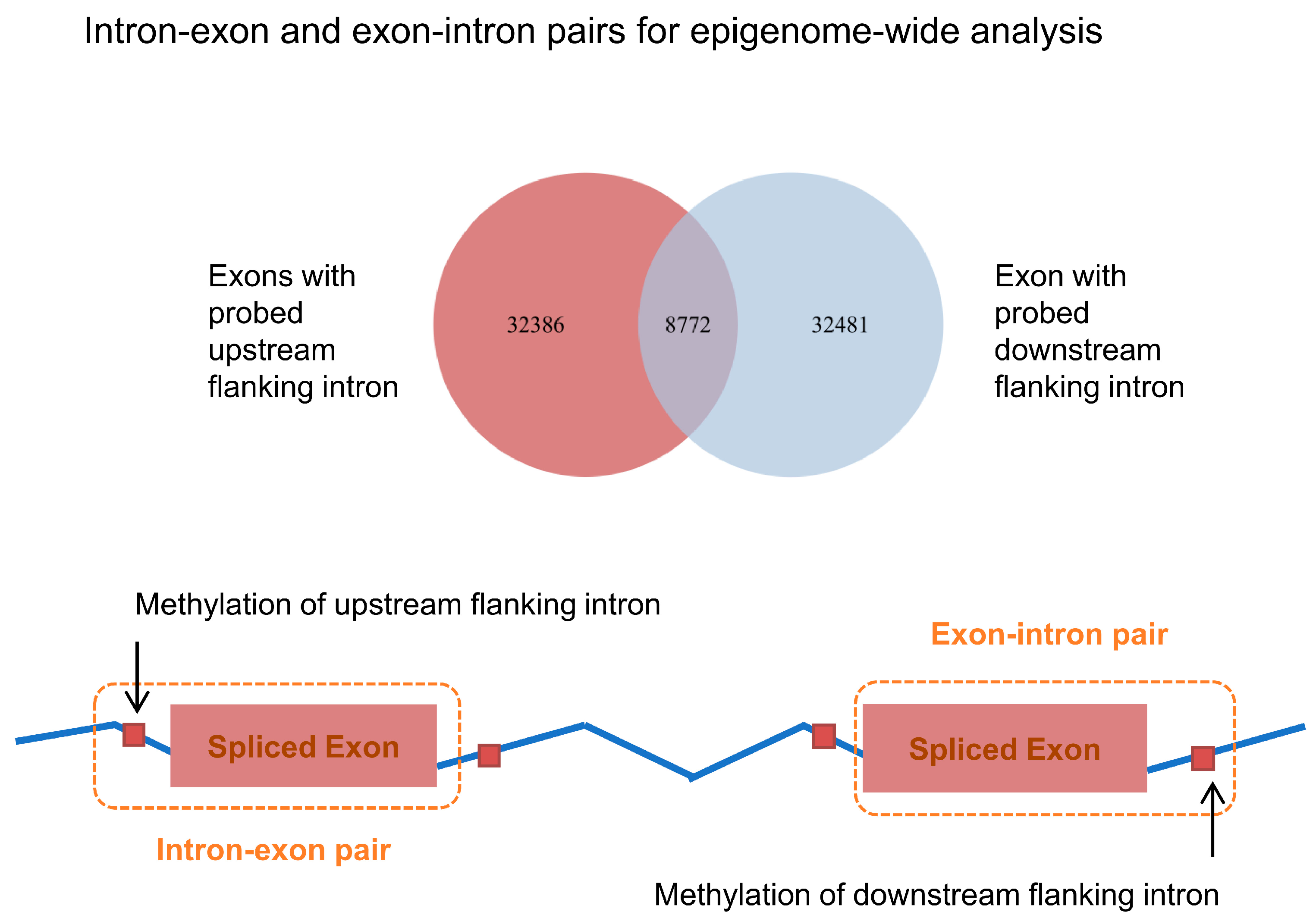
2.6. Genome-Wide Analysis of Neighboring Intron and Exon
2.7. Methylome-Transcriptome Correlation
3. Results
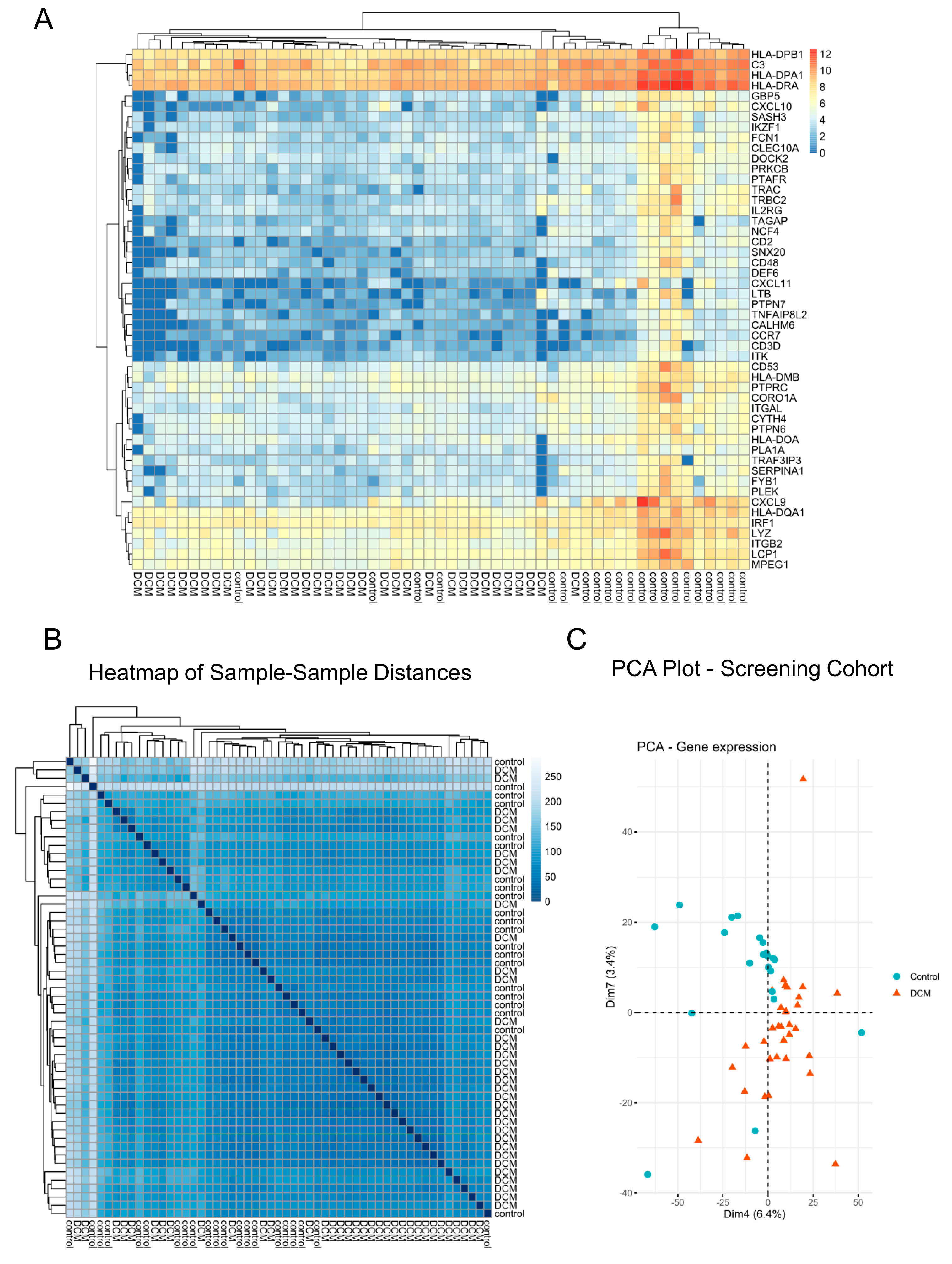
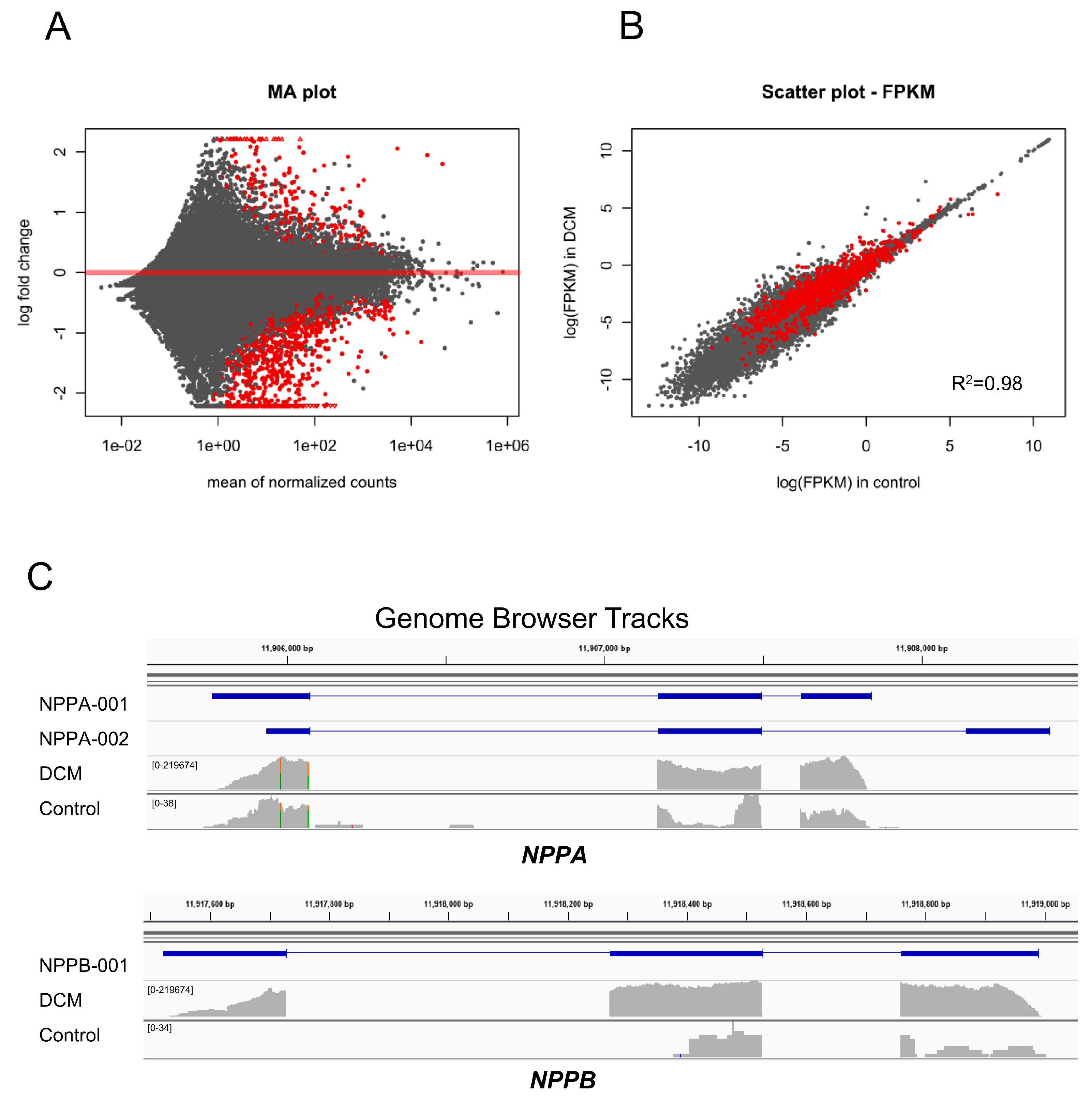
3.1. DCM-Related Reconnection of mRNA Expression
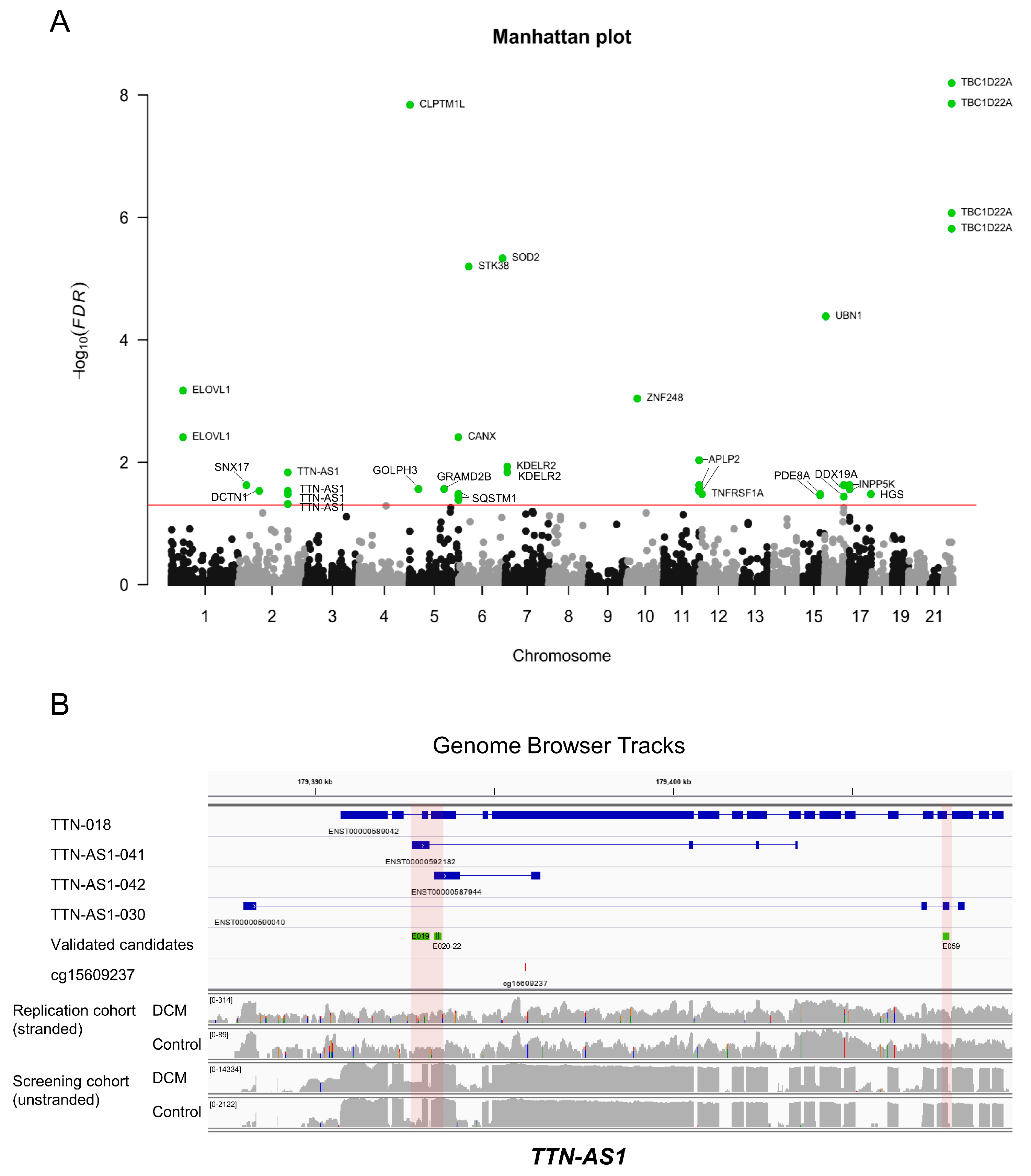
3.2. Epigenome-Wide Linkage of DNA Methylation and Inclusion of Exon
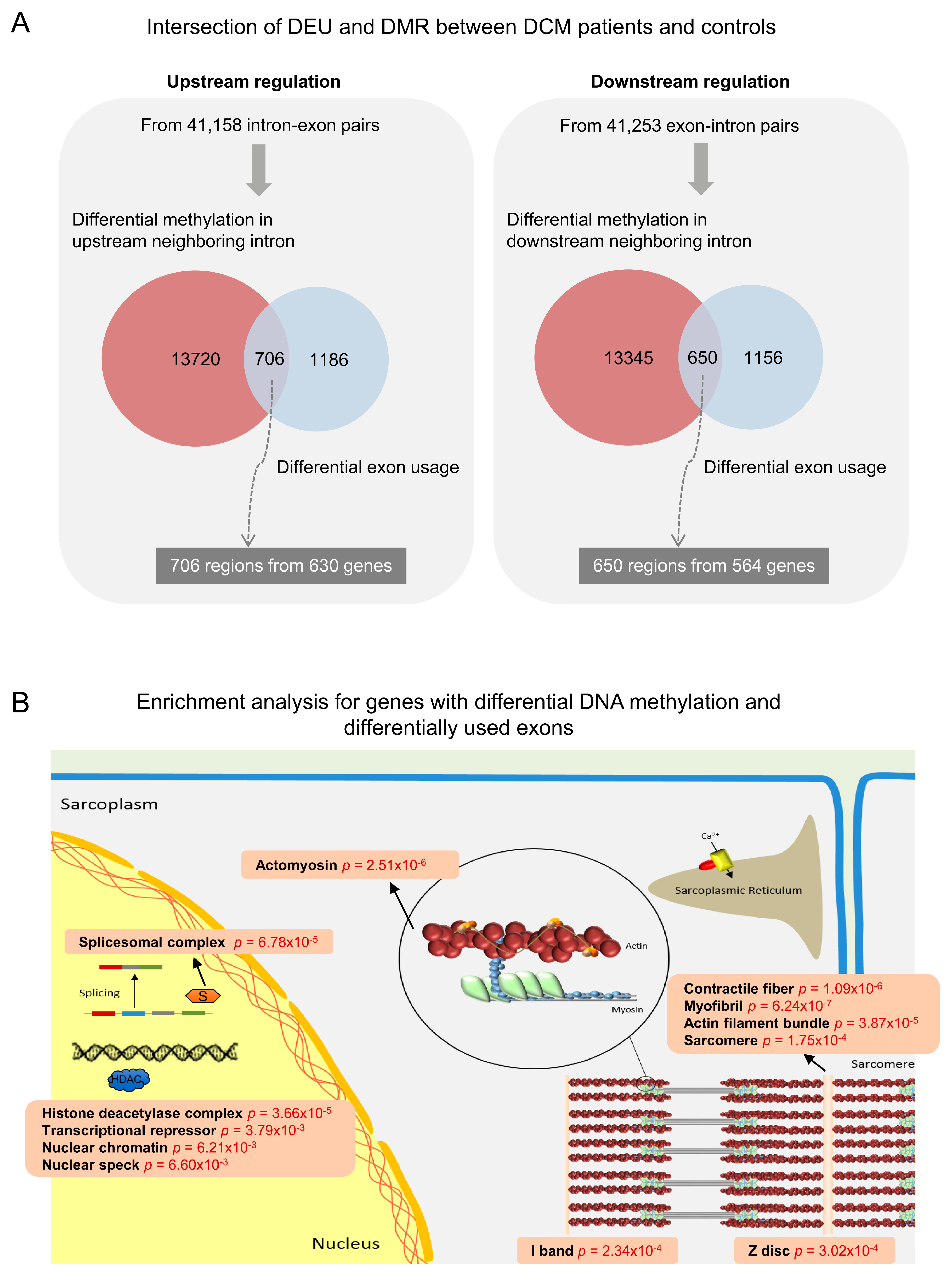
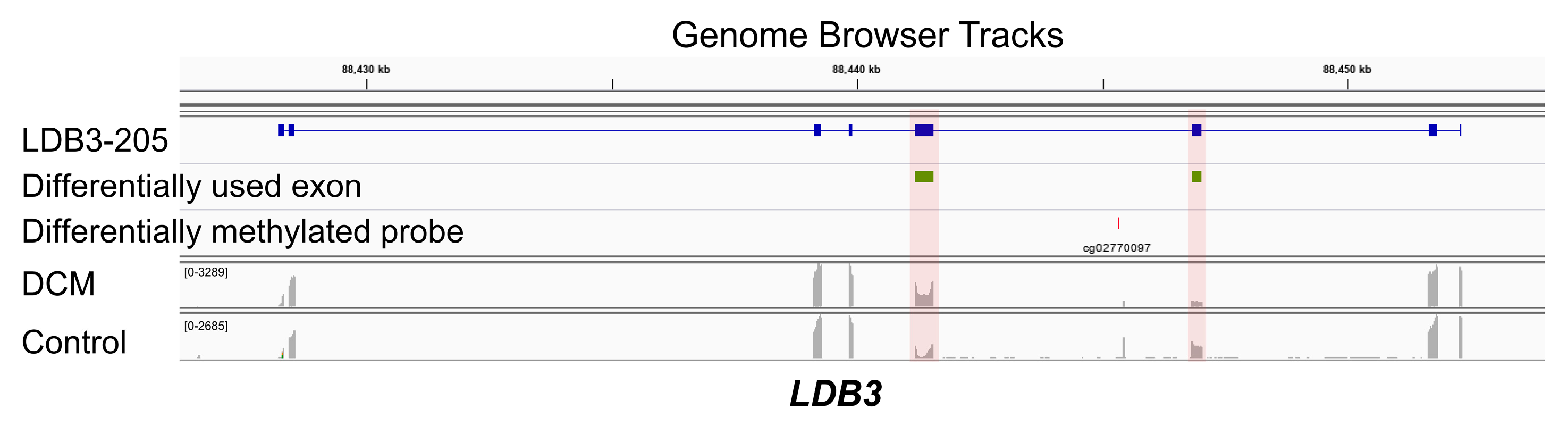
3.3. Co-Occurrence of Differential Exon Usage and Differential DNA Methylation between DCM and Control
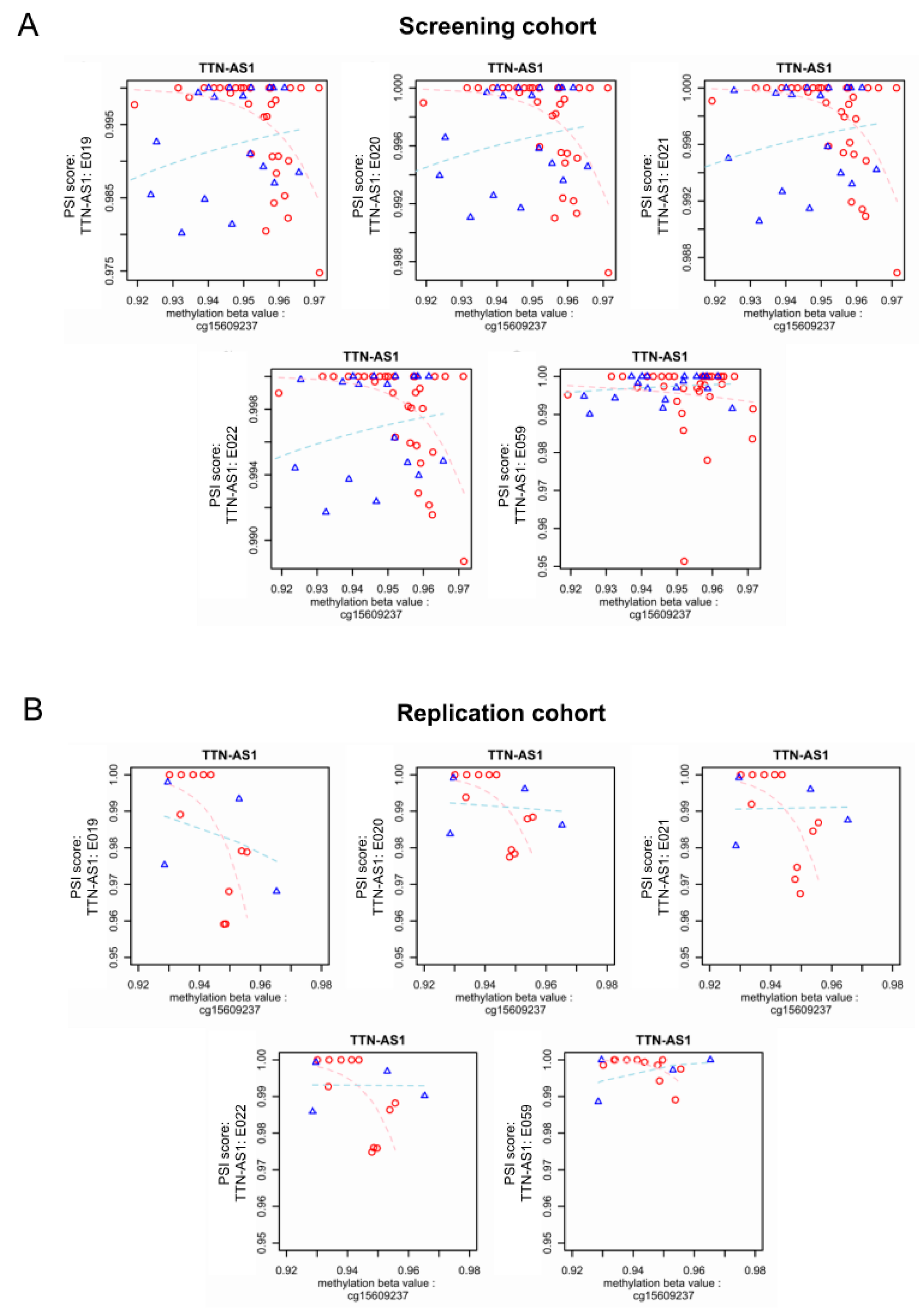
3.4. Epigenome-Wide Association of Intronic DNA Methylation and Splicing in DCM vs. Control
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hershberger, R.E.; Hedges, D.J.; Morales, A. Dilated cardiomyopathy: The complexity of a diverse genetic architecture. Nat. Rev. Cardiol. 2013, 10, 531–547. [Google Scholar] [CrossRef] [PubMed]
