Involvement of the Endocrine-Disrupting Chemical Bisphenol A (BPA) in Human Placentation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Syncytialisation Using 8-Bromo-cAMP and Treatment with Bisphenol A (BPA)
2.3. RNA Extraction, cDNA Synthesis, and Quantitative Reverse Transcription PCR (qRT-PCR)
2.4. Western Blotting
2.5. Immunofluorescence
2.6. Statistical Analysis
3. Results
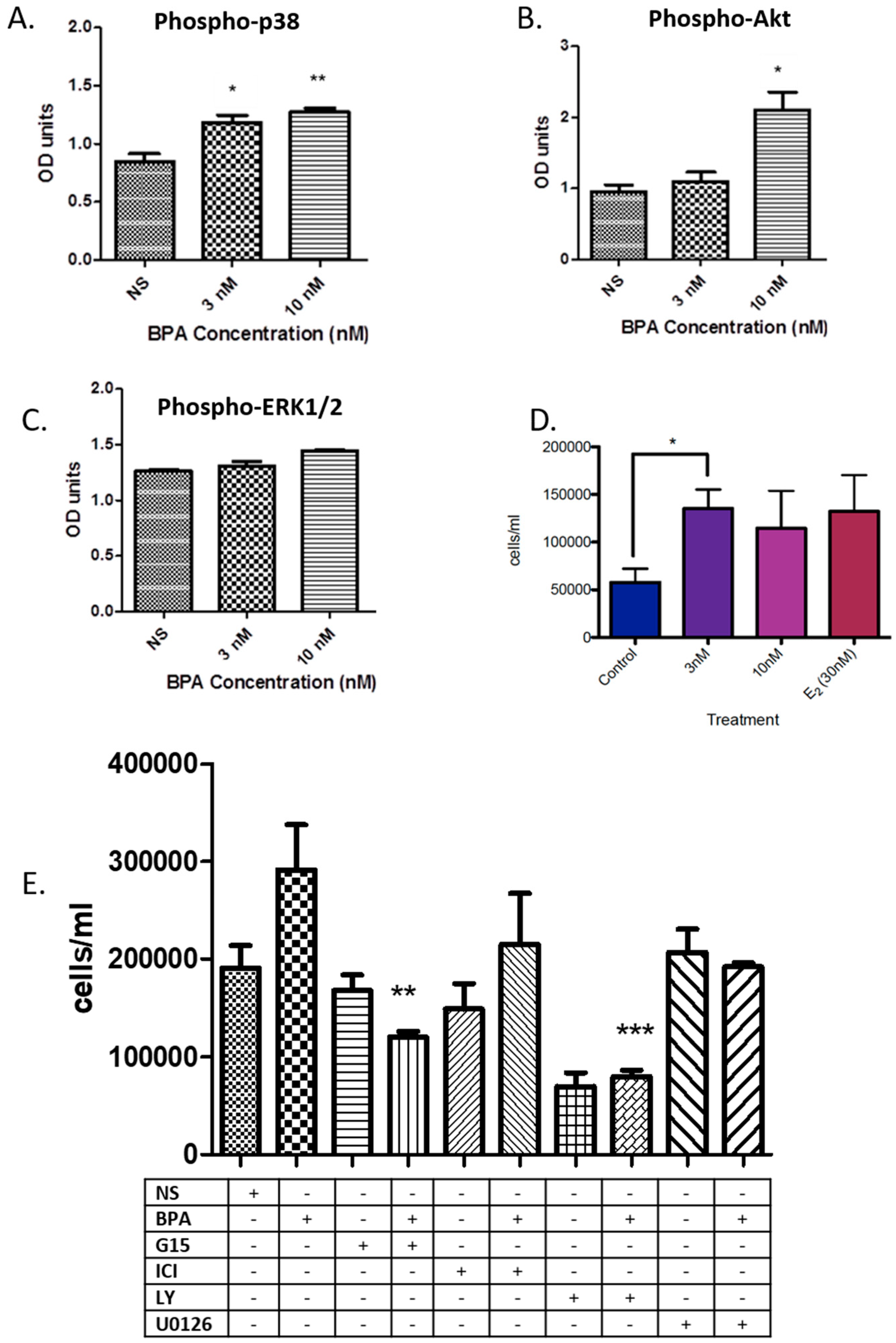
3.1. BPA Effects on Phosphorylation of Key Kinases and Cell Number
3.2. Gene Microarray Analyses Assessing the Effects of BPA in Nonsyncytialised BeWo Cells
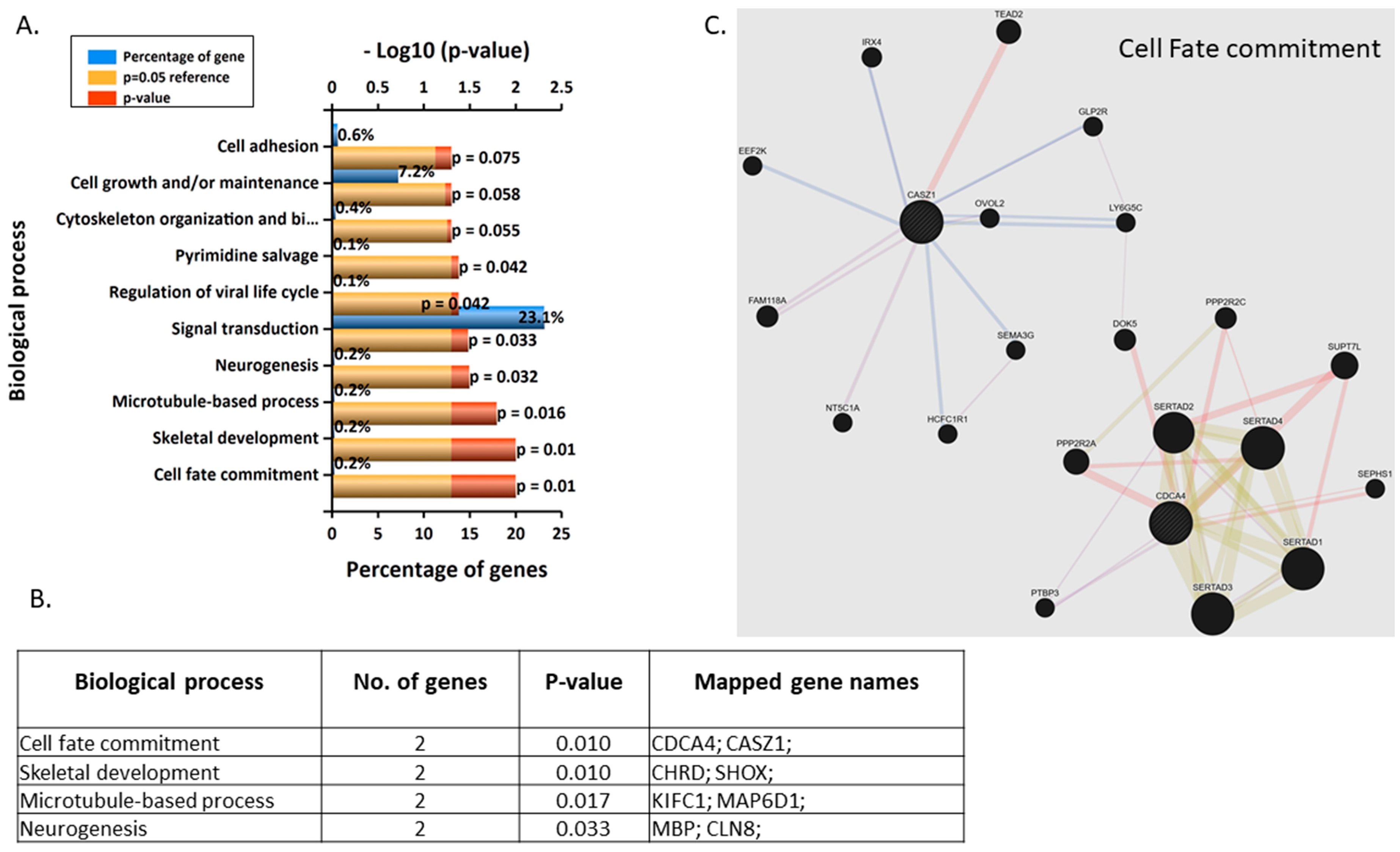
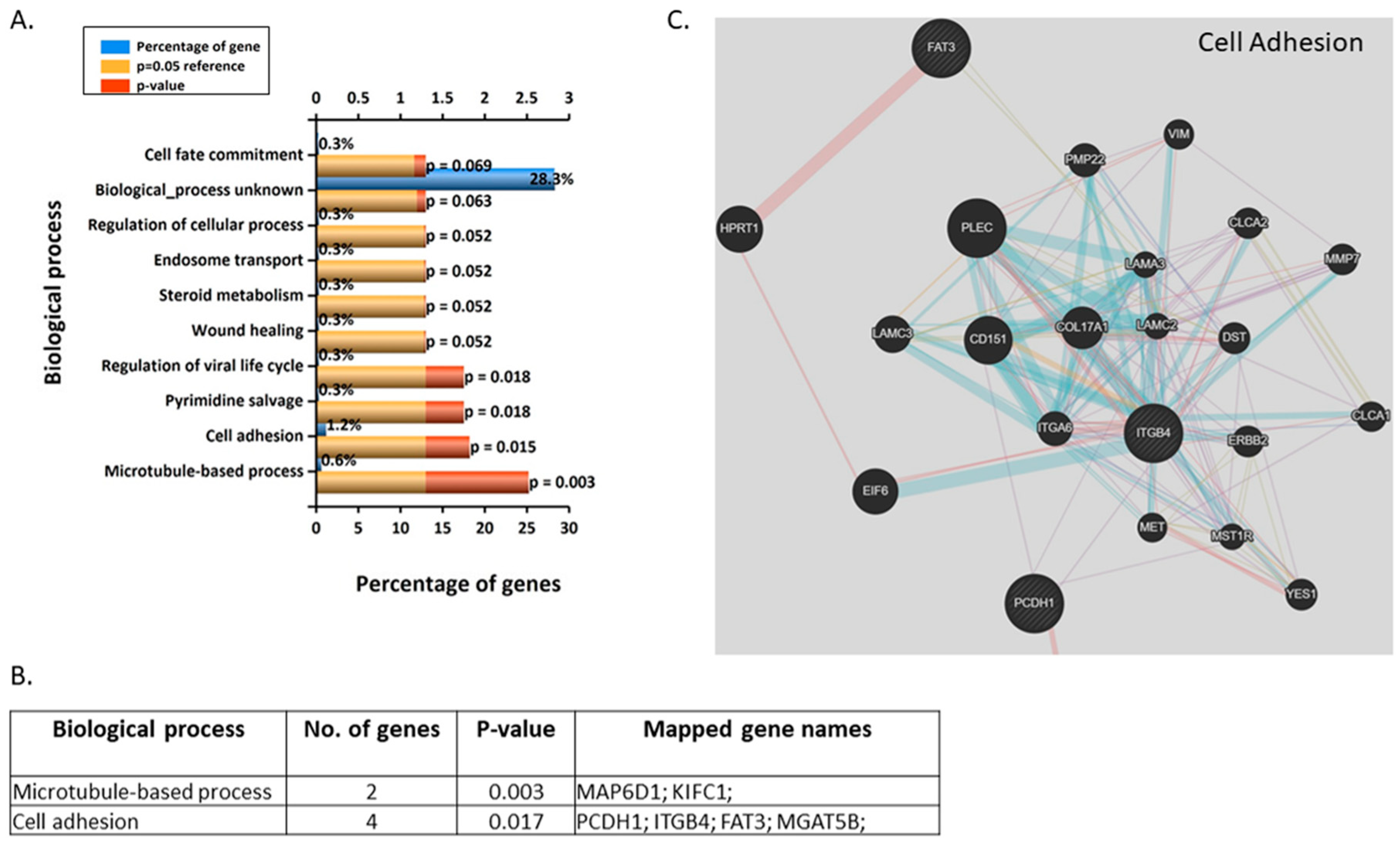
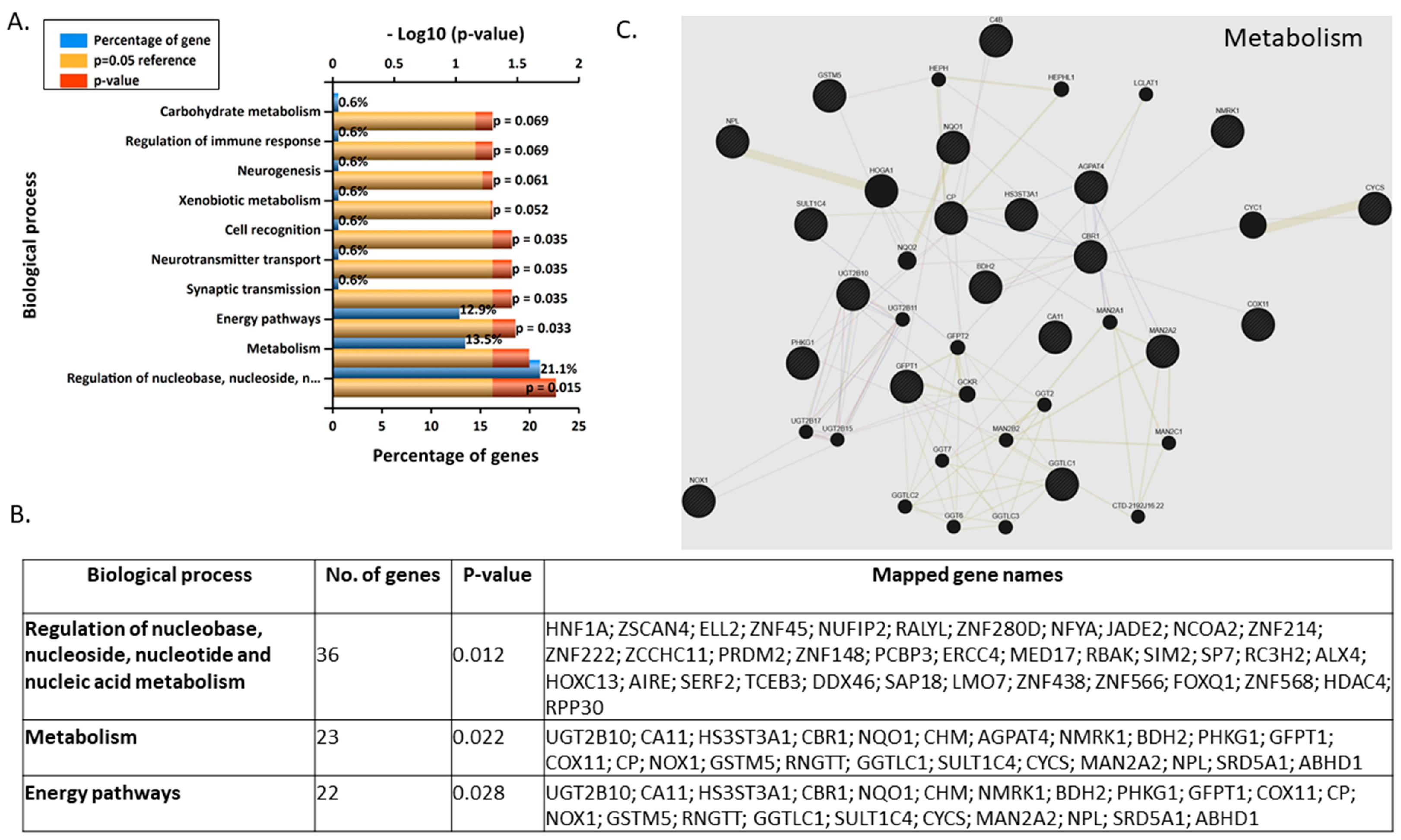
3.3. Validation and Enrichment Analyses on Gene Microarrays
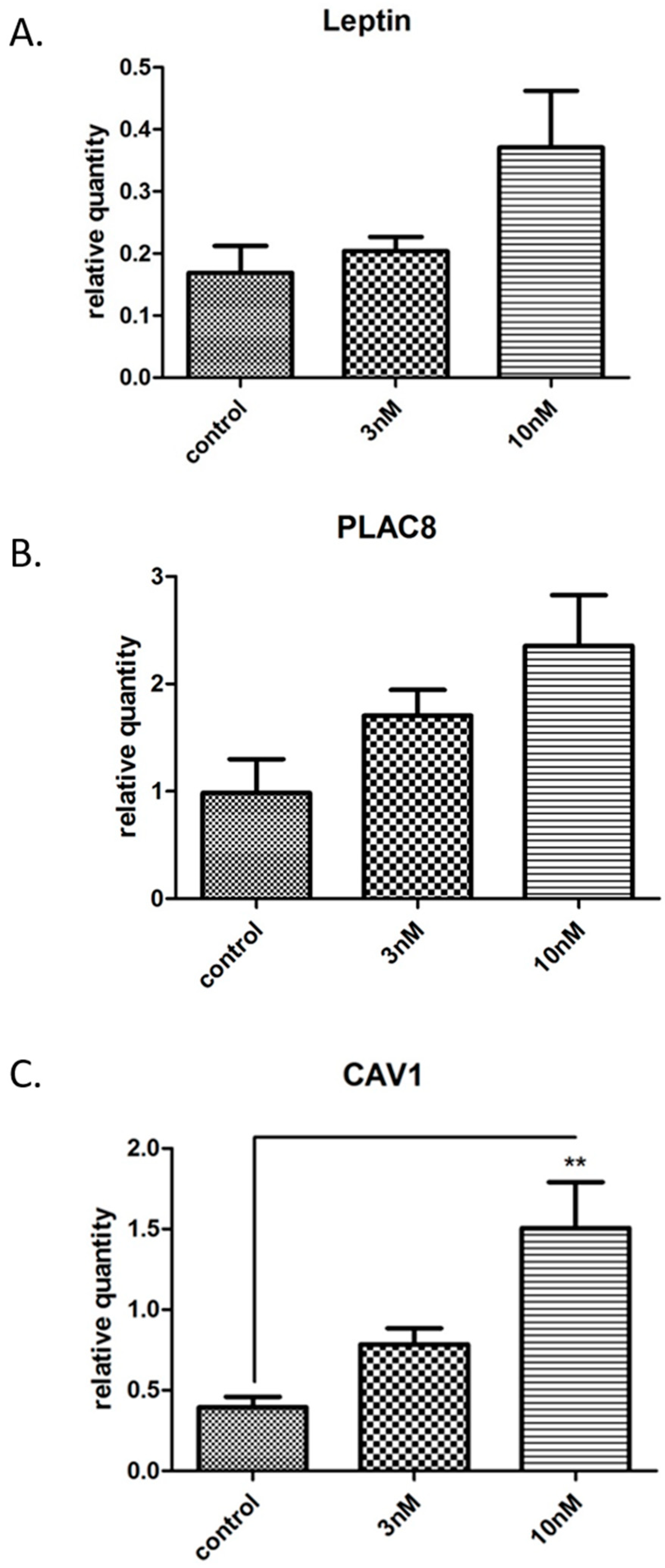
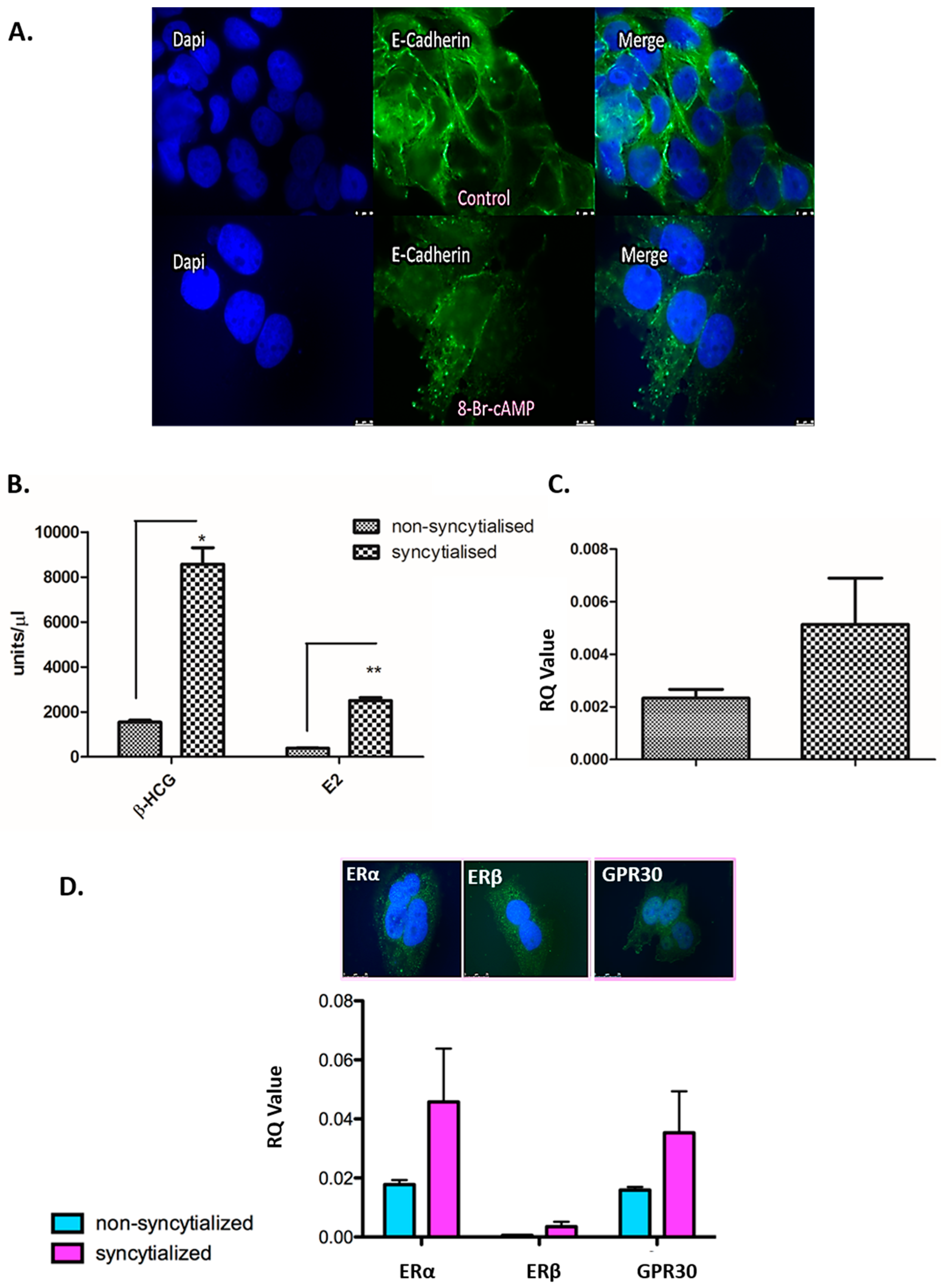
3.4. Validation of the in Vitro Syncytialisation Model of BeWo Cells
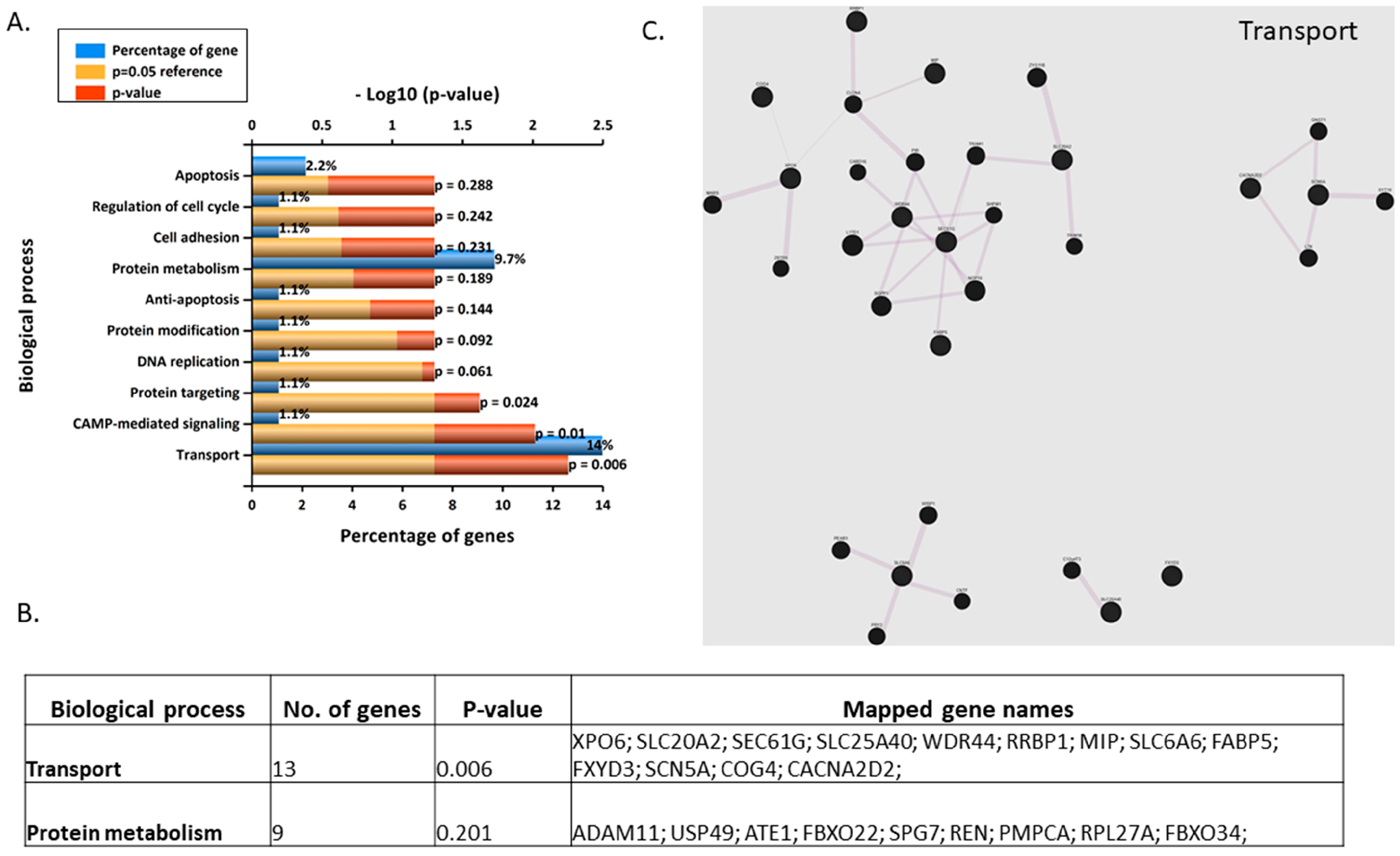
3.5. Gene Microarray Analyses Assessing the in Vitro Effects of BPA in Syncytialised BeWo Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gore, A.C.; Crews, D.; Doan, L.L.; Merrill, M.L.; Patisaul, H.; Zota, A. Introduction to Endocrine Disrupting Chemicals (EDCs): A Guide for Public Interest Organizations and Policy-Makers. Endocr. Rev. 2014. Available online: https://www.endocrine.org/-/media/endosociety/files/advocacy-and-outreach/important-documents/introduction-to-endocrine-disrupting-chemicals.pdf (accessed on 31 January 2020).
- Gore, A.C.; Chappell, V.A.; Fenton, S.E.; Flaws, J.A.; Nadal, A.; Prins, G.S.; Zoeller, R.T. EDC-2: The Endocrine Society’s second scientific statement on endocrine-disrupting chemicals. Endocr. Rev. 2015, 36, 1–150. [Google Scholar] [CrossRef]
- Montes-Grajales, D.; Fennix-Agudelo, M.; Miranda-Castro, W. Occurrence of personal care products as emerging chemicals of concern in water resources: A review. Sci. Total Environ. 2017, 595, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Brieño-Enríquez, M.A.; Robles, P.; Camats-Tarruella, N.; García-Cruz, R.; Roig, I.; Cabero, L.; Caldés, M.G. Human meiotic progression and recombination are affected by bisphenol A exposure during In Vitro human oocyte development. Hum. Reprod. 2011, 26, 2807–2818. [Google Scholar] [CrossRef]
