Long Non-Coding RNAs in Biliary Tract Cancer—An Up-to-Date Review
Abstract
1. Introduction
1.1. Biliary Tract Cancer
1.2. Long Non-Coding RNAs
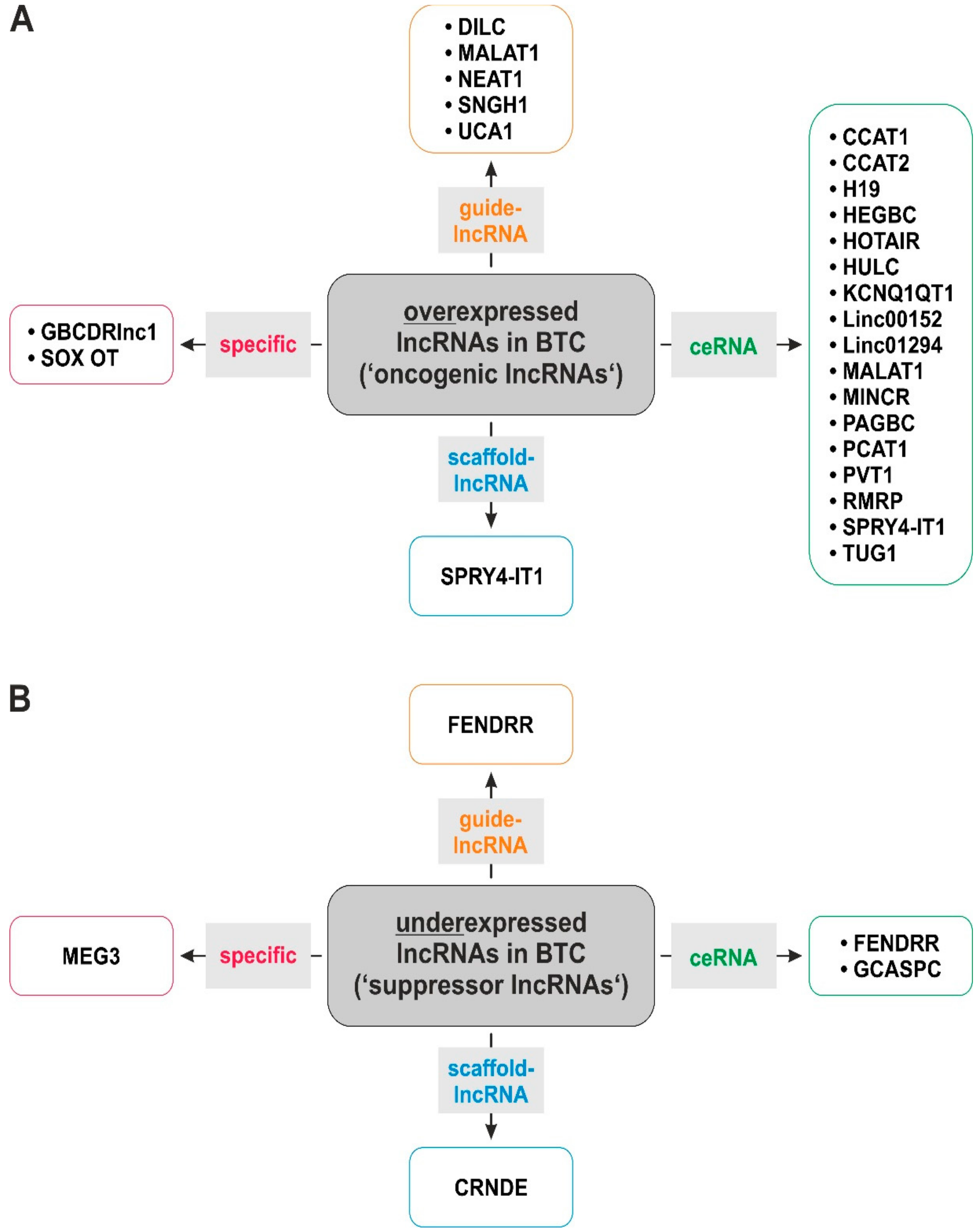
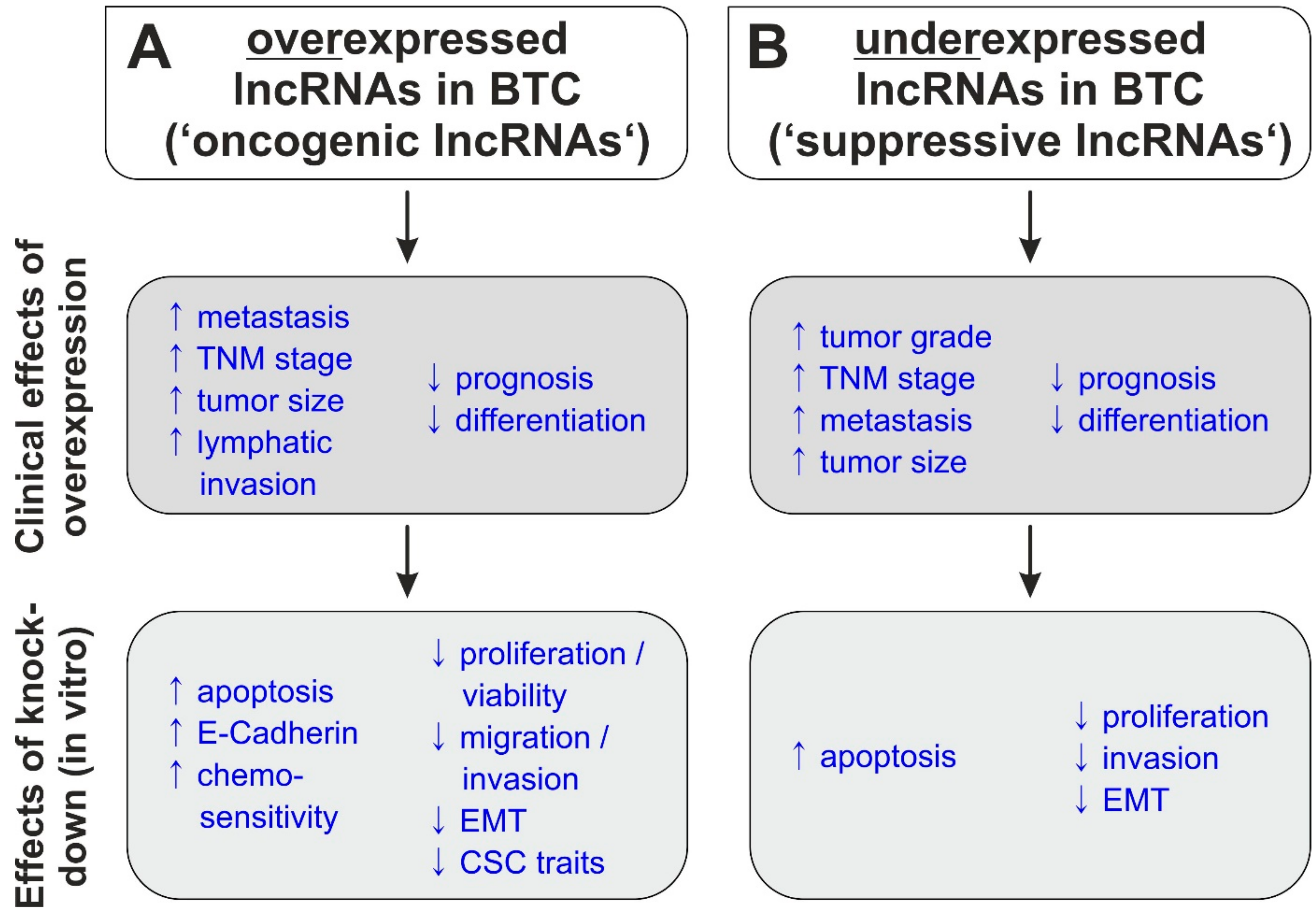
2. LncRNAs in Biliary Tract Cancer
3. DILC
4. EPIC1
5. CCAT 1
6. LINC00152
7. PAGBC
8. H19
9. PVT1
10. GBCDRlnc1
11. NEAT1
12. SNHG1
13. MALAT1
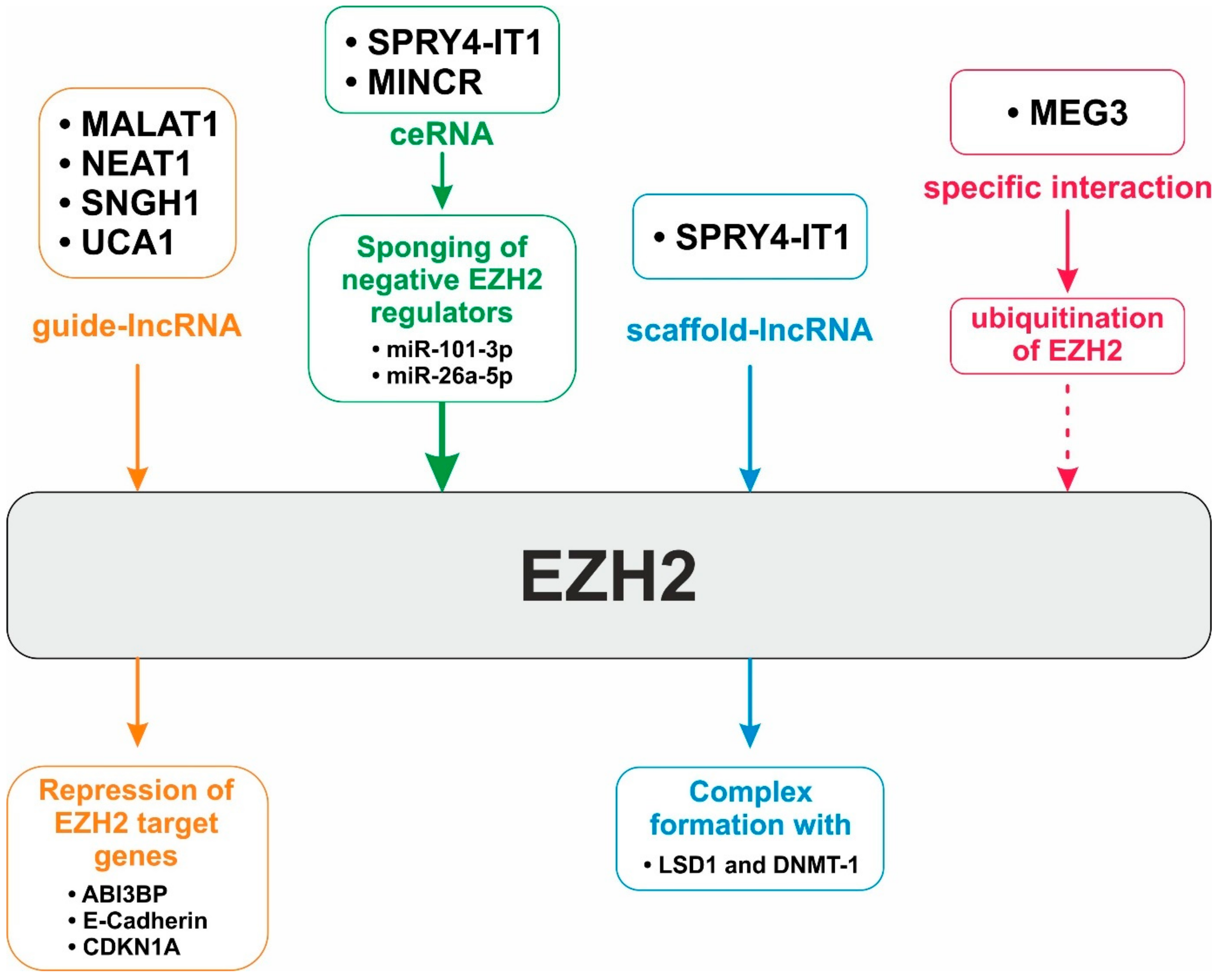
14. EZH2—A Major Target of LncRNA-based Regulation
15. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zhu, A.X.; Hong, T.S.; Hezel, A.F.; Kooby, D.A. Current management of gallbladder carcinoma. Oncologist 2010, 15, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, S.; Khan, S.A.; Hallemeier, C.L.; Kelley, R.K.; Gores, G.J. Cholangiocarcinoma—Evolving concepts and therapeutic strategies. Nat. Rev. Clin. Oncol. 2018, 15, 95–111. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, V.; Bragazzi, M.C.; Carpino, G.; Torrice, A.; Fraveto, A.; Gentile, R.; Pasqualino, V.; Melandro, F.; Aliberti, C.; Bastianelli, C.; et al. Cholangiocarcinoma: Increasing burden of classifications. Hepatobiliary Surg. Nutr. 2013, 2, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Kendall, T.; Verheij, J.; Gaudio, E.; Evert, M.; Guido, M.; Goeppert, B.; Carpino, G. Anatomical, histomorphological and molecular classification of cholangiocarcinoma. Liver Int. 2019, 39 (Suppl. 1), 7–18. [Google Scholar] [CrossRef]
- Patel, T. Increasing incidence and mortality of primary intrahepatic cholangiocarcinoma in the United States. Hepatology 2001, 33, 1353–1357. [Google Scholar] [CrossRef]
- Sripa, B.; Kaewkes, S.; Sithithaworn, P.; Mairiang, E.; Laha, T.; Smout, M.; Pairojkul, C.; Bhudhisawasdi, V.; Tesana, S.; Thinkamrop, B.; et al. Liver fluke induces cholangiocarcinoma. PLoS Med. 2007, 4, e201. [Google Scholar] [CrossRef]
- Yamamoto, S.; Kubo, S.; Hai, S.; Uenishi, T.; Yamamoto, T.; Shuto, T.; Takemura, S.; Tanaka, H.; Yamazaki, O.; Hirohashi, K.; et al. Hepatitis C virus infection as a likely etiology of intrahepatic cholangiocarcinoma. Cancer Sci. 2004, 95, 592–595. [Google Scholar] [CrossRef]
- Sripa, B.; Tangkawattana, S.; Sangnikul, T. The Lawa model: A sustainable, integrated opisthorchiasis control program using the EcoHealth approach in the Lawa Lake region of Thailand. Parasitol. Int. 2017, 66, 346–354. [Google Scholar] [CrossRef]
- Chapman, R.W. Risk factors for biliary tract carcinogenesis. Ann. Oncol. 1999, 10 (Suppl. 4), 308–311. [Google Scholar] [CrossRef]
- Shaib, Y.H.; El-Serag, H.B.; Davila, J.A.; Morgan, R.; McGlynn, K.A. Risk factors of intrahepatic cholangiocarcinoma in the United States: A case-control study. Gastroenterology 2005, 128, 620–626. [Google Scholar] [CrossRef]
- Nagorney, D.M.; Donohue, J.H.; Farnell, M.B.; Schleck, C.D.; Ilstrup, D.M. Outcomes after curative resections of cholangiocarcinoma. Arch. Surg. 1993, 128, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Rakic, M.; Patrlj, L.; Kopljar, M.; Klicek, R.; Kolovrat, M.; Loncar, B.; Busic, Z. Gallbladder cancer. Hepatobiliary Surg. Nutr. 2014, 3, 221–226. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Valle, J.; Wasan, H.; Palmer, D.H.; Cunningham, D.; Anthoney, A.; Maraveyas, A.; Madhusudan, S.; Iveson, T.; Hughes, S.; Pereira, S.P.; et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N. Engl. J. Med. 2010, 362, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Coburn, N.G.; Cleary, S.P.; Tan, J.C.; Law, C.H. Surgery for gallbladder cancer: A population-based analysis. J. Am. Coll. Surg. 2008, 207, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Moik, F.; Riedl, J.M.; Winder, T.; Terbuch, A.; Rossmann, C.H.; Szkandera, J.; Bauernhofer, T.; Kasparek, A.K.; Schaberl-Moser, R.; Reicher, A.; et al. Benefit of second-line systemic chemotherapy for advanced biliary tract cancer: A propensity score analysis. Sci. Rep. 2019, 9, 5548. [Google Scholar] [CrossRef]
- Kessler, S.M.; Lederer, E.; Laggai, S.; Golob-Schwarzl, N.; Hosseini, K.; Petzold, J.; Schweiger, C.; Reihs, R.; Keil, M.; Hoffmann, J.; et al. IMP2/IGF2BP2 expression, but not IMP1 and IMP3, predicts poor outcome in patients and high tumor growth rate in xenograft models of gallbladder cancer. Oncotarget 2017, 8, 89736–89745. [Google Scholar] [CrossRef]
- Khandelwal, A.; Malhotra, A.; Jain, M.; Vasquez, K.M.; Jain, A. The emerging role of long non-coding RNA in gallbladder cancer pathogenesis. Biochimie 2017, 132, 152–160. [Google Scholar] [CrossRef]
- Ling, H.; Vincent, K.; Pichler, M.; Fodde, R.; Berindan-Neagoe, I.; Slack, F.J.; Calin, G.A. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene 2015, 34, 5003–5011. [Google Scholar] [CrossRef]
- Khandelwal, A.; Bacolla, A.; Vasquez, K.M.; Jain, A. Long non-coding RNA: A new paradigm for lung cancer. Mol. Carcinog. 2015, 54, 1235–1251. [Google Scholar] [CrossRef]
- Brown, C.J.; Ballabio, A.; Rupert, J.L.; Lafreniere, R.G.; Grompe, M.; Tonlorenzi, R.; Willard, H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 1991, 349, 38–44. [Google Scholar] [CrossRef]
- Ohtsuka, M.; Ling, H.; Ivan, C.; Pichler, M.; Matsushita, D.; Goblirsch, M.; Stiegelbauer, V.; Shigeyasu, K.; Zhang, X.; Chen, M.; et al. H19 Noncoding RNA, an Independent Prognostic Factor, Regulates Essential Rb-E2F and CDK8-beta-Catenin Signaling in Colorectal Cancer. EBioMedicine 2016, 13, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010, 3, ra8. [Google Scholar] [CrossRef] [PubMed]
- Grote, P.; Herrmann, B.G. The long non-coding RNA Fendrr links epigenetic control mechanisms to gene regulatory networks in mammalian embryogenesis. RNA Biol. 2013, 10, 1579–1585. [Google Scholar] [CrossRef] [PubMed]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Chen, B.; Yu, M.; Chang, Q.; Lu, Y.; Thakur, C.; Ma, D.; Yi, Z.; Chen, F. Mdig de-represses H19 large intergenic non-coding RNA (lincRNA) by down-regulating H3K9me3 and heterochromatin. Oncotarget 2013, 4, 1427–1437. [Google Scholar] [CrossRef]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990, 10, 28–36. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Prensner, J.R.; Chinnaiyan, A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011, 1, 391–407. [Google Scholar] [CrossRef]
- Fernandes, J.C.R.; Acuna, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Noncoding RNA 2019, 5, 17. [Google Scholar] [CrossRef]
- Clark, M.B.; Johnston, R.L.; Inostroza-Ponta, M.; Fox, A.H.; Fortini, E.; Moscato, P.; Dinger, M.E.; Mattick, J.S. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012, 22, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, H.; Fang, S.; Kang, Y.; Wu, W.; Hao, Y.; Li, Z.; Bu, D.; Sun, N.; Zhang, M.Q.; et al. NONCODE 2016: An informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2015, 44, D203–D208. [Google Scholar] [CrossRef] [PubMed]
- Louro, R.; Smirnova, A.S.; Verjovski-Almeida, S. Long intronic noncoding RNA transcription: Expression noise or expression choice? Genomics 2009, 93, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Nie, L.; Wu, H.J.; Hsu, J.M.; Chang, S.S.; Labaff, A.M.; Li, C.W.; Wang, Y.; Hsu, J.L.; Hung, M.C. Long non-coding RNAs: Versatile master regulators of gene expression and crucial players in cancer. Am. J. Transl. Res. 2012, 4, 127–150. [Google Scholar] [PubMed]
- Kravchenko, J.E.; Rogozin, I.B.; Koonin, E.V.; Chumakov, P.M. Transcription of mammalian messenger RNAs by a nuclear RNA polymerase of mitochondrial origin. Nature 2005, 436, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Pichler, M.; Rodriguez-Aguayo, C.; Nam, S.Y.; Dragomir, M.P.; Bayraktar, R.; Anfossi, S.; Knutsen, E.; Ivan, C.; Fuentes-Mattei, E.; Lee, S.K.; et al. Therapeutic potential of FLANC, a novel primate-specific long non-coding RNA in colorectal cancer. Gut 2020. [Google Scholar] [CrossRef]
- Benetatos, L.; Vartholomatos, G.; Hatzimichael, E. MEG3 imprinted gene contribution in tumorigenesis. Int. J. Cancer 2011, 129, 773–779. [Google Scholar] [CrossRef]