- McNally, E.M.; Mestroni, L. Dilated Cardiomyopathy: Genetic Determinants and Mechanisms. Circ. Res. 2017, 121, 731–748. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, S.K.; Arnett, D.K.; Assimes, T.L.; Basson, C.T.; Chakravarti, A.; Ellinor, P.T.; Engler, M.B.; Goldmuntz, E.; Herrington, D.M.; Hershberger, R.E.; et al. Genetics and genomics for the prevention and treatment of cardiovascular disease: Update: A scientific statement from the American Heart Association. Circulation 2013, 128, 2813–2851. [Google Scholar] [CrossRef] [PubMed]
- Towbin, J.A.; Bowles, N.E. The failing heart. Nature 2002, 415, 227–233. [Google Scholar] [CrossRef]
- Kayvanpour, E.; Sedaghat-Hamedani, F.; Amr, A.; Lai, A.; Haas, J.; Holzer, D.B.; Frese, K.S.; Keller, A.; Jensen, K.; Katus, H.A.; et al. Genotype-phenotype associations in dilated cardiomyopathy: Meta-analysis on more than 8000 individuals. Clin. Res. Cardiol. 2017, 106, 127–139. [Google Scholar] [CrossRef]
- Herman, D.S.; Lam, L.; Taylor, M.R.; Wang, L.; Teekakirikul, P.; Christodoulou, D.; Conner, L.; DePalma, S.R.; McDonough, B.; Sparks, E.; et al. Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med. 2012, 366, 619–628. [Google Scholar] [CrossRef]
- Paldino, A.; De Angelis, G.; Merlo, M.; Gigli, M.; Dal Ferro, M.; Severini, G.M.; Mestroni, L.; Sinagra, G. Genetics of Dilated Cardiomyopathy: Clinical Implications. Curr. Cardiol. Rep. 2018, 20, 83. [Google Scholar] [CrossRef]
- Meder, B.; Haas, J.; Sedaghat-Hamedani, F.; Kayvanpour, E.; Frese, K.; Lai, A.; Nietsch, R.; Scheiner, C.; Mester, S.; Bordalo, D.M.; et al. Epigenome-Wide Association Study Identifies Cardiac Gene Patterning and a Novel Class of Biomarkers for Heart Failure. Circulation 2017, 136, 1528–1544. [Google Scholar] [CrossRef]