- Koch, C.A.; Diamanti-Kandarakis, E. Introduction to endocrine disrupting chemicals—Is it time to act? Rev. Endocr. Metab. Disord. 2015, 16, 269–270. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Miodovnik, A.; Engel, S.M.; Zhu, C.; Ye, X.; Soorya, L.V.; Silva, M.J.; Wolff, M.S. Endocrine disruptors and childhood social impairment. Neurotoxicol. Teratol. 2010, 32, 261–267. [Google Scholar] [CrossRef]
- Rahman, M.S.; Kwon, W.S.; Lee, J.S.; Yoon, S.J.; Ryu, B.Y.; Pang, M.G. Bisphenol-A affects male fertility via fertility-related proteins in spermatozoa. Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef]
- Snyder, R.W.; Maness, S.C.; Gaido, K.W.; Welsch, F.; Sumner, S.C.J.; Fennell, T.R. Metabolism and disposition of Bisphenol A in female rats. Toxicol. Appl. Pharmacol. 2000, 168, 225–234. [Google Scholar] [CrossRef]
- Watkins, D.J.; Sánchez, B.N.; Téllez-Rojo, M.M.; Lee, J.M.; Mercado-García, A.; Blank-Goldenberg, C.; Watkins, D.J. Phthalate and Bisphenol A exposure during in utero windows of susceptibility in relation to reproductive hormones and pubertal development in girls. Environ. Res. 2017, 159, 143–151. [Google Scholar] [CrossRef]
- Wozniak, A.L.; Bulayeva, N.N.; Watson, C.S. Xenoestrogens at picomolar to nanomolar concentrations trigger membrane estrogen receptor-alpha-mediated Ca2+ fluxes and prolactin release in GH3/B6 pituitary tumor cells. Environ. Health Perspect. 2005, 113, 431–439. [Google Scholar] [CrossRef]
- Yolton, K.; Xu, Y.; Strauss, D.; Altaye, M.; Calafat, A.M.; Khoury, J. Prenatal exposure to Bisphenol A and phthalates and infant neurobehavior. Neurotoxicol. Teratol. 2011, 33, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Tyl, R.W. Abbreviated assessment of Bisphenol A toxicology literature. Semin. Fetal Neonatal Med. 2014, 19, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Peretz, J.; Vrooman, L.; Ricke, W.A.; Hunt, P.A.; Ehrlich, S.; Hauser, R.; Flaws, J.A. Bisphenol A and reproductive health: Update of experimental and human evidence, 2007–2013. Environ. Health Perspect. 2014, 122, 775–786. [Google Scholar] [CrossRef] [PubMed]
- Muhamad, M.S.; Salim, M.R.; Lau, W.J.; Yusop, Z. A review on Bisphenol A occurrences, health effects and treatment process via membrane technology for drinking water. Environ. Sci. Pollut. Res. 2016, 23, 11549–11567. [Google Scholar] [CrossRef] [PubMed]