- Bhat, S.A.; Ahmad, S.M.; Mumtaz, P.T.; Malik, A.A.; Dar, M.A.; Urwat, U.; Shah, R.A.; Ganai, N.A. Long non-coding RNAs: Mechanism of action and functional utility. Noncoding RNA Res. 2016, 1, 43–50. [Google Scholar] [CrossRef]
- Scheer, S.; Zaph, C. The Lysine Methyltransferase G9a in Immune Cell Differentiation and Function. Front. Immunol. 2017, 8, 429. [Google Scholar] [CrossRef]
- Huang, M.-D.; Chen, W.-M.; Qi, F.-Z.; Xia, R.; Sun, M.; Xu, T.-P.; Yin, L.; Zhang, E.-B.; De, W.; Shu, Y.-Q. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell apoptosis by epigenetic silencing of KLF2. J. Hematol. Oncol. 2015, 8, 50-50. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Rigoutsos, I.; Lee, S.K.; Nam, S.Y.; Anfossi, S.; Pasculli, B.; Pichler, M.; Jing, Y.; Rodriguez-Aguayo, C.; Telonis, A.G.; Rossi, S.; et al. N-BLR, a primate-specific non-coding transcript leads to colorectal cancer invasion and migration. Genome Biol. 2017, 18, 98. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, H.; Li, G.; Hu, J.; Liu, X.; Lin, L. LncRNA ANRIL promotes cell growth, migration and invasion of hepatocellular carcinoma cells via sponging miR-144. Anticancer Drugs 2019, 30, 1013–1021. [Google Scholar] [CrossRef]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16. [Google Scholar] [CrossRef]
- An, Y.; Furber, K.L.; Ji, S. Pseudogenes regulate parental gene expression via ceRNA network. J. Cell. Mol. Med. 2017, 21, 185–192. [Google Scholar] [CrossRef]
- Wang, X.; Hu, K.B.; Zhang, Y.Q.; Yang, C.J.; Yao, H.H. Comprehensive analysis of aberrantly expressed profiles of lncRNAs, miRNAs and mRNAs with associated ceRNA network in cholangiocarcinoma. Cancer Biomark. 2018, 23, 549–559. [Google Scholar] [CrossRef]
- Gibb, E.A.; Brown, C.J.; Lam, W.L. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer 2011, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhou, C.; Li, R.; Deng, Y.; Zhao, L.; Zhai, W. Long Noncoding RNA AFAP1-AS1 Promoted Tumor Growth and Invasion in Cholangiocarcinoma. Cell. Physiol. Biochem. 2017, 42, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Wang, S.H.; Cai, Q.; Zhang, M.D.; Yang, Y.; Ding, J. Overexpression of LncRNA AFAP1-AS1 predicts poor prognosis and promotes cells proliferation and invasion in gallbladder cancer. Biomed. Pharmacother. 2016, 84, 1249–1255. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Shen, E.D.; Liao, M.M.; Hu, Y.B.; Wu, K.; Yang, P.; Zhou, L.; Chen, W.D. Expression and mechanisms of long non-coding RNA genes MEG3 and ANRIL in gallbladder cancer. Tumour Biol. 2016, 37, 9875–9886. [Google Scholar] [CrossRef] [PubMed]
- Angenard, G.; Merdrignac, A.; Louis, C.; Edeline, J.; Coulouarn, C. Expression of long non-coding RNA ANRIL predicts a poor prognosis in intrahepatic cholangiocarcinoma. Dig. Liver Dis. 2019, 51, 1337–1343. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.L.; Li, A.J.; Hu, Z.Y.; Shang, F.S.; Wu, M.C. Coexpression of the carbamoylphosphate synthase 1 gene and its long noncoding RNA correlates with poor prognosis of patients with intrahepatic cholangiocarcinoma. Mol. Med. Rep. 2015, 12, 7915–7926. [Google Scholar] [CrossRef]
- Jiang, X.M.; Li, Z.L.; Li, J.L.; Zheng, W.Y.; Li, X.H.; Cui, Y.F.; Sun, D.J. LncRNA CCAT1 as the unfavorable prognostic biomarker for cholangiocarcinoma. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 1242–1247. [Google Scholar]
- Zhang, S.; Xiao, J.; Chai, Y.; Du, Y.Y.; Liu, Z.; Huang, K.; Zhou, X.; Zhou, W. LncRNA-CCAT1 Promotes Migration, Invasion, and EMT in Intrahepatic Cholangiocarcinoma Through Suppressing miR-152. Dig. Dis. Sci. 2017, 62, 3050–3058. [Google Scholar] [CrossRef]
- Ma, M.Z.; Chu, B.F.; Zhang, Y.; Weng, M.Z.; Qin, Y.Y.; Gong, W.; Quan, Z.W. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015, 6, e1583. [Google Scholar] [CrossRef]
- Bai, J.G.; Tang, R.F.; Shang, J.F.; Qi, S.; Yu, G.D.; Sun, C. Upregulation of long noncoding RNA CCAT2 indicates a poor prognosis and promotes proliferation and metastasis in intrahepatic cholangiocarcinoma. Mol. Med. Rep. 2018, 17, 5328–5335. [Google Scholar] [CrossRef]
- Liang, C.; Yang, P.; Han, T.; Wang, R.Y.; Xing, X.L.; Si, A.F.; Ma, Q.Y.; Chen, Z.; Li, H.Y.; Zhang, B. Long non-coding RNA DILC promotes the progression of gallbladder carcinoma. Gene 2019, 694, 102–110. [Google Scholar] [CrossRef]
- Li, Y.; Cai, Q.; Li, W.; Feng, F.; Yang, L. Long non-coding RNA EPIC1 promotes cholangiocarcinoma cell growth. Biochem. Biophys. Res. Commun. 2018, 504, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Wang, S.; Jin, L.; Weng, M.; Zhou, D.; Wang, J.; Tang, Z.; Quan, Z. Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol. Cancer 2019, 18, 82. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-T.; Ye, H.; Wei, P.-P.; Han, B.-W.; He, B.; Chen, Z.-H.; Chen, Y.-Q. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J. Hematol. Oncol. 2016, 9, 117. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Wu, X.C.; Zhang, M.D.; Weng, M.Z.; Zhou, D.; Quan, Z.W. Long noncoding RNA H19 contributes to gallbladder cancer cell proliferation by modulated miR-194-5p targeting AKT2. Tumour Biol. 2016, 37, 9721–9730. [Google Scholar] [CrossRef]
- Wang, S.H.; Ma, F.; Tang, Z.H.; Wu, X.C.; Cai, Q.; Zhang, M.D.; Weng, M.Z.; Zhou, D.; Wang, J.D.; Quan, Z.W. Long non-coding RNA H19 regulates FOXM1 expression by competitively binding endogenous miR-342-3p in gallbladder cancer. J. Exp. Clin. Cancer Res. 2016, 35, 160. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, Z.; Jiang, X.; Cui, Y. Overexpression of long noncoding RNA H19 indicates a poor prognosis for cholangiocarcinoma and promotes cell migration and invasion by affecting epithelial-mesenchymal transition. Biomed. Pharmacother. 2017, 92, 17–23. [Google Scholar] [CrossRef]
- Hu, Y.P.; Jin, Y.P.; Wu, X.S.; Yang, Y.; Li, Y.S.; Li, H.F.; Xiang, S.S.; Song, X.L.; Jiang, L.; Zhang, Y.J.; et al. LncRNA-HGBC stabilized by HuR promotes gallbladder cancer progression by regulating miR-502-3p/SET/AKT axis. Mol. Cancer 2019, 18, 167. [Google Scholar] [CrossRef]
- Yang, L.; Gao, Q.; Wu, X.; Feng, F.; Xu, K. Long noncoding RNA HEGBC promotes tumorigenesis and metastasis of gallbladder cancer via forming a positive feedback loop with IL-11/STAT3 signaling pathway. J. Exp. Clin. Cancer Res. 2018, 37, 186. [Google Scholar] [CrossRef]
- Qin, W.; Kang, P.; Xu, Y.; Leng, K.; Li, Z.; Huang, L.; Gao, J.; Cui, Y.; Zhong, X. Long non-coding RNA HOTAIR promotes tumorigenesis and forecasts a poor prognosis in cholangiocarcinoma. Sci. Rep. 2018, 8, 12176. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.Z.; Li, C.X.; Zhang, Y.; Weng, M.Z.; Zhang, M.D.; Qin, Y.Y.; Gong, W.; Quan, Z.W. Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol. Cancer 2014, 13, 156. [Google Scholar] [CrossRef]
- Zhang, P.; Cao, P.; Zhu, X.; Pan, M.; Zhong, K.; He, R.; Li, Y.; Jiao, X.; Gao, Y. Upregulation of long non-coding RNA HOXA-AS2 promotes proliferation and induces epithelial-mesenchymal transition in gallbladder carcinoma. Oncotarget 2017, 8, 33137–33143. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Li, Y.; Kong, H.; Dai, S.; Qian, H. Dysregulation of KCNQ1OT1 promotes cholangiocarcinoma progression via miR-140-5p/SOX4 axis. Arch. Biochem. Biophys. 2018, 658, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Wang, Z.; Wang, S.; Weng, M.; Zhou, D.; Li, C.; Wang, J.; Chen, E.; Quan, Z. Long non-coding RNA LINC00152 promotes gallbladder cancer metastasis and epithelial-mesenchymal transition by regulating HIF-1alpha via miR-138. Open Biol. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Wang, Z.Q.; Wang, S.H.; Li, C.; Zhu, Z.G.; Quan, Z.W.; Zhang, W.J. Upregulation of long non-coding RNA LINC00152 by SP1 contributes to gallbladder cancer cell growth and tumor metastasis via PI3K/AKT pathway. Am. J. Transl. Res. 2016, 8, 4068–4081. [Google Scholar] [PubMed]