- Pepin, M.E.; Drakos, S.; Ha, C.M.; Tristani-Firouzi, M.; Selzman, C.H.; Fang, J.C.; Wende, A.R.; Wever-Pinzon, O. DNA methylation reprograms cardiac metabolic gene expression in end-stage human heart failure. Am. J. Physiol. Heart Circ. Physiol. 2019, 317, H674–H684. [Google Scholar] [CrossRef]
- Ito, E.; Miyagawa, S.; Fukushima, S.; Yoshikawa, Y.; Saito, S.; Saito, T.; Harada, A.; Takeda, M.; Kashiyama, N.; Nakamura, Y.; et al. Histone modification is correlated with reverse left ventricular remodeling in nonischemic dilated cardiomyopathy. Ann. Thorac. Surg. 2017, 104, 1531–1539. [Google Scholar] [CrossRef]
- Deo, R.C. Alternative Splicing, Internal Promoter, Nonsense-Mediated Decay, or All Three: Explaining the Distribution of Truncation Variants in Titin. Circ. Cardiovasc. Genet. 2016, 9, 419–425. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sedaghat-Hamedani, F.; Haas, J.; Zhu, F.; Geier, C.; Kayvanpour, E.; Liss, M.; Lai, A.; Frese, K.; Pribe-Wolferts, R.; Amr, A.; et al. Clinical genetics and outcome of left ventricular non-compaction cardiomyopathy. Eur. Heart J. 2017, 38, 3449–3460. [Google Scholar] [CrossRef] [PubMed]
- Beqqali, A.; Bollen, I.A.; Rasmussen, T.B.; van den Hoogenhof, M.M.; van Deutekom, H.W.; Schafer, S.; Haas, J.; Meder, B.; Sørensen, K.E.; van Oort, R.J.; et al. A mutation in the glutamate-rich region of RNA-binding motif protein 20 causes dilated cardiomyopathy through missplicing of titin and impaired Frank-Starling mechanism. Cardiovasc. Res. 2016, 112, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Haas, J.; Mester, S.; Lai, A.; Frese, K.S.; Sedaghat-Hamedani, F.; Kayvanpour, E.; Rausch, T.; Nietsch, R.; Boeckel, J.N.; Carstensen, A.; et al. Genomic structural variations lead to dysregulation of important coding and non-coding RNA species in dilated cardiomyopathy. EMBO Mol. Med. 2018, 10, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. Available online: https://rpkgs.datanovia.com/factoextra/index.html (accessed on 1 March 2020).