- Erler, C.; Novak, J. Bisphenol A exposure: Human risk and health policy background of Bisphenol A. J. Pediatr. Nurs. 2010, 25, 400–407. [Google Scholar] [CrossRef]
- Michałowicz, J. Bisphenol A—Sources, toxicity and biotransformation. Environ. Toxicol. Pharmacol. 2014, 37, 738–758. [Google Scholar] [CrossRef]
- Richter, C.A.; Birnbaum, L.S.; Farabollini, F.; Newbold, R.R.; Rubin, B.S.; Talsness, C.E.; vom Saal, F.S. In Vivo effects of Bisphenol A in laboratory rodent studies. Birth Defects Res. B Dev. Reprod. Toxicol. 2007, 24, 199–224. [Google Scholar] [CrossRef]
- Delfosse, V.; Grimaldi, M.; Pons, J.L.; Boulahtouf, A.; le Maire, A.; Cavailles, V.; Labesse, G.; Bourgouet, W.; Balaguer, P. Structural and mechanistic insights inot bisphenols action provide guidelines for risk assessment and discovery of Bisphenol A substitutes. Proc. Natl. Acad. Sci. USA 2012, 109, 14930–14935. [Google Scholar] [CrossRef]
- Matsushima, A.; Kakuta, Y.; Teramoto, T.; Koshiba, T.; Liu, X.; Okada, H.; Tokunaga, T.; Kawabata, S.; Kimura, M.; Shimohigashi, Y. Structural evidence for endocrine disruptor Bisphenol A binding to human nuclear receptor ERR gamma. J. Biochem. 2007, 142, 517–524. [Google Scholar] [CrossRef]
- Liu, X.; Matsushima, A.; Nakamura, M.; Costa, T.; Nose, T.; Shimohigashi, Y. Fine spatial assembly for construction of the phenol-binding pocket to capture Bisphenol A in the human nuclear receptor estrogen-related receptor γ. J. Biochem. 2012, 151, 403–415. [Google Scholar] [CrossRef]
- Balakrishnan, B.; Henare, K.; Thorstensen, E.B.; Ponnampalam, A.P.; Mitchell, M.D. Transfer of Bisphenol A across the human placenta. Am. J. Obstet. Ginecol. 2010, 202, 393.e1–393.e7. [Google Scholar] [CrossRef] [PubMed]
- Schönfelder, G.; Wittfoht, W.; Hopp, H.; Talsness, C.E.; Paul, M.; Chahoud, I. Parent Bisphenol A accumulation in the human maternal-fetal-placental unit. Environ. Health Perspect. 2002, 110, A703–A707. [Google Scholar] [CrossRef] [PubMed]
- Ejaredar, M.; Lee, Y.; Roberts, D.J.; Sauve, R.; Dewey, D. Bisphenol A exposure and children’s behavior: A systematic review. J. Expo. Sci. Environ. Epidemiol. 2017, 27, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Gupta, P.; Chandolia, A.; Alam, I. Bisphenol A: A threat to human health? J. Environ. Health 2015, 77, 20–26. [Google Scholar] [PubMed]