- Zhang, D.; Li, H.; Xie, J.; Jiang, D.; Cao, L.; Yang, X.; Xue, P.; Jiang, X. Long noncoding RNA LINC01296 promotes tumor growth and progression by sponging miR-5095 in human cholangiocarcinoma. Int. J. Oncol. 2018, 52, 1777–1786. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.C.; Wang, S.H.; Ou, H.H.; Zhu, B.; Zhu, Y.; Zhang, Q.; Yang, Y.; Li, H. The NmrA-like family domain containing 1 pseudogene Loc344887 is amplified in gallbladder cancer and promotes epithelial-mesenchymal transition. Chem. Biol. Drug Des. 2017, 90, 456–463. [Google Scholar] [CrossRef]
- Wang, S.H.; Zhang, W.J.; Wu, X.C.; Weng, M.Z.; Zhang, M.D.; Cai, Q.; Zhou, D.; Wang, J.D.; Quan, Z.W. The lncRNA MALAT1 functions as a competing endogenous RNA to regulate MCL-1 expression by sponging miR-363-3p in gallbladder cancer. J. Cell. Mol. Med. 2016, 20, 2299–2308. [Google Scholar] [CrossRef]
- Lin, N.; Yao, Z.; Xu, M.; Chen, J.; Lu, Y.; Yuan, L.; Zhou, S.; Zou, X.; Xu, R. Long noncoding RNA MALAT1 potentiates growth and inhibits senescence by antagonizing ABI3BP in gallbladder cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 244. [Google Scholar] [CrossRef]
- Wang, C.; Mao, Z.P.; Wang, L.; Wu, G.H.; Zhang, F.H.; Wang, D.Y.; Shi, J.L. Long non-coding RNA MALAT1 promotes cholangiocarcinoma cell proliferation and invasion by activating PI3K/Akt pathway. Neoplasma 2017, 64, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Zhang, W.J.; Wu, X.C.; Zhang, M.D.; Weng, M.Z.; Zhou, D.; Wang, J.D.; Quan, Z.W. Long non-coding RNA Malat1 promotes gallbladder cancer development by acting as a molecular sponge to regulate miR-206. Oncotarget 2016, 7, 37857–37867. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.S.; Wang, X.A.; Wu, W.G.; Hu, Y.P.; Li, M.L.; Ding, Q.; Weng, H.; Shu, Y.J.; Liu, T.Y.; Jiang, L.; et al. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol. Ther. 2014, 15, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Yang, Y.; Wu, X.C.; Zhang, M.D.; Weng, M.Z.; Zhou, D.; Wang, J.D.; Quan, Z.W. Long non-coding RNA MINCR promotes gallbladder cancer progression through stimulating EZH2 expression. Cancer Lett. 2016, 380, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Parasramka, M.; Yan, I.K.; Wang, X.; Nguyen, P.; Matsuda, A.; Maji, S.; Foye, C.; Asmann, Y.; Patel, T. BAP1 dependent expression of long non-coding RNA NEAT-1 contributes to sensitivity to gemcitabine in cholangiocarcinoma. Mol. Cancer 2017, 16, 22. [Google Scholar] [CrossRef]
- Zhang, C.; Li, J.Y.; Tian, F.Z.; Zhao, G.; Hu, H.; Ma, Y.F.; Yang, Y.L. Long Noncoding RNA NEAT1 Promotes Growth and Metastasis of Cholangiocarcinoma Cells. Oncol. Res. 2018, 26, 879–888. [Google Scholar] [CrossRef]
- Wu, X.S.; Wang, F.; Li, H.F.; Hu, Y.P.; Jiang, L.; Zhang, F.; Li, M.L.; Wang, X.A.; Jin, Y.P.; Zhang, Y.J.; et al. LncRNA-PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO Rep. 2017, 18, 1837–1853. [Google Scholar] [CrossRef]
- Xu, Y.; Jiang, X.; Cui, Y. Upregulated long noncoding RNA PANDAR predicts an unfavorable prognosis and promotes tumorigenesis in cholangiocarcinoma. Oncol. Targets Ther. 2017, 10, 2873–2883. [Google Scholar] [CrossRef]
- Zhang, F.; Wan, M.; Xu, Y.; Li, Z.; Leng, K.; Kang, P.; Cui, Y.; Jiang, X. Long noncoding RNA PCAT1 regulates extrahepatic cholangiocarcinoma progression via the Wnt/beta-catenin-signaling pathway. Biomed. Pharmacother. 2017, 94, 55–62. [Google Scholar] [CrossRef]
- Chen, J.; Yu, Y.; Li, H.; Hu, Q.; Chen, X.; He, Y.; Xue, C.; Ren, F.; Ren, Z.; Li, J.; et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol. Cancer 2019, 18, 33. [Google Scholar] [CrossRef]
- Tang, L.; Wang, Y.; Wang, H.; Xu, B.; Ji, H.; Xu, G.; Ge, X.; Li, Q.; Miao, L. Long noncoding-RNA component of mitochondrial RNA processing endoribonuclease is involved in the progression of cholangiocarcinoma by regulating microRNA-217. Cancer Sci. 2019, 110, 2166–2179. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Zhang, M.D.; Wu, X.C.; Weng, M.Z.; Zhou, D.; Quan, Z.W. Overexpression of LncRNA-ROR predicts a poor outcome in gallbladder cancer patients and promotes the tumor cells proliferation, migration, and invasion. Tumour Biol. 2016, 37, 12867–12875. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, M.; Wang, N.; Li, Q.; Yang, J.; Yan, S.; He, X.; Ji, G.; Miao, L. Epigenetic silencing of tumor suppressor gene CDKN1A by oncogenic long non-coding RNA SNHG1 in cholangiocarcinoma. Cell Death Dis. 2018, 9, 746. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, X.; Du, X.; Zhang, H.; Wu, Z.; Ren, K.; Han, X. The Interaction Between lncRNA SNHG1 and miR-140 in Regulating Growth and Tumorigenesis via the TLR4/NF-kappaB Pathway in Cholangiocarcinoma. Oncol. Res. 2019, 27, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.X.; Wong, H.; Xu, F.; Liu, Z.; Ran, L.; Jiang, R.D. IRF4-induced upregulation of lncRNA SOX2-OT promotes cell proliferation and metastasis in cholangiocarcinoma by regulating SOX2 and PI3K/AKT signaling. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8169–8178. [Google Scholar] [CrossRef]
- Li, Z.; Li, J.; Ji, D.; Leng, K.; Xu, Y.; Huang, L.; Jiang, X.; Cui, Y. Overexpressed long noncoding RNA Sox2ot predicts poor prognosis for cholangiocarcinoma and promotes cell proliferation and invasion. Gene 2018, 645, 131–136. [Google Scholar] [CrossRef]
- Xu, Y.; Yao, Y.; Jiang, X.; Zhong, X.; Wang, Z.; Li, C.; Kang, P.; Leng, K.; Ji, D.; Li, Z.; et al. SP1-induced upregulation of lncRNA SPRY4-IT1 exerts oncogenic properties by scaffolding EZH2/LSD1/DNMT1 and sponging miR-101-3p in cholangiocarcinoma. J. Exp. Clin. Cancer Res. 2018, 37, 81. [Google Scholar] [CrossRef]
- Yang, L.; Cheng, X.; Ge, N.; Guo, W.; Feng, F.; Wan, F. Long non-coding RNA SPRY4-IT1 promotes gallbladder carcinoma progression. Oncotarget 2017, 8, 3104–3110. [Google Scholar] [CrossRef]
- Yao, Y.; Sun, Y.; Jiang, Y.; Qu, L.; Xu, Y. Enhanced expression of lncRNA TP73-AS1 predicts adverse phenotypes for cholangiocarcinoma and exerts oncogenic properties in vitro and in vivo. Biomed. Pharmacother. 2018, 106, 260–266. [Google Scholar] [CrossRef]
- Ma, F.; Wang, S.H.; Cai, Q.; Jin, L.Y.; Zhou, D.; Ding, J.; Quan, Z.W. Long non-coding RNA TUG1 promotes cell proliferation and metastasis by negatively regulating miR-300 in gallbladder carcinoma. Biomed. Pharmacother. 2017, 88, 863–869. [Google Scholar] [CrossRef]
- Cai, Q.; Jin, L.; Wang, S.; Zhou, D.; Wang, J.; Tang, Z.; Quan, Z. Long non-coding RNA UCA1 promotes gallbladder cancer progression by epigenetically repressing p21 and E-cadherin expression. Oncotarget 2017, 8, 47957–47968. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shen, S.; Liu, H.; Wang, Y.; Wang, J.; Ni, X.; Ai, Z.; Pan, H.; Liu, H.; Shao, Y. Long non-coding RNA CRNDE promotes gallbladder carcinoma carcinogenesis and as a scaffold of DMBT1 and C-IAP1 complexes to activating PI3K-AKT pathway. Oncotarget 2016, 7, 72833–72844. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Lu, M.; Zhou, Y.; Li, G.; Liu, Z. LncRNA FENDRR represses proliferation, migration and invasion through suppression of survivin in cholangiocarcinoma cells. Cell Cycle 2019, 18, 889–897. [Google Scholar] [CrossRef]
- Zhang, L.; Geng, Z.; Meng, X.; Meng, F.; Wang, L. Screening for key lncRNAs in the progression of gallbladder cancer using bioinformatics analyses. Mol. Med. Rep. 2018, 17, 6449–6455. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.Z.; Zhang, Y.; Weng, M.Z.; Wang, S.H.; Hu, Y.; Hou, Z.Y.; Qin, Y.Y.; Gong, W.; Zhang, Y.J.; Kong, X.; et al. Long Noncoding RNA GCASPC, a Target of miR-17-3p, Negatively Regulates Pyruvate Carboxylase-Dependent Cell Proliferation in Gallbladder Cancer. Cancer Res. 2016, 76, 5361–5371. [Google Scholar] [CrossRef]
- Ma, M.Z.; Kong, X.; Weng, M.Z.; Zhang, M.D.; Qin, Y.Y.; Gong, W.; Zhang, W.J.; Quan, Z.W. Long non-coding RNA-LET is a positive prognostic factor and exhibits tumor-suppressive activity in gallbladder cancer. Mol. Carcinog. 2015, 54, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Cai, Q.; Wang, S.; Wang, S.; Mondal, T.; Wang, J.; Quan, Z. Long noncoding RNA MEG3 regulates LATS2 by promoting the ubiquitination of EZH2 and inhibits proliferation and invasion in gallbladder cancer. Cell Death Dis. 2018, 9, 1017. [Google Scholar] [CrossRef]