- Siren, J.; Valimaki, N.; Makinen, V. Indexing Graphs for Path Queries with Applications in Genome Research. IEEE ACM Trans. Comput. Biol. Bioinform. 2014, 11, 375–388. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Anders, S.; Reyes, A.; Huber, W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012, 22, 2008–2017. [Google Scholar] [CrossRef]
- Schafer, S.; Miao, K.; Benson, C.C.; Heinig, M.; Cook, S.A.; Hubner, N. Alternative Splicing Signatures in RNA-seq Data: Percent Spliced in (PSI). Curr. Protoc. Hum. Genet. 2015, 87, 11–16. [Google Scholar] [CrossRef]
- Lawrence, M.; Huber, W.; Pagès, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.; Carey, V. Software for Computing and Annotating Genomic Ranges. PLoS Computat. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- GeneTrail2 v1.6 Statistica analysis and molecular signatures. Available online: https://genetrail2.bioinf.uni-sb.de/ (accessed on 23 May 2018).
- Daniel, S.; Tim, K.; Patrick, T.; Lara, S.; Christina, B.; Nicole, L.; Andreas, G.; Michael, K.; Manfred, G.; Norbert, G.; et al. Multi-omics Enrichment Analysis using the GeneTrail2 Web Service. Bioinformatics 2016, 32, 1502–1508. [Google Scholar]
- Bush, W.S.; Moore, J.H. Chapter 11: Genome-wide association studies. PLoS Comput. Biol. 2012, 8, e1002822. [Google Scholar] [CrossRef]
- Chanock, S.J.; Manolio, T.; Boehnke, M.; Boerwinkle, E.; Hunter, D.J.; Thomas, G.; Hirschhorn, J.N.; Abecasis, G.; Altshuler, D.; Bailey-Wilson, J.E.; et al. Replicating genotype-phenotype associations. Nature 2007, 447, 655–660. [Google Scholar]
- Diedenhofen, B.; Musch, J. cocor: A comprehensive solution for the statistical comparison of correlations. PLoS ONE 2015, 10, e0121945. [Google Scholar] [CrossRef]
- Do, C.; Shearer, A.; Suzuki, M.; Terry, M.B.; Gelernter, J.; Greally, J.M.; Tycko, B. Genetic-epigenetic interactions in cis: A major focus in the post-GWAS era. Genome Biol. 2017, 18, 120. [Google Scholar] [CrossRef]
- Wang, X.; Tucker, N.R.; Rizki, G.; Mills, R.; Krijger, P.H.; de Wit, E.; Subramanian, V.; Bartell, E.; Nguyen, X.X.; Ye, J.; et al. Discovery and validation of sub-threshold genome-wide association study loci using epigenomic signatures. Elife 2016, 5, e10557. [Google Scholar] [CrossRef]
- Pastinen, T.; Hudson, T.J. Cis-acting regulatory variation in the human genome. Science 2004, 306, 647–650. [Google Scholar] [CrossRef]
- Wan, J.; Oliver, V.F.; Zhu, H.; Zack, D.J.; Qian, J.; Merbs, S.L. Integrative analysis of tissue-specific methylation and alternative splicing identifies conserved transcription factor binding motifs. Nucleic Acids Res. 2013, 41, 8503–8514. [Google Scholar] [CrossRef]
- Beyer, A.L.; Osheim, Y.N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988, 2, 754–765. [Google Scholar] [CrossRef]
- Shukla, S.; Kavak, E.; Gregory, M.; Imashimizu, M.; Shutinoski, B.; Kashlev, M.; Oberdoerffer, P.; Sandberg, R.; Oberdoerffer, S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 2011, 479, 74–79. [Google Scholar] [CrossRef]
- Maunakea, A.K.; Chepelev, I.; Cui, K.; Zhao, K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013, 23, 1256–1269. [Google Scholar] [CrossRef]
- Lev Maor, G.; Yearim, A.; Ast, G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015, 31, 274–280. [Google Scholar] [CrossRef]
- Yearim, A.; Gelfman, S.; Shayevitch, R.; Melcer, S.; Glaich, O.; Mallm, J.P.; Nissim-Rafinia, M.; Cohen, A.H.S.; Rippe, K.; Meshorer, E.; et al. HP1 is involved in regulating the global impact of DNA methylation on alternative splicing. Cell Rep. 2015, 10, 1122–1134. [Google Scholar] [CrossRef]
- Luco, R.F.; Allo, M.; Schor, I.E.; Kornblihtt, A.R.; Misteli, T. Epigenetics in alternative pre-mRNA splicing. Cell 2011, 144, 16–26. [Google Scholar] [CrossRef]
- Vire, E.; Brenner, C.; Deplus, R.; Blanchon, L.; Fraga, M.; Didelot, C.; Morey, L.; Van Eynde, A.; Bernard, D.; Vanderwinden, J.M.; et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006, 439, 871–874. [Google Scholar] [CrossRef]
- Schlesinger, Y.; Straussman, R.; Keshet, I.; Farkash, S.; Hecht, M.; Zimmerman, J.; Eden, E.; Yakhini, Z.; Ben-Shushan, E.; Reubinoff, B.E.; et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007, 39, 232–236. [Google Scholar] [CrossRef]
- Bird, A. Molecular biology. Methylation talk between histones and DNA. Science 2001, 294, 2113–2115. [Google Scholar] [CrossRef]