- Zhang, Y.; Wei, F.; Zhang, J.; Hao, L.; Jiang, J.; Dang, L.; Jiang, L. Bisphenol A and estrogen induce proliferation of human thyroid tumor cells via an estrogen-receptor-dependent pathway. Arch. Biochem. Biophys. 2017, 633, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Berger, R.G.; Foster, W.G.; deCatanzaro, D. Bisphenol-A exposure during the period of blastocyst implantation alters uterine morphology and perturbs measures of estrogen and progesterone receptor expression in mice. Birth Defects Res. B Dev. Reprod. Toxicol. 2010, 30, 393–400. [Google Scholar] [CrossRef]
- Huo, W.; Xia, W.; Wan, Y.; Zhang, B.; Zhou, A.; Zhang, Y.; Xu, S. Maternal urinary Bisphenol A levels and infant low birth weight: A nested case-control study of the Health Baby Cohort in China. Environ. Int. 2015, 85, 96–103. [Google Scholar] [CrossRef]
- Machtinger, R.; Orvieto, R. Bisphenol A, oocyte maturation, implantation, and IVF outcome: Review of animal and human data. Reprod. BioMed Online 2014, 29, 404–410. [Google Scholar] [CrossRef]
- Rajakumar, C.; Guan, H.; Langlois, D.; Cernea, M.; Yang, K. Bisphenol A disrupts gene expression in human placental trophoblast cells. Birth Defects Res. B Dev. Reprod. Toxicol. 2015, 53, 39–44. [Google Scholar] [CrossRef]
- Robins, J.C.; Marsit, C.J.; Padbury, J.F.; Sharma, S.S. Endocrine disruptors, environmental oxygen, epigenetics and pregnancy. Front. Biosci. 2011, 3, 690–700. [Google Scholar]
- Guida, M.; Troisi, J.; Ciccone, C.; Granozio, G.; Cosimato, C.; Sardo, A.D.S.; Di Carlo, C. Bisphenol A and congenital developmental defects in humans. Mutat. Res. 2015, 774, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, R.E.; George, J.D.; Price, C.J.; Tyl, R.W.; Marr, M.C.; Kimmel, C.A. The developmental toxicity of Bisphenol A in rats and mice. Fundam. Appl. Toxicol. 1987, 8, 1–582. [Google Scholar] [CrossRef]
- Bigsby, R.; Chapin, R.E.; Daston, G.P.; Davis, B.J.; Gorski, J.; Gray, L.E.; vom Saal, F.S. Evaluating the effects of endocrine disruptors on endocrine function during development. Environ. Health Perspect. 1999, 107 (Suppl. 4), 613–618. [Google Scholar] [PubMed]