- Mayr, C.; Neureiter, D.; Wagner, A.; Pichler, M.; Kiesslich, T. The role of polycomb repressive complexes in biliary tract cancer. Expert Opin. Ther. Targets 2015, 19, 363–375. [Google Scholar] [CrossRef]
- Kreso, A.; Dick, J.E. Evolution of the cancer stem cell model. Cell Stem Cell 2014, 14, 275–291. [Google Scholar] [CrossRef]
- Yu, D.; Shin, H.-S.; Lee, Y.S.; Lee, Y.C. miR-106b modulates cancer stem cell characteristics through TGF-β/Smad signaling in CD44-positive gastric cancer cells. Lab. Investig. 2014, 94, 1370–1381. [Google Scholar] [CrossRef]
- Wang, X.; Sun, W.; Shen, W.; Xia, M.; Chen, C.; Xiang, D.; Ning, B.; Cui, X.; Li, H.; Li, X.; et al. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J. Hepatol. 2016, 64, 1283–1294. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cai, H.-B.; Chen, L.-L.; Zhao, W.-J.; Li, P.; Wang, Z.-Q.; Li, Z. STAT3 correlates with stem cell-related transcription factors in cervical cancer. J. Huazhong Univ. Sci. Technol. Med. Sci. 2015, 35, 891–897. [Google Scholar] [CrossRef] [PubMed]
- S Franco, S.; Szczesna, K.; Iliou, M.S.; Al-Qahtani, M.; Mobasheri, A.; Kobolák, J.; Dinnyés, A. In vitro models of cancer stem cells and clinical applications. BMC Cancer 2016, 16, 738-738. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C.; Ocker, M.; Ritter, M.; Pichler, M.; Neureiter, D.; Kiesslich, T. Biliary tract cancer stem cells—Translational options and challenges. World J. Gastroenterol. 2017, 23, 2470–2482. [Google Scholar] [CrossRef]
- Dick, J.E. Looking ahead in cancer stem cell research. Nat. Biotechnol. 2009, 27, 44–46. [Google Scholar] [CrossRef]
- Mayr, C.; Wagner, A.; Stoecklinger, A.; Jakab, M.; Illig, R.; Berr, F.; Pichler, M.; Di Fazio, P.; Ocker, M.; Neureiter, D.; et al. 3-Deazaneplanocin A May Directly Target Putative Cancer Stem Cells in Biliary Tract Cancer. Anticancer Res. 2015, 35, 4697–4705. [Google Scholar]
- Mayr, C.; Wagner, A.; Loeffelberger, M.; Bruckner, D.; Jakab, M.; Berr, F.; Di Fazio, P.; Ocker, M.; Neureiter, D.; Pichler, M.; et al. The BMI1 inhibitor PTC-209 is a potential compound to halt cellular growth in biliary tract cancer cells. Oncotarget 2016, 7, 745–758. [Google Scholar] [CrossRef]
- Kemmerling, R.; Alinger, B.; Dietze, O.; Bösmüller, H.C.; Ocker, M.; Wolkersdörfer, G.W.; Berr, F.; Neureiter, D.; Kiesslich, T. Association of stem cell marker expression pattern and survival in human biliary tract cancer. Int. J. Oncol. 2012, 41, 511–522. [Google Scholar] [CrossRef]
- Beyreis, M.; Gaisberger, M.; Jakab, M.; Neureiter, D.; Helm, K.; Ritter, M.; Kiesslich, T.; Mayr, C. The Cancer Stem Cell Inhibitor Napabucasin (BBI608) Shows General Cytotoxicity in Biliary Tract Cancer Cells and Reduces Cancer Stem Cell Characteristics. Cancers 2019, 11, 276. [Google Scholar] [CrossRef]
- Tarlow, B.D.; Pelz, C.; Naugler, W.E.; Wakefield, L.; Wilson, E.M.; Finegold, M.J.; Grompe, M. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell 2014, 15, 605–618. [Google Scholar] [CrossRef]
- Kokuryo, T.; Yokoyama, Y.; Nagino, M. Recent advances in cancer stem cell research for cholangiocarcinoma. J. Hepatobiliary Pancreat. Sci. 2012, 19, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Iwahashi, S.; Utsunomiya, T.; Shimada, M.; Saito, Y.; Morine, Y.; Imura, S.; Ikemoto, T.; Mori, H.; Hanaoka, J.; Bando, Y. High expression of cancer stem cell markers in cholangiolocellular carcinoma. Surg. Today 2013, 43, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Yasuchika, K.; Suemori, H.; Nakatsuji, N.; Ikai, I.; Uemoto, S. Alpha-fetoprotein producing cells act as cancer progenitor cells in human cholangiocarcinoma. Cancer Lett. 2010, 294, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Malato, Y.; Calvisi, D.F.; Naqvi, S.; Razumilava, N.; Ribback, S.; Gores, G.J.; Dombrowski, F.; Evert, M.; Chen, X.; et al. Cholangiocarcinomas can originate from hepatocytes in mice. J. Clin. Investig. 2012, 122, 2911–2915. [Google Scholar] [CrossRef]
- Wang, M.; Xiao, J.; Shen, M.; Yahong, Y.; Tian, R.; Zhu, F.; Jiang, J.; Du, Z.; Hu, J.; Liu, W.; et al. Isolation and characterization of tumorigenic extrahepatic cholangiocarcinoma cells with stem cell-like properties. Int. J. Cancer 2011, 128, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.J.; Gao, J.; Wang, M.; Wang, X.; Tian, R.; Zhu, F.; Shen, M.; Qin, R.Y. CD133(+) gallbladder carcinoma cells exhibit self-renewal ability and tumorigenicity. World J. Gastroenterol. 2011, 17, 2965–2971. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Liu, C.; Zhao, Q.; Lu, J.; Ding, X.; Luo, A.; He, J.; Wang, G.; Li, Y.; Cai, Z.; et al. Long non-coding RNA CCAT2 promotes oncogenesis in triple-negative breast cancer by regulating stemness of cancer cells. Pharmacol. Res. 2020, 152, 104628. [Google Scholar] [CrossRef]
- Zhan, Y.; Chen, Z.; He, S.; Gong, Y.; He, A.; Li, Y.; Zhang, L.; Zhang, X.; Fang, D.; Li, X.; et al. Long non-coding RNA SOX2OT promotes the stemness phenotype of bladder cancer cells by modulating SOX2. Mol. Cancer 2020, 19, 25. [Google Scholar] [CrossRef]
- Gu, L.-Q.; Xing, X.-L.; Cai, H.; Si, A.-F.; Hu, X.-R.; Ma, Q.-Y.; Zheng, M.-L.; Wang, R.-Y.; Li, H.-Y.; Zhang, X.-P. Long non-coding RNA DILC suppresses cell proliferation and metastasis in colorectal cancer. Gene 2018, 666, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Ji, J.; Budhu, A.; Forgues, M.; Yang, W.; Wang, H.-Y.; Jia, H.; Ye, Q.; Qin, L.-X.; Wauthier, E.; et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology 2009, 136, 1012–1024. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.L.; Yadav, A.; Gupta, A.; Tulsayan, S.; Kumar, V.; Misra, S.; Kumar, A.; Mittal, B. Association of genetic variants of cancer stem cell gene CD44 haplotypes with gallbladder cancer susceptibility in North Indian population. Tumour Biol. 2014, 35, 2583–2589. [Google Scholar] [CrossRef]
- Yadav, A.; Gupta, A.; Rastogi, N.; Agrawal, S.; Kumar, A.; Kumar, V.; Mittal, B. Association of cancer stem cell markers genetic variants with gallbladder cancer susceptibility, prognosis, and survival. Tumour Biol. 2016, 37, 1835–1844. [Google Scholar] [CrossRef] [PubMed]
- Sebio, A.; Kahn, M.; Lenz, H.-J. The potential of targeting Wnt/β-catenin in colon cancer. Expert Opin. Ther. Targets 2014, 18, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, B.; Zhang, M.; Guo, W.; Wu, Z.; Wang, Y.; Jia, L.; Li, S.; Xie, W.; Yang, D. lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer. Cancer Cell 2018, 33, 706–720. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Lu, H.Y.; Xia, Y.H.; Jiang, A.G.; Lv, Y.X. Long non-coding RNA EPIC1 promotes human lung cancer cell growth. Biochem. Biophys. Res. Commun. 2018, 503, 1342–1348. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Liu, P.; Fu, Q.; Liu, C.; Luo, Q.; Zhang, X.; Cheng, L.; Qin, T.; Zhang, H. Long noncoding RNA EPIC1 interacts with YAP1 to regulate the cell cycle and promote the growth of pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2020, 522, 978–985. [Google Scholar] [CrossRef]
- Zeller, K.I.; Jegga, A.G.; Aronow, B.J.; O’Donnell, K.A.; Dang, C.V. An integrated database of genes responsive to the Myc oncogenic transcription factor: Identification of direct genomic targets. Genome Biol. 2003, 4, R69. [Google Scholar] [CrossRef]
- Bretones, G.; Delgado, M.D.; León, J. Myc and cell cycle control. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2015, 1849, 506–516. [Google Scholar] [CrossRef]
- Carroll, P.A.; Freie, B.W.; Mathsyaraja, H.; Eisenman, R.N. The MYC transcription factor network: Balancing metabolism, proliferation and oncogenesis. Front. Med. 2018, 12, 412–425. [Google Scholar] [CrossRef]
- Nissan, A.; Stojadinovic, A.; Mitrani-Rosenbaum, S.; Halle, D.; Grinbaum, R.; Roistacher, M.; Bochem, A.; Dayanc, B.E.; Ritter, G.; Gomceli, I.; et al. Colon cancer associated transcript-1: A novel RNA expressed in malignant and pre-malignant human tissues. Int. J. Cancer 2012, 130, 1598–1606. [Google Scholar] [CrossRef]