- Kelly, T.K.; Liu, Y.; Lay, F.D.; Liang, G.; Berman, B.P.; Jones, P.A. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 2012, 22, 2497–2506. [Google Scholar] [CrossRef]
- Chodavarapu, R.K.; Feng, S.; Bernatavichute, Y.V.; Chen, P.Y.; Stroud, H.; Yu, Y.; Hetzel, J.A.; Kuo, F.; Kim, J.; Cokus, S.J.; et al. Relationship between nucleosome positioning and DNA methylation. Nature 2010, 466, 388–392. [Google Scholar] [CrossRef]
- Hashimshony, T.; Zhang, J.; Keshet, I.; Bustin, M.; Cedar, H. The role of DNA methylation in setting up chromatin structure during development. Nat. Genet. 2003, 34, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.E.; Meissner, A.; Lander, E.S. The mammalian epigenome. Cell 2007, 128, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Zhang, M.Q.; Zhang, X. Histone methylation marks play important roles in predicting the methylation status of CpG islands. Biochem. Biophys. Res. Commun. 2008, 374, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Cedar, H.; Bergman, Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat. Rev. Genet. 2009, 10, 295–304. [Google Scholar] [CrossRef]
- Gelfman, S.; Ast, G. When epigenetics meets alternative splicing: The roles of DNA methylation and GC architecture. Epigenomics 2013, 5, 351–353. [Google Scholar] [CrossRef]
- Ounzain, S.; Crippa, S.; Pedrazzini, T. Small and long non-coding RNAs in cardiac homeostasis and regeneration. Biochim. Biophys. Acta 2013, 1833, 923–933. [Google Scholar] [CrossRef]
- Calore, M.; De Windt, L.J.; Rampazzo, A. Genetics meets epigenetics: Genetic variants that modulate noncoding RNA in cardiovascular diseases. J. Mol. Cell. Cardiol. 2015, 89, 27–34. [Google Scholar] [CrossRef]
- Xie, Z.; Xia, W.; Hou, M. Long intergenic noncoding RNAp21 mediates cardiac senescence via the Wnt/betacatenin signaling pathway in doxorubicin-induced cardiotoxicity. Mol. Med. Rep. 2018, 17, 2695–2704. [Google Scholar]
- Thum, T.; Condorelli, G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ. Res. 2015, 116, 751–762. [Google Scholar] [CrossRef]
- Korostowski, L.; Sedlak, N.; Engel, N. The Kcnq1ot1 long non-coding RNA affects chromatin conformation and expression of Kcnq1, but does not regulate its imprinting in the developing heart. PLoS Genet. 2012, 8, e1002956. [Google Scholar] [CrossRef]
- Long, Q.Q.; Wang, H.; Gao, W.; Fan, Y.; Li, Y.F.; Ma, Y.; Yang, Y.; Shi, H.J.; Chen, B.R.; Meng, H.Y.; et al. Long Noncoding RNA Kcna2 Antisense RNA Contributes to Ventricular Arrhythmias via Silencing Kcna2 in Rats with Congestive Heart Failure. J. Am. Heart Assoc. 2017, 6, e005965. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Li, W.; Lin, C.H.; Yang, J.; Shang, C.; Nurnberg, S.T.; Jin, K.K.; Xu, W.; Lin, C.Y.; Lin, C.J.; et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014, 514, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Duan, Y.; Liu, T.; Wang, X.; Lv, W.; Wang, M.; Wang, J.; Liu, L. LncRNA TTN-AS1 promotes migration, invasion, and epithelial mesenchymal transition of lung adenocarcinoma via sponging miR-142-5p to regulate CDK5. Cell Death Dis. 2019, 10, 573. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Wang, R.; Yue, Q.; Hao, M. Long non-coding RNA TTN-AS1 promotes cell growth and metastasis in cervical cancer via miR-573/E2F3. Biochem. Biophys. Res. Commun. 2018, 503, 2956–2962. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Zhang, S.; Wang, Y.; Wang, Y.; Nice, E.; Guo, C.; Zhang, E.; Yu, L.; Li, M.; Liu, C.; et al. Functional Role of a Novel Long Noncoding RNA TTN-AS1 in Esophageal Squamous Cell Carcinoma Progression and Metastasis. Clin. Cancer Res. 2018, 24, 486–498. [Google Scholar] [CrossRef]
- Dong, M.M.; Peng, S.J.; Yuan, Y.N.; Luo, H.P. LncRNA TTN-AS1 contributes to gastric cancer progression by acting as a competing endogenous RNA of miR-376b-3p. Neoplasma 2019, 66, 564–575. [Google Scholar] [CrossRef]
- Cui, Z.; Luo, Z.; Lin, Z.; Shi, L.; Hong, Y.; Yan, C. Long non-coding RNA TTN-AS1 facilitates tumorigenesis of papillary thyroid cancer through modulating the miR-153-3p/ZNRF2 axis. J. Gene Med. 2019, 21, e3083. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, F.; Xiong, Y.; Cheng, X.; Qiu, Z.; Song, R. LncRNA TTN-AS1 sponges miR-376a-3p to promote colorectal cancer progression via upregulating KLF15. Life Sci. 2020, 244, 116936. [Google Scholar] [CrossRef]