- Betancourt, A.M.; Mobley, J.A.; Russo, J.; Lamartiniere, C.A. Proteomic analysis in mammary glands of rat offspring exposed in utero to bisphenol A. Int. J. Proteom. 2010, 73, 1241–1253. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, Y.; Xu, F.; Wei, X.; Zhang, J.; Wang, C.; Wang, Q. The rapid effect of Bisphenol-A on long-term potentiation in hippocampus involves estrogen receptors and ERK activation. J. Neural Transplant. Plast. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, R.; Zanatta, A.P.; Cavalari, F.C.; do Nascimento, M.A.W.; Delalande-Lecapitaine, C.; Bouraïma-Lelong, H.; Silva, F.R.M.B. Acute effect of Bisphenol A: Signaling pathways on calcium influx in immature rat testes. Birth Defects Res. B Dev. Reprod. Toxicol. 2018, 77, 94–102. [Google Scholar] [CrossRef]
- Lei, B.; Peng, W.; Xu, G.; Wu, M.; Wen, Y.; Xu, J.; Wang, Y. Activation of G protein-coupled receptor 30 by thiodiphenol promotes proliferation of estrogen receptor α-positive breast cancer cells. Chemosphere 2017, 169, 204–211. [Google Scholar] [CrossRef]
- Wang, C.; Fu, W.; Quan, C.; Yan, M.; Liu, C.; Qi, S.; Yang, K. The role of Pten/Akt signaling pathway involved in BPA-induced apoptosis of rat Sertoli cells. Res. J. Environ. Toxicol. 2015, 30, 793–802. [Google Scholar] [CrossRef]
- Orendi, K.; Gauster, M.; Moser, G.; Meiri, H.; Huppertz, B. The choriocarcinoma cell line BeWo: Syncytial fusion and expression of syncytium-specific proteins. Reproduction 2010, 140, 759–766. [Google Scholar] [CrossRef]
- Pattillo, R.A.; Gey, G.O. The establishment trophoblastic of a cell line of human hormone-synthesizing trophoblastic cells In Vitro. Cancer Res. 1968, 28, 1231–1236. [Google Scholar]
- Wice, B.; Menton, D.; Geuze, H.; Schwartz, A.L. Modulators of cyclic AMP metabolism induce syncytiotrophoblast formation in vitro. Exp. Cell Res. 1990, 186, 306–316. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Chisanga, D.; Alessandro, R.; Ang, C.-S.; Askenase, P.; Mathivanan, S. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J. Extracell. Vesicles 2017, 6, 1321455. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Chudasama, D.; Bo, V.; Hall, M.; Anikin, V.; Jeyaneethi, J.; Gregory, J.; Karteris, E. Identification of cancer biomarkers of prognostic value using specific gene regulatory networks (GRN): A novel role of RAD51AP1 for ovarian and lung cancers. Carcinogenesis 2018, 39, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Rebut-Bonneton, C.; Boutemy-Roulier, S.; Evain-Brion, D. Modulation of pp60c-src activity and cellular localization during differentiation of human trophoblast cells in culture. J. Cell Sci. 1993, 105 Pt 3, 629–636. [Google Scholar]
- García-Arévalo, M.; Alonso-Magdalena, P.; Servitja, J.-M.; Boronat-Belda, T.; Merino, B.; Villar-Pazos, S.; Nadal, A. Maternal exposure to Bisphenol-A during pregnancy increases pancreatic β-Cell growth during early life in male mice offspring. Endocrinology 2016, 157, 4158–4171. [Google Scholar] [CrossRef]
- Huang, D.; Wu, J.; Su, X.; Yan, H.; Sun, Z. Effects of low dose of Bisphenol A on the proliferation and mechanism of primary cultured prostate epithelial cells in rodents. Oncol. Lett. 2017, 14, 2635–2642. [Google Scholar] [CrossRef]
- Jeong, J.S.; Nam, K.T.; Lee, B.; Pamungkas, A.D.; Song, D.; Kim, M.; Lim, K.-M. Low-Dose Bisphenol A increases bile duct proliferation in juvenile rats: A possible evidence for risk of liver cancer in the exposed population? Biomol. Ther. (Seoul) 2017, 25, 545–552. [Google Scholar] [CrossRef]
- Sheng, Z.-G.; Huang, W.; Liu, Y.-X.; Zhu, B.-Z. Bisphenol A at a low concentration boosts mouse spermatogonial cell proliferation by inducing the G protein-coupled receptor 30 expression. Toxicol. Appl. Pharmacol. 2013, 267, 88–94. [Google Scholar] [CrossRef]
- Shi, X.-Y.; Wang, Z.; Liu, L.; Feng, L.-M.; Li, N.; Liu, S.; Gao, H. Low concentrations of Bisphenol A promote human ovarian cancer cell proliferation and glycolysis-based metabolism through the estrogen receptor-α pathway. Chemosphere 2017, 185, 361–367. [Google Scholar] [CrossRef]
- Ponniah, M.; Billett, E.E.; De Girolamo, L.A. Bisphenol A increases BeWo trophoblast survival in stress-induced paradigms through regulation of oxidative stress and apoptosis. Chem. Res. Toxicol. 2015, 28, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Takai, Y.; Tsutsumi, O.; Ikezuki, Y.; Hiroi, H.; Osuga, Y.; Momoeda, M.; Taketani, Y. Estrogen receptor-mediated effects of a xenoestrogen, bisphenol A, on preimplantation mouse embryos. Biochem. Biophys. Res. Commun. 2000, 270, 918–921. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Lu, J.; Zhang, Y.-Z.; Zhang, M.; Liu, T.; Qu, X.-L. Effect of Bisphenol A on invasion ability of human trophoblastic cell line BeWo. Int. J. Clin. Exp. Pathol. 2015, 8, 14355–14364. [Google Scholar] [PubMed]