- Wang, C.; Chen, F.; Fan, Z.; Yao, C.; Xiao, L. LncRNA CCAT1/miR-490-3p/MAPK1/c-Myc Positive Feedback Loop Drives Progression of Acute Myeloid Leukemia. J. Biochem. 2019. [Google Scholar] [CrossRef]
- Tang, T.; Guo, C.; Xia, T.; Zhang, R.; Zen, K.; Pan, Y.; Jin, L. LncCCAT1 Promotes Breast Cancer Stem Cell Function through Activating WNT/beta-catenin Signaling. Theranostics 2019, 9, 7384–7402. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, G.; Ma, Y.; Qu, H. lncRNA CCAT1 contributes to the growth and invasion of gastric cancer via targeting miR-219-1. J. Cell. Biochem. 2019, 120, 19457–19468. [Google Scholar] [CrossRef]
- Li, R.; Liu, J.; Qi, J. Knockdown of long non-coding RNA CCAT1 suppresses proliferation and EMT of human cervical cancer cell lines by down-regulating Runx2. Exp. Mol. Pathol. 2020, 113, 104380. [Google Scholar] [CrossRef]
- Jiao, K.; Jiang, W.; Zhao, C.; Su, D.; Zhang, H. Bmi-1 in gallbladder carcinoma: Clinicopathology and mechanism of regulation of human gallbladder carcinoma proliferation. Oncol. Lett. 2019, 18, 1365–1371. [Google Scholar] [CrossRef]
- Huang, S.; Li, X.; Zhu, H. MicroRNA-152 Targets Phosphatase and Tensin Homolog to Inhibit Apoptosis and Promote Cell Migration of Nasopharyngeal Carcinoma Cells. Med. Sci. Monit. 2016, 22, 4330–4337. [Google Scholar] [CrossRef][Green Version]
- Yamasaki, T.; Seki, N.; Yoshino, H.; Itesako, T.; Hidaka, H.; Yamada, Y.; Tatarano, S.; Yonezawa, T.; Kinoshita, T.; Nakagawa, M.; et al. MicroRNA-218 inhibits cell migration and invasion in renal cell carcinoma through targeting caveolin-2 involved in focal adhesion pathway. J. Urol. 2013, 190, 1059–1068. [Google Scholar] [CrossRef]
- Mathew, L.K.; Skuli, N.; Mucaj, V.; Lee, S.S.; Zinn, P.O.; Sathyan, P.; Imtiyaz, H.Z.; Zhang, Z.; Davuluri, R.V.; Rao, S.; et al. miR-218 opposes a critical RTK-HIF pathway in mesenchymal glioblastoma. Proc. Natl. Acad. Sci. USA 2014, 111, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, Q.; Hann, S.S. The functions and oncogenic roles of CCAT1 in human cancer. Biomed. Pharmacother. 2019, 115, 108943. [Google Scholar] [CrossRef] [PubMed]
- Neumann, O.; Kesselmeier, M.; Geffers, R.; Pellegrino, R.; Radlwimmer, B.; Hoffmann, K.; Ehemann, V.; Schemmer, P.; Schirmacher, P.; Lorenzo Bermejo, J.; et al. Methylome analysis and integrative profiling of human HCCs identify novel protumorigenic factors. Hepatology 2012, 56, 1817–1827. [Google Scholar] [CrossRef]
- Zhou, J.; Zhi, X.; Wang, L.; Wang, W.; Li, Z.; Tang, J.; Wang, J.; Zhang, Q.; Xu, Z. Linc00152 promotes proliferation in gastric cancer through the EGFR-dependent pathway. J. Exp. Clin. Cancer Res. 2015, 34, 135. [Google Scholar] [CrossRef] [PubMed]
- Yue, B.; Cai, D.; Liu, C.; Fang, C.; Yan, D. Linc00152 Functions as a Competing Endogenous RNA to Confer Oxaliplatin Resistance and Holds Prognostic Values in Colon Cancer. Mol. Ther. 2016, 24, 2064–2077. [Google Scholar] [CrossRef] [PubMed]
- Hers, I.; Vincent, E.E.; Tavare, J.M. Akt signalling in health and disease. Cell. Signal. 2011, 23, 1515–1527. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.P.; Liu, X.X.; Xia, R.; Yin, L.; Kong, R.; Chen, W.M.; Huang, M.D.; Shu, Y.Q. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene 2015, 34, 5648–5661. [Google Scholar] [CrossRef]
- Huang, Z.; Huang, L.; Shen, S.; Li, J.; Lu, H.; Mo, W.; Dang, Y.; Luo, D.; Chen, G.; Feng, Z. Sp1 cooperates with Sp3 to upregulate MALAT1 expression in human hepatocellular carcinoma. Oncol. Rep. 2015, 34, 2403–2412. [Google Scholar] [CrossRef]
- Guan, H.; Cai, J.; Zhang, N.; Wu, J.; Yuan, J.; Li, J.; Li, M. Sp1 is upregulated in human glioma, promotes MMP-2-mediated cell invasion and predicts poor clinical outcome. Int. J. Cancer 2012, 130, 593–601. [Google Scholar] [CrossRef]
- Yeh, Y.M.; Chuang, C.M.; Chao, K.C.; Wang, L.H. MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1alpha. Int. J. Cancer 2013, 133, 867–878. [Google Scholar] [CrossRef]
- Song, T.; Zhang, X.; Wang, C.; Wu, Y.; Cai, W.; Gao, J.; Hong, B. MiR-138 suppresses expression of hypoxia-inducible factor 1alpha (HIF-1alpha) in clear cell renal cell carcinoma 786-O cells. Asian Pac. J. Cancer Prev. 2011, 12, 1307–1311. [Google Scholar]
- Iyer, N.V.; Kotch, L.E.; Agani, F.; Leung, S.W.; Laughner, E.; Wenger, R.H.; Gassmann, M.; Gearhart, J.D.; Lawler, A.M.; Yu, A.Y.; et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998, 12, 149–162. [Google Scholar] [CrossRef]
- Zang, C.; Nie, F.-Q.; Wang, Q.; Sun, M.; Li, W.; He, J.; Zhang, M.; Lu, K.-H. Long non-coding RNA LINC01133 represses KLF2, P21 and E-cadherin transcription through binding with EZH2, LSD1 in non small cell lung cancer. Oncotarget 2016, 7, 11696–11707. [Google Scholar] [CrossRef]
- Kong, J.; Sun, W.; Li, C.; Wan, L.; Wang, S.; Wu, Y.; Xu, E.; Zhang, H.; Lai, M. Long non-coding RNA LINC01133 inhibits epithelial-mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016, 380, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Änkö, M.-L. Regulation of gene expression programmes by serine–arginine rich splicing factors. Semin. Cell Dev. Biol. 2014, 32, 11–21. [Google Scholar] [CrossRef]
- Zeng, H.-F.; Qiu, H.-Y.; Feng, F.-B. Long Noncoding RNA LINC01133 Functions as an miR-422a Sponge to Aggravate the Tumorigenesis of Human Osteosarcoma. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2018, 26, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, T.; Taniguchi, K.; Matsuhashi, N.; Tajirika, T.; Futamura, M.; Takai, T.; Akao, Y.; Yoshida, K. MiR-133b inhibits growth of human gastric cancer cells by silencing pyruvate kinase muscle-splicer polypyrimidine tract-binding protein 1. Cancer Sci. 2016, 107, 1767–1775. [Google Scholar] [CrossRef]
- Ramezani-Rad, P.; Geng, H.; Hurtz, C.; Chan, L.N.; Chen, Z.; Jumaa, H.; Melnick, A.; Paietta, E.; Carroll, W.L.; Willman, C.L.; et al. SOX4 enables oncogenic survival signals in acute lymphoblastic leukemia. Blood 2013, 121, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Dong, W.; Meng, X.; Liu, H.; Liao, H.; Liu, S. MiR-511 inhibits growth and metastasis of human hepatocellular carcinoma cells by targeting PIK3R3. Tumour Biol. 2015, 36, 4453–4459. [Google Scholar] [CrossRef] [PubMed]
- Pachnis, V.; Belayew, A.; Tilghman, S.M. Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proc. Natl. Acad. Sci. USA 1984, 81, 5523–5527. [Google Scholar] [CrossRef]
- Zhu, M.; Chen, Q.; Liu, X.; Sun, Q.; Zhao, X.; Deng, R.; Wang, Y.; Huang, J.; Xu, M.; Yan, J.; et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014, 281, 3766–3775. [Google Scholar] [CrossRef]
- Zhang, Y.; Shields, T.; Crenshaw, T.; Hao, Y.; Moulton, T.; Tycko, B. Imprinting of human H19: Allele-specific CpG methylation, loss of the active allele in Wilms tumor, and potential for somatic allele switching. Am. J. Hum. Genet. 1993, 53, 113–124. [Google Scholar] [PubMed]
- Zhang, L.; Yang, F.; Yuan, J.-h.; Yuan, S.-x.; Zhou, W.-p.; Huo, X.-s.; Xu, D.; Bi, H.-s.; Wang, F.; Sun, S.-h. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis 2013, 34, 577–586. [Google Scholar] [CrossRef]
- Zhao, H.-J.; Ren, L.-L.; Wang, Z.-H.; Sun, T.-T.; Yu, Y.-N.; Wang, Y.-C.; Yan, T.-T.; Zou, W.; He, J.; Zhang, Y.; et al. MiR-194 Deregulation Contributes To Colorectal Carcinogenesis via Targeting AKT2 Pathway. Theranostics 2014, 4, 1193–1208. [Google Scholar] [CrossRef]
- Li, X.-R.; Chu, H.-J.; Lv, T.; Wang, L.; Kong, S.-F.; Dai, S.-Z. miR-342-3p suppresses proliferation, migration and invasion by targeting FOXM1 in human cervical cancer. FEBS Lett. 2014, 588, 3298–3307. [Google Scholar] [CrossRef]
- Wang, Z.; Ahmad, A.; Li, Y.; Banerjee, S.; Kong, D.; Sarkar, F.H. Forkhead box M1 transcription factor: A novel target for cancer therapy. Cancer Treat. Rev. 2010, 36, 151–156. [Google Scholar] [CrossRef]