- Zhu, Y.; Yang, Z.; Luo, X.H.; Xu, P. Long noncoding RNA TTN-AS1 promotes the proliferation and migration of prostate cancer by inhibiting miR-1271 level. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10678–10684. [Google Scholar]
- Roberts, A.M.; Ware, J.S.; Herman, D.S.; Schafer, S.; Baksi, J.; Bick, A.G.; Buchan, R.J.; Walsh, R.; John, S.; Wilkinson, S.; et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci. Transl. Med. 2015, 7, 270ra6. [Google Scholar] [CrossRef]
- Schafer, S.; de Marvao, A.; Adami, E.; Fiedler, L.R.; Ng, B.; Khin, E.; Rackham, O.J.; Van Heesch, S.; Pua, C.J.; Kui, M.; et al. Titin-truncating variants affect heart function in disease cohorts and the general population. Nat. Genet. 2017, 49, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Gramlich, M.; Pane, L.S.; Zhou, Q.; Chen, Z.; Murgia, M.; Schötterl, S.; Goedel, A.; Metzger, K.; Brade, T.; Parrotta, E.; et al. Antisense-mediated exon skipping: A therapeutic strategy for titin-based dilated cardiomyopathy. EMBO Mol. Med. 2015, 7, 562–576. [Google Scholar] [CrossRef] [PubMed]
- Hahn, J.K.; Neupane, B.; Pradhan, K.; Zhou, Q.; Testa, L.; Pelzl, L.; Maleck, C.; Gawaz, M.; Gramlich, M. The assembly and evaluation of antisense oligonucleotides applied in exon skipping for titin-based mutations in dilated cardiomyopathy. J. Mol. Cell. Cardiol. 2019, 131, 12–19. [Google Scholar] [CrossRef] [PubMed]
| Variables | Group | Coefficient | p Values | |
|---|---|---|---|---|
| Without adjustment | Methylation level in upstream flanking intron & | DCM | 0.310220 | <2 × 10−16 |
| Control | 0.319190 | <2 × 10−16 | ||
| Methylation level in downstream flanking intron $ | DCM | 0.333570 | <2 × 10−16 | |
| Control | 0.361230 | <2 × 10−16 | ||
| Adjusted by distance between exon and intron # | Methylation level in upstream flanking intron & | DCM | 0.315900 | <2 × 10−16 |
| Control | 0.325100 | <2 × 10−16 | ||
| Methylation level in downstream flanking intron $ | DCM | 0.336900 | <2 × 10−16 | |
| Control | 0.365900 | <2 × 10−16 |
| Odds Ratio | p Value | |||
|---|---|---|---|---|
| Variables | DCM | Control | DCM | Control |
| cg15609237 vs. TTN-AS1:E019 | 0.71 (0.08–3.02) | 1.18 (1.11–1.25) | 0.71 | 0.000018 |
| cg15609237 vs. TTN-AS1:E020 | 0.73 (0.06–3.46) | 1.17 (1.11–1.24) | 0.76 | 0.000017 |
| cg15609237 vs. TTN-AS1:E021 | 0.72 (0.07–3.17) | 1.18 (1.11–1.25) | 0.73 | 0.000024 |
| cg15609237 vs. TTN-AS1:E022 | 0.72 (0.07–3.12) | 1.18 (1.11–1.25) | 0.71 | 0.000013 |
| cg15609237 vs. TTN-AS1:E059 | 0.78 (0.14–2.53) | 1.18 (1.12–1.24) | 0.74 | 0.000006 |
| Odds Ratio | p Value | |||
|---|---|---|---|---|
| Variables | DCM | Control | DCM | Control |
| cg15609237 vs. TTN-AS1:E019 | 0.34 (0.13–0.77) | 4.75 (0.62-Inf) | 0.01 | 0.011 |
| cg15609237 vs. TTN-AS1:E020 | 0.35 (0.13–0.75) | 5.34 (0.85-Inf) | 0.01 | 0.003 |
| cg15609237 vs. TTN-AS1:E021 | 0.35 (0.13–0.78) | 5.07 (1.01-Inf) | 0.01 | 0.007 |
| cg15609237 vs. TTN-AS1:E022 | 0.36 (0.14–0.78) | 5.84 (0.92–73.01) | 0.01 | 0.007 |
| cg15609237 vs. TTN-AS1:E059 | 0.78 (0.14–2.53) | 7.83 (3.37–73.01) | 0.25 | 0.112 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gi, W.-T.; Haas, J.; Sedaghat-Hamedani, F.; Kayvanpour, E.; Tappu, R.; Lehmann, D.H.; Shirvani Samani, O.; Wisdom, M.; Keller, A.; Katus, H.A.; et al. Epigenetic Regulation of Alternative mRNA Splicing in Dilated Cardiomyopathy. J. Clin. Med. 2020, 9, 1499. https://doi.org/10.3390/jcm9051499
Gi W-T, Haas J, Sedaghat-Hamedani F, Kayvanpour E, Tappu R, Lehmann DH, Shirvani Samani O, Wisdom M, Keller A, Katus HA, et al. Epigenetic Regulation of Alternative mRNA Splicing in Dilated Cardiomyopathy. Journal of Clinical Medicine. 2020; 9(5):1499. https://doi.org/10.3390/jcm9051499
Chicago/Turabian StyleGi, Weng-Tein, Jan Haas, Farbod Sedaghat-Hamedani, Elham Kayvanpour, Rewati Tappu, David Hermann Lehmann, Omid Shirvani Samani, Michael Wisdom, Andreas Keller, Hugo A. Katus, and et al. 2020. "Epigenetic Regulation of Alternative mRNA Splicing in Dilated Cardiomyopathy" Journal of Clinical Medicine 9, no. 5: 1499. https://doi.org/10.3390/jcm9051499
APA StyleGi, W.-T., Haas, J., Sedaghat-Hamedani, F., Kayvanpour, E., Tappu, R., Lehmann, D. H., Shirvani Samani, O., Wisdom, M., Keller, A., Katus, H. A., & Meder, B. (2020). Epigenetic Regulation of Alternative mRNA Splicing in Dilated Cardiomyopathy. Journal of Clinical Medicine, 9(5), 1499. https://doi.org/10.3390/jcm9051499