- Narciso, L.; Letta, F.; Romagnoli, R.; Paulesu, L.; Mantovani, A.; Tait, S. Effects of Bisphenol A on endogenous retroviral envelopes expression and trophoblast fusion in BeWo cells. Reprod. Toxicol. 2019, 89, 35–44. [Google Scholar] [CrossRef]
- Welshons, W.V.; Nagel, S.C.; Vom Saal, F.S. Large effects from small exposures. III. Endocrine mechanisms mediating effects of bisphenol A at levels of human exposure. Endocrinology 2006, 147, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Welshons, W.V.; Thayer, K.A.; Judy, B.M.; Taylor, J.A.; Curran, E.M.; vom Saal, F.S. Large effects from small exposures. I. Mechanisms for endocrine-disrupting chemicals with estrogenic activity. Environ. Health Perspect. 2003, 111, 994–1006. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, N.; Bouskine, A.; Fenichel, P. Bisphenol A promotes testicular seminoma cell proliferation through GPER/GPR30. Int. J. Cancer 2012, 130, 241–242. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, I.; Oriente, F.; D’Esposito, V.; Liguoro, D.; Liguoro, P.; Ambrosio, M.R.; Cabaro, S.; D’Andrea, F.; Beguinot, F.; Formisano, P.; et al. Low-Dose Bisphenol A regulates inflammatory cytokines through GPR30 in mammary adipose cells. J. Mol. Endocrinol. 2019, 63, 273–283. [Google Scholar] [CrossRef]
- Herz, C.; Tran, H.T.T.; Schlotz, N.; Michels, K.; Lamy, E. Low-Dose levels of Bisphenol A inhibit telomerase via ER/GPR30-ERK signalling, impar DNA integrity and reduce cell proliferation in primary PBMC. Sci. Rep. 2017, 7, 16631. [Google Scholar] [CrossRef]
- Wang, C.; Li, Z.; Han, H.; Luo, G.; Zhou, B.; Wang, S.; Wang, J. Impairment of object recognition memory by maternal Bisphenol A exposure is associated with inhibition of Akt and ERK/CREB/BDNF pathway in the male offspring hippocampus. Toxicology 2016, 341–343, 56–64. [Google Scholar] [CrossRef]
- Vatish, M.; Tesfa, L.; Grammatopoulos, D.; Yamada, E.; Bastie, C.C.; Pessin, J.E. Inhibition of Akt activity and calcium channel function coordinately drive cell-cell fusion in the BeWo choreocarcinoma placental cell line. PLoS ONE 2012, 7, e29353. [Google Scholar] [CrossRef][Green Version]