- Liu, L.; Wu, J.; Guo, Y.; Xie, W.; Chen, B.; Zhang, Y.; Li, S.; Hua, Y.; Peng, B.; Shen, S. Overexpression of FoxM1 predicts poor prognosis of intrahepatic cholangiocarcinoma. Aging 2018, 10, 4120–4140. [Google Scholar] [CrossRef] [PubMed]
- Karin, M.; Clevers, H. Reparative inflammation takes charge of tissue regeneration. Nature 2016, 529, 307–315. [Google Scholar] [CrossRef]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef]
- Yongvanit, P.; Pinlaor, S.; Bartsch, H. Oxidative and nitrative DNA damage: Key events in opisthorchiasis-induced carcinogenesis. Parasitol. Int. 2012, 61, 130–135. [Google Scholar] [CrossRef]
- Kawanishi, S.; Hiraku, Y.; Pinlaor, S.; Ma, N. Oxidative and nitrative DNA damage in animals and patients with inflammatory diseases in relation to inflammation-related carcinogenesis. Biol. Chem. 2006, 387, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Di Virgilio, F. New pathways for reactive oxygen species generation in inflammation and potential novel pharmacological targets. Curr. Pharm. Des. 2004, 10, 1647–1652. [Google Scholar] [CrossRef]
- Fava, G.; Lorenzini, I. Molecular pathogenesis of cholangiocarcinoma. Int. J. Hepatol. 2012, 2012, 630543-630543. [Google Scholar] [CrossRef] [PubMed]
- Pinlaor, S.; Hiraku, Y.; Yongvanit, P.; Tada-Oikawa, S.; Ma, N.; Pinlaor, P.; Sithithaworn, P.; Sripa, B.; Murata, M.; Oikawa, S.; et al. iNOS-dependent DNA damage via NF-kappaB expression in hamsters infected with Opisthorchis viverrini and its suppression by the antihelminthic drug praziquantel. Int. J. Cancer 2006, 119, 1067–1072. [Google Scholar] [CrossRef] [PubMed]
- Thanan, R.; Pairojkul, C.; Pinlaor, S.; Khuntikeo, N.; Wongkham, C.; Sripa, B.; Ma, N.; Vaeteewoottacharn, K.; Furukawa, A.; Kobayashi, H.; et al. Inflammation-related DNA damage and expression of CD133 and Oct3/4 in cholangiocarcinoma patients with poor prognosis. Free Radic. Biol. Med. 2013, 65, 1464–1472. [Google Scholar] [CrossRef] [PubMed]
- Rosen, H.R.; Winkle, P.J.; Kendall, B.J.; Diehl, D.L. Biliary Interleukin-6 and Tumor Necrosis Factor-α in Patients Undergoing Endoscopic Retrograde Cholangiopancreatography. Dig. Dis. Sci. 1997, 42, 1290–1294. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.Y.; Ye, H.; Han, B.W.; Wang, W.T.; Wei, P.P.; He, B.; Li, X.J.; Chen, Y.Q. Genome-wide screen identified let-7c/miR-99a/miR-125b regulating tumor progression and stem-like properties in cholangiocarcinoma. Oncogene 2016, 35, 3376–3386. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-Y.; Kang, J.W.; Song, X.; Kim, B.K.; Yoo, Y.D.; Kwon, Y.T.; Lee, Y.J. Role of the IL-6-JAK1-STAT3-Oct-4 pathway in the conversion of non-stem cancer cells into cancer stem-like cells. Cell. Signal. 2013, 25, 961–969. [Google Scholar] [CrossRef]
- Goydos, J.; Brumfield, A.; Frezza, E.; Booth, A.; Lotze, M.; Carty, S. Marked elevation of serum interleukin-6 in patients with cholangiocarcinoma: Validation of utility as a clinical marker. Ann. Surg. 1998, 227, 398–404. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Wolf, A.; Agnihotri, S.; Micallef, J.; Mukherjee, J.; Sabha, N.; Cairns, R.; Hawkins, C.; Guha, A. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J. Exp. Med. 2011, 208, 313–326. [Google Scholar] [CrossRef]
- Guo, W.; Qiu, Z.; Wang, Z.; Wang, Q.; Tan, N.; Chen, T.; Chen, Z.; Huang, S.; Gu, J.; Li, J.; et al. MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology 2015, 62, 1132–1144. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Chen, W.G.; Li, X.W. MicroRNA-143 acts as a tumor suppressor by targeting hexokinase 2 in human prostate cancer. Am. J. Cancer Res. 2015, 5, 2056–2063. [Google Scholar] [PubMed]
- Peschiaroli, A.; Giacobbe, A.; Formosa, A.; Markert, E.K.; Bongiorno-Borbone, L.; Levine, A.J.; Candi, E.; D’Alessandro, A.; Zolla, L.; Finazzi Agro, A.; et al. miR-143 regulates hexokinase 2 expression in cancer cells. Oncogene 2013, 32, 797–802. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, L.H.; Jacobsen, A.; Frankel, L.B.; Wen, J.; Krogh, A.; Lund, A.H. MicroRNA-143 down-regulates Hexokinase 2 in colon cancer cells. BMC Cancer 2012, 12, 232. [Google Scholar] [CrossRef] [PubMed]
- Marin, J.J.G.; Lozano, E.; Herraez, E.; Asensio, M.; Di Giacomo, S.; Romero, M.R.; Briz, O.; Serrano, M.A.; Efferth, T.; Macias, R.I.R. Chemoresistance and chemosensitization in cholangiocarcinoma. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1444–1453. [Google Scholar] [CrossRef] [PubMed]
- Tiemin, P.; Peng, X.; Qingfu, L.; Yan, W.; Junlin, X.; Zhefeng, H.; Ming, Z.; Desen, L.; Qinghui, M. Dysregulation of the miR-148a-GLUT1 axis promotes the progression and chemoresistance of human intrahepatic cholangiocarcinoma. Oncogenesis 2020, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Marin, J.J.G.; Lozano, E.; Briz, O.; Al-Abdulla, R.; Serrano, M.A.; Macias, R.I.R. Molecular Bases of Chemoresistance in Cholangiocarcinoma. Curr. Drug Targets 2017, 18, 889–900. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J. Autophagy revisited: A conversation with Christian de Duve. Autophagy 2008, 4, 740–743. [Google Scholar] [CrossRef]
- Ravanan, P.; Srikumar, I.F.; Talwar, P. Autophagy: The spotlight for cellular stress responses. Life Sci. 2017, 188, 53–67. [Google Scholar] [CrossRef]
- Liang, X.H.; Jackson, S.; Seaman, M.; Brown, K.; Kempkes, B.; Hibshoosh, H.; Levine, B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999, 402, 672–676. [Google Scholar] [CrossRef]
- Eccles, D.M.; Cranston, G.; Steel, C.M.; Nakamura, Y.; Leonard, R.C. Allele losses on chromosome 17 in human epithelial ovarian carcinoma. Oncogene 1990, 5, 1599–1601. [Google Scholar]
- Mizushima, N.; Levine, B.; Cuervo, A.M.; Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 2008, 451, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Eskelinen, E.-L.; Saftig, P. Autophagy: A lysosomal degradation pathway with a central role in health and disease. Biochim. Biophys. Acta 2009, 1793, 664–673. [Google Scholar] [CrossRef]
- Wang, Z.; Shi, X.; Li, Y.; Fan, J.; Zeng, X.; Xian, Z.; Wang, Z.; Sun, Y.; Wang, S.; Song, P.; et al. Blocking autophagy enhanced cytotoxicity induced by recombinant human arginase in triple-negative breast cancer cells. Cell Death Dis. 2014, 5, e1563-e1563. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carew, J.S.; Nawrocki, S.T.; Cleveland, J.L. Modulating autophagy for therapeutic benefit. Autophagy 2007, 3, 464–467. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.K.; Palanisamy, K.; Sun, K.T.; Yu, S.H.; Yu, T.M.; Li, C.H.; Lin, F.Y.; Chou, A.K.; Wang, G.J.; Chen, K.B.; et al. The functional interplay of lncRNA EGOT and HuR regulates hypoxia-induced autophagy in renal tubular cells. J. Cell. Biochem. 2020. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Zhao, X.; Lu, K.; Cheng, G. LncRNA BDNF-AS promotes autophagy and apoptosis in MPTP-induced Parkinson’s disease via ablating microRNA-125b-5p. Brain Res. Bull. 2020. [Google Scholar] [CrossRef]
- Huang, W.; Huang, F.; Lei, Z.; Luo, H. LncRNA SNHG11 Promotes Proliferation, Migration, Apoptosis, and Autophagy by Regulating hsa-miR-184/AGO2 in HCC. Onco. Targets Ther. 2020, 13, 413–421. [Google Scholar] [CrossRef]
- Shukla, S.K.; Singh, G.; Shahi, K.S.; Pant, P. Staging, Treatment, and Future Approaches of Gallbladder Carcinoma. J. Gastrointest. Cancer 2018, 49, 9–15. [Google Scholar] [CrossRef]
- Caldow Pilgrim, C.H.; Groeschl, R.T.; Quebbeman, E.J.; Gamblin, T.C. Recent advances in systemic therapies and radiotherapy for gallbladder cancer. Surg. Oncol. 2013, 22, 61–67. [Google Scholar] [CrossRef]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 and Autophagy. Methods Mol. Biol. 2008, 445, 77–88. [Google Scholar] [CrossRef]
- Scopes, R.K. Studies with a reconstituted muscle glycolytic system. The rate and extent of creatine phosphorylation by anaerobic glycolysis. Biochem. J. 1973, 134, 197–208. [Google Scholar] [CrossRef]
- Qian, X.; Li, X.; Lu, Z. Protein kinase activity of the glycolytic enzyme PGK1 regulates autophagy to promote tumorigenesis. Autophagy 2017, 13, 1246–1247. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Li, X.; Cai, Q.; Zhang, C.; Yu, Q.; Jiang, Y.; Lee, J.-H.; Hawke, D.; Wang, Y.; Xia, Y.; et al. Phosphoglycerate Kinase 1 Phosphorylates Beclin1 to Induce Autophagy. Mol. Cell 2017, 65, 917–931.e916. [Google Scholar] [CrossRef] [PubMed]
- Malouf, G.G.; Zhang, J.; Yuan, Y.; Compérat, E.; Rouprêt, M.; Cussenot, O.; Chen, Y.; Thompson, E.J.; Tannir, N.M.; Weinstein, J.N.; et al. Characterization of long non-coding RNA transcriptome in clear-cell renal cell carcinoma by next-generation deep sequencing. Mol. Oncol. 2015, 9, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Klec, C.; Prinz, F.; Pichler, M. Involvement of the long noncoding RNA NEAT1 in carcinogenesis. Mol. Oncol. 2019, 13, 46–60. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-X.; Yin, Y.; Cheng, J.-W.; Huang, A.; Hu, B.; Zhang, X.; Sun, Y.-F.; Wang, J.; Wang, Y.-P.; Ji, Y.; et al. BAP1 acts as a tumor suppressor in intrahepatic cholangiocarcinoma by modulating the ERK1/2 and JNK/c-Jun pathways. Cell Death Dis. 2018, 9, 1036. [Google Scholar] [CrossRef] [PubMed]
- McCabe, M.T.; Ott, H.M.; Ganji, G.; Korenchuk, S.; Thompson, C.; Van Aller, G.S.; Liu, Y.; Graves, A.P.; Della Pietra, A., III; Diaz, E.; et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 2012, 492, 108–112. [Google Scholar] [CrossRef]
- Zhang, H.; Zhou, D.; Ying, M.; Chen, M.; Chen, P.; Chen, Z.; Zhang, F. Expression of Long Non-Coding RNA (lncRNA) Small Nucleolar RNA Host Gene 1 (SNHG1) Exacerbates Hepatocellular Carcinoma Through Suppressing miR-195. Med. Sci. Monit. 2016, 22, 4820–4829. [Google Scholar] [CrossRef]
- Yang, H.; Wang, S.; Kang, Y.J.; Wang, C.; Xu, Y.; Zhang, Y.; Jiang, Z. Long non-coding RNA SNHG1 predicts a poor prognosis and promotes colon cancer tumorigenesis. Oncol. Rep. 2018, 40, 261–271. [Google Scholar] [CrossRef]
- Li, X.; Zheng, H. LncRNA SNHG1 influences cell proliferation, migration, invasion, and apoptosis of non-small cell lung cancer cells via the miR-361-3p/FRAT1 axis. Thorac. Cancer 2020, 11, 295–304. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Z.; Xiong, L.; Guo, C.; Jiang, T.; Zeng, L.; Li, G.; Wang, J. SNHG1 lncRNA negatively regulates miR-199a-3p to enhance CDK7 expression and promote cell proliferation in prostate cancer. Biochem. Biophys. Res. Commun. 2017, 487, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.; Jiang, S.; Chung, N.; Alikhan, A.; Ni, C.; Lee, C.C.; Hornyak, T.J. EZH2-dependent suppression of a cellular senescence phenotype in melanoma cells by inhibition of p21/CDKN1A expression. Mol. Cancer Res. 2011, 9, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhang, W.; Tang, H.; Qian, H.; Yang, J.; Zhu, Z.; Ren, P.; Lu, B. Septin 2 accelerates the progression of biliary tract cancer and is negatively regulated by mir-140-5p. Gene 2016, 589, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Zhang, Z.; Li, X.; Ren, Q.; Wang, Q. Long non-coding RNA TTN-AS1 promotes breast cancer cell migration and invasion via sponging miR-140-5p. Oncol. Lett. 2020, 19, 1255–1260. [Google Scholar] [CrossRef]
- Li, J.L.; Luo, P. MiR-140-5p and miR-92a-3p suppress the cell proliferation, migration and invasion and promoted apoptosis in Wilms’ tumor by targeting FRS2. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 97–108. [Google Scholar] [CrossRef]
- Woods, D.C.; White, Y.A.; Dau, C.; Johnson, A.L. TLR4 activates NF-kappaB in human ovarian granulosa tumor cells. Biochem. Biophys. Res. Commun. 2011, 409, 675–680. [Google Scholar] [CrossRef]
- Lee, H.; Herrmann, A.; Deng, J.H.; Kujawski, M.; Niu, G.; Li, Z.; Forman, S.; Jove, R.; Pardoll, D.M.; Yu, H. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell 2009, 15, 283–293. [Google Scholar] [CrossRef]
- He, W.; Liu, Q.; Wang, L.; Chen, W.; Li, N.; Cao, X. TLR4 signaling promotes immune escape of human lung cancer cells by inducing immunosuppressive cytokines and apoptosis resistance. Mol. Immunol. 2007, 44, 2850–2859. [Google Scholar] [CrossRef]
- Zhang, X.; Hamblin, M.H.; Yin, K.-J. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017, 14, 1705–1714. [Google Scholar] [CrossRef]
- Ying, L.; Chen, Q.; Wang, Y.; Zhou, Z.; Huang, Y.; Qiu, F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol. Biosyst. 2012, 8, 2289–2294. [Google Scholar] [CrossRef]
- Yang, L.; Bai, H.S.; Deng, Y.; Fan, L. High MALAT1 expression predicts a poor prognosis of cervical cancer and promotes cancer cell growth and invasion. Eur. Rev. Med Pharmacol. Sci. 2015, 19, 3187–3193. [Google Scholar] [PubMed]
- Xu, C.; Yang, M.; Tian, J.; Wang, X.; Li, Z. MALAT-1: A long non-coding RNA and its important 3′ end functional motif in colorectal cancer metastasis. Int. J. Oncol. 2011, 39, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Tang, K.; Liu, P.; Chen, K.; Hu, J.; Zeng, J.; Xiao, W.; Yu, G.; Yao, W.; Zhou, H.; et al. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget 2015, 6, 38005–38015. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Ooi, H.S.; Wu, J.; Chen, J.; Zhang, X.; Tan, S.; Yu, Q.; Li, Y.-Y.; Kang, Y.; Li, H.; et al. MALAT1 long ncRNA promotes gastric cancer metastasis by suppressing PCDH10. Oncotarget 2016, 7, 12693–12703. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Wu, Y.; Tan, D.; Meng, H.; Wang, K.; Bai, Y.; Yang, K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2015, 34, 7. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.K.; Hu, P.P.; Xu, F. Prognostic significance of long non-coding RNA MALAT1 for predicting the recurrence and metastasis of gallbladder cancer and its effect on cell proliferation, migration, invasion, and apoptosis. J. Cell. Biochem. 2018, 119, 3099–3110. [Google Scholar] [CrossRef]
- Ou, Y.; Zhai, D.; Wu, N.; Li, X. Downregulation of miR-363 increases drug resistance in cisplatin-treated HepG2 by dysregulating Mcl-1. Gene 2015, 572, 116–122. [Google Scholar] [CrossRef]
- Keklikoglou, I.; Hosaka, K.; Bender, C.; Bott, A.; Koerner, C.; Mitra, D.; Will, R.; Woerner, A.; Muenstermann, E.; Wilhelm, H.; et al. MicroRNA-206 functions as a pleiotropic modulator of cell proliferation, invasion and lymphangiogenesis in pancreatic adenocarcinoma by targeting ANXA2 and KRAS genes. Oncogene 2015, 34, 4867–4878. [Google Scholar] [CrossRef]
- Latini, F.R.M.; Hemerly, J.P.; Freitas, B.C.G.; Oler, G.; Riggins, G.J.; Cerutti, J.M. ABI3 ectopic expression reduces in vitro and in vivocell growth properties while inducing senescence. BMC Cancer 2011, 11, 11. [Google Scholar] [CrossRef]
- Völkel, P.; Dupret, B.; Le Bourhis, X.; Angrand, P.-O. Diverse involvement of EZH2 in cancer epigenetics. Am. J. Transl. Res. 2015, 7, 175–193. [Google Scholar]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef] [PubMed]
- Sauvageau, M.; Sauvageau, G. Polycomb group proteins: Multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 2010, 7, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Collett, K.; Eide, G.E.; Arnes, J.; Stefansson, I.M.; Eide, J.; Braaten, A.; Aas, T.; Otte, A.P.; Akslen, L.A. Expression of enhancer of zeste homologue 2 is significantly associated with increased tumor cell proliferation and is a marker of aggressive breast cancer. Clin. Cancer Res. 2006, 12, 1168–1174. [Google Scholar] [CrossRef] [PubMed]
- Varambally, S.; Dhanasekaran, S.M.; Zhou, M.; Barrette, T.R.; Kumar-Sinha, C.; Sanda, M.G.; Ghosh, D.; Pienta, K.J.; Sewalt, R.G.; Otte, A.P.; et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002, 419, 624–629. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, J.; Sasaki, M.; Sato, Y.; Itatsu, K.; Harada, K.; Zen, Y.; Ikeda, H.; Nimura, Y.; Nagino, M.; Nakanuma, Y. Histone deacetylase inhibitor (SAHA) and repression of EZH2 synergistically inhibit proliferation of gallbladder carcinoma. Cancer Sci. 2010, 101, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.C.; Yang, Z.L. Overexpression of EZH2 and loss of expression of PTEN is associated with invasion, metastasis, and poor progression of gallbladder adenocarcinoma. Pathol. Res. Pract. 2011, 207, 472–478. [Google Scholar] [CrossRef]
- Sasaki, M.; Yamaguchi, J.; Itatsu, K.; Ikeda, H.; Nakanuma, Y. Over-expression of polycomb group protein EZH2 relates to decreased expression of p16 INK4a in cholangiocarcinogenesis in hepatolithiasis. J. Pathol. 2008, 215, 175–183. [Google Scholar] [CrossRef]
- Yamaguchi, J.; Sasaki, M.; Harada, K.; Zen, Y.; Sato, Y.; Ikeda, H.; Itatsu, K.; Yokoyama, Y.; Ando, H.; Ohta, T.; et al. Papillary hyperplasia of the gallbladder in pancreaticobiliary maljunction represents a senescence-related lesion induced by lysolecithin. Lab. Investig. 2009, 89, 1018–1031. [Google Scholar] [CrossRef]
- Visser, S.; Yang, X. LATS tumor suppressor: A new governor of cellular homeostasis. Cell Cycle 2010, 9, 3892–3903. [Google Scholar] [CrossRef]
- Lee, G.L.; Dobi, A.; Srivastava, S. Prostate cancer: Diagnostic performance of the PCA3 urine test. Nat. Rev. Urol. 2011, 8, 123–124. [Google Scholar] [CrossRef]
- He, C.; Zheng, S.; Luo, Y.; Wang, B. Exosome Theranostics: Biology and Translational Medicine. Theranostics 2018, 8, 237–255. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, R.; Wang, Y.; Zhu, W.; Zhao, D.; Wang, X.; Yang, H.; Gurley, E.C.; Chen, W.; Hylemon, P.B.; et al. Cholangiocyte-Derived Exosomal lncRNA H19 Promotes Macrophage Activation and Hepatic Inflammation under Cholestatic Conditions. Cells 2020, 9, 190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, Z.; Trottier, J.; Barbier, O.; Wang, L. Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology 2017, 65, 604–615. [Google Scholar] [CrossRef]
- Sato, K.; Glaser, S.; Francis, H.; Alpini, G. Concise Review: Functional Roles and Therapeutic Potentials of Long Non-coding RNAs in Cholangiopathies. Front. Med. 2020, 7. [Google Scholar] [CrossRef]
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.-C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Schmidt, H.H.; Coelho, T.; Berk, J.L.; et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018, 379, 11–21. [Google Scholar] [CrossRef] [PubMed]
| LncRNA | Tissue | Clinical Effects | Ref | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Prognosis | Tumor Size | TNM Status | Differentiation | Metastasis | Lymphatic Invasion | ||||
| Up-Regulated | AFAP-AS1 | CCA/GBC | ↓ | ↑ | ↑ | [53,54] | |||
| ANRIL | IHC | ↓ | [55,56] | ||||||
| CPS1-IT1 | IHC | ↓ | [57] | ||||||
| CCAT1 | IHC | ↓ | ↑ | ↓ | ↑ | [58,59,60] | |||
| CCAT2 | IHC | ↓ | ↑ | ↑ | [61] | ||||
| DILC | GBC | * | [62] | ||||||
| EPIC1 | CCA | * | [63] | ||||||
| GBCDRlnc1 | GBC | ↑ | [64] | ||||||
| H19 | GBC | ↓ | ↑ | ↑ | [65,66,67,68] | ||||
| HEGBC | GBC | ↓ | [69,70] | ||||||
| HOTAIR | CCA | ↓ | ↑ | ↑ | [71,72] | ||||
| HOXA-AS2 | GBC | ↑ | ↑ | [73] | |||||
| HULC | CCA | * | [65] | ||||||
| KCNQ1QT1 | CCA | ↓ | [74] | ||||||
| Linc00152 | GBC | ↓ | ↑ | ↑ | [75,76] | ||||
| Linc01296 | CCA | ↓ | ↑ | [77] | |||||
| Loc344887 | GBC | ↑ | [78] | ||||||
| MALAT1 | GBC | ↓ | ↑ | ↑ | [79,80,81,82,83] | ||||
| MINCR | GBC | ↓ | ↑ | ↑ | [84] | ||||
| NEAT1 | CCA | * | [85,86] | ||||||
| PAGBC | GBC | ↑ | ↑ | [87] | |||||
| PANDAR | CCA | ↓ | ↑ | ↑ | [88] | ||||
| PCAT1 | EHC | * | [89] | ||||||
| PVT1 | GBC | ↓ | ↑ | ↑ | [90] | ||||
| RMRP | CCA | * | [91] | ||||||
| ROR | GBC | ↓ | ↑ | ↑ | [92] | ||||
| SNHG1 | CCA | * | [93,94] | ||||||
| SOX OT2 | CCA | ↓ | ↑ | ↑ | [95,96] | ||||
| SPRY4-IT1 | CCA/GBC | ↓ | ↑ | [97,98] | |||||
| TP73-AS1 | CCA | ↑ | ↑ | [99] | |||||
| TUG1 | GBC | ↑ | [100] | ||||||
| UCA1 | GBC | ↓ | ↑ | ↑ | ↑ | [101] | |||
| Down- Regulated | CRNDE | GBC | * | [102] | |||||
| FENDRR | CCA | * | [103,104] | ||||||
| GCASPC | GBC | ↓ | ↑ | [105] | |||||
| LET1 | GBC | ↓ | ↓ | ↑ | [106] | ||||
| MEG3 | GBC | ↓ | ↑ | ↑ | [107] | ||||
| oncogenic lncRNAs | lncRNA | Molecular Mechanism | Predicted Targets | Effects of Knockdown | Ref | ||||||||||||
| ↓ | ↑ | ||||||||||||||||
| ceRNA | scaffold | guide lncRNA | specific | not described | proliferation | cell viability | migration | invasion | EMT | CSC traits | chemosensitivity | apoptosis | E-Cadherin | ||||
| AFAP-AS1 | x | x | x | x | x | [53,54] | |||||||||||
| ANRIL | x | x | x | x | [55,56] | ||||||||||||
| CPS1-IT1 | x | x | x | [57] | |||||||||||||
| CCAT1 | x | miR-152, miR-218-5p | x | x | x | [58,59,60] | |||||||||||
| CCAT2 | x | x | x | x | x | [61] | |||||||||||
| DILC | x | x | x | [62] | |||||||||||||
| EPIC1 | x | x | x | x | [63] | ||||||||||||
| GBCDRlnc1 | x | PGK1 | x | [64] | |||||||||||||
| H19 | x | miR-342-2p, miR-195-5p, let7a/b | x | x | x | x | [65,66,67,68] | ||||||||||
| HEGBC | x | miR-502-3p | x | x | x | [69,70] | |||||||||||
| HOTAIR | x | miR-130a | x | x | x | x | x | [71,72] | |||||||||
| HOXA-AS2 | x | x | x | x | x | x | [73] | ||||||||||
| HULC | x | miR-372, miR-373 | x | x | [65] | ||||||||||||
| KCNQ1QT1 | x | miR-140-5p | x | x | x | [74] | |||||||||||
| Linc00152 | x | miR-138 | x | x | x | x | x | [75,76] | |||||||||
| Linc01296 | x | miR-5095 | x | x | x | x | [77] | ||||||||||
| oncogenic lncRNAs (continued) | lncRNA | Molecular Mechanism | Predicted Targets | Effects of Knockdown | Ref | ||||||||||||
| ↓ | ↑ | ||||||||||||||||
| ceRNA | scaffold | guide lncRNA | specific | not described | proliferation | cell viability | migration | invasion | EMT | CSC traits | chemosensitivity | apoptosis | E-Cadherin | ||||
| Loc344887 | x | x | x | x | x | [78] | |||||||||||
| MALAT1 | x | x | miR-206, miR-363-3p/EZH2 | x | x | x | x | x | x | [79,80,81,82,83] | |||||||
| MINCR | x | miR-26a-5p | x | x | x | x | [84] | ||||||||||
| NEAT1 | x | EZH2 | x | x | x | x | x | [85,86] | |||||||||
| PAGBC | x | miR-133, miR-511 | x | x | [87] | ||||||||||||
| PANDAR | x | x | x | x | x | x | x | [88] | |||||||||
| PCAT1 | x | miR-122 | x | x | x | x | [89] | ||||||||||
| PVT1 | x | miR-143 | x | x | x | [90] | |||||||||||
| RMRP | x | miR-217 | x | x | [91] | ||||||||||||
| ROR | x | x | x | x | x | [92] | |||||||||||
| SNHG1 | x | EZH2 | x | x | x | x | [93,94] | ||||||||||
| SOX2 OT | x | x | [95,96] | ||||||||||||||
| SPRY4-IT1 | x | x | miR-101-3p; EZH2/LSD1/DNMT1 | x | x | x | x | x | [97,98] | ||||||||
| TP73-AS1 | x | x | x | [99] | |||||||||||||
| TUG1 | x | miR-300 | x | x | [100] | ||||||||||||
| UCA1 | x | EZH2 | x | [101] | |||||||||||||
| tumor-suppressive lncRNAs | lncRNA | Molecular Mechanism | Predicted Targets | Effects of Overexpression | Ref | ||||||||||||
| ↓ | ↑ | ||||||||||||||||
| ceRNA | scaffold | guide lncRNA | specific | not described | proliferation | invasion | EMT | specific | - | - | - | apoptosis | - | ||||
| CRNDE | x | DMBT/c-IAP1 | x | [102] | |||||||||||||
| FENDRR | x | x | miR-18b-5p; Survivin | x | [103,104] | ||||||||||||
| GCASPC | x | miR-17-3p | x | [105] | |||||||||||||
| LET1 | x | x | x | [106] | |||||||||||||
| MEG3 | x | EZH2 | x | x | x | x | [107] | ||||||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bekric, D.; Neureiter, D.; Ritter, M.; Jakab, M.; Gaisberger, M.; Pichler, M.; Kiesslich, T.; Mayr, C. Long Non-Coding RNAs in Biliary Tract Cancer—An Up-to-Date Review. J. Clin. Med. 2020, 9, 1200. https://doi.org/10.3390/jcm9041200
Bekric D, Neureiter D, Ritter M, Jakab M, Gaisberger M, Pichler M, Kiesslich T, Mayr C. Long Non-Coding RNAs in Biliary Tract Cancer—An Up-to-Date Review. Journal of Clinical Medicine. 2020; 9(4):1200. https://doi.org/10.3390/jcm9041200
Chicago/Turabian StyleBekric, Dino, Daniel Neureiter, Markus Ritter, Martin Jakab, Martin Gaisberger, Martin Pichler, Tobias Kiesslich, and Christian Mayr. 2020. "Long Non-Coding RNAs in Biliary Tract Cancer—An Up-to-Date Review" Journal of Clinical Medicine 9, no. 4: 1200. https://doi.org/10.3390/jcm9041200
APA StyleBekric, D., Neureiter, D., Ritter, M., Jakab, M., Gaisberger, M., Pichler, M., Kiesslich, T., & Mayr, C. (2020). Long Non-Coding RNAs in Biliary Tract Cancer—An Up-to-Date Review. Journal of Clinical Medicine, 9(4), 1200. https://doi.org/10.3390/jcm9041200