- Zachariades, E.; Foster, H.; Goumenou, A.; Thomas, P.; Rand-Weaver, M.; Karteris, E. Expression of membrane and nuclear progesterone receptors in two human placental choriocarcinoma cell lines (JEG-3 and BeWo): Effects of syncytialization. Int. J. Mol. Med. 2011, 27, 767–774. [Google Scholar]
- Chou, W.C.; Lee, P.H.; Tan, Y.Y.; Lin, H.C.; Yang, C.W.; Chen, K.H.; Chuang, C.Y. An integrative transcriptomic analysis reveals Bisphenol A exposure-induced dysregulation of microRNA expression in human endometrial cells. Toxicol. In Vitro 2017, 41, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Hui, L.; Li, H.; Lu, G.; Chen, Z.; Sun, W.; Shi, Y.; Fu, Z.; Huang, B.; Zhu, X.; Lu, W.; et al. Low dose of Bisphenol A modulates ovarian cancer gene expression profile and promotes epithelial to mesenchymal transition via canonical wnt pathway. Toxicol. Sci. 2018, 164, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Asterholm, I.W.; Mundy, D.I.; Weng, J.; Anderson, R.G.W.; Scherer, P.E. Altered mitochondrial function and metabolic inflexibility associated with loss of caveolin-1. Cell Metab. 2012, 15, 171–185. [Google Scholar] [PubMed]
- Ding, L.; Zeng, Q.; Wu, J.; Li, D.; Wang, H.; Lu, W.; Xu, G. Caveolin-1 regulates oxidative stress-induced senescence in nucleus pulposus cells primarily via the p53/p21 signaling pathway In Vitro. Mol. Med. Rep. 2017, 16, 9521–9527. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Rojo, M.A.; Gongora, M.; Fitzsimmons, R.L.; Martel, N.; Martin, S.D.; Nixon, S.J.; Parton, R.G. Caveolin-1 is necessary for hepatic oxidative lipid metabolism: Evidence for crosstalk between caveolin-1 and bile acid signaling. Cell Rep. 2013, 4, 238–247. [Google Scholar] [CrossRef]
- Li, M.; Chen, D.; Huang, H.; Wang, J.; Wan, X.; Xu, C.; Li, Y. Caveolin1 protects against diet induced hepatic lipid accumulation in mice. PLoS ONE 2017, 12, e0178748. [Google Scholar] [CrossRef]
- Parton, R.G.; Simons, K. The multiple faces of caveolae. Nat. Rev. Mol. Cell Biol. 2007, 8, 185–194. [Google Scholar] [CrossRef]
- Smart, E.J.; Graf, G.A.; McNiven, M.A.; Sessa, W.C.; Engelman, J.A.; Scherer, P.E.; Lisanti, M.P. Caveolins, liquid-ordered domains, and signal transduction. Mol. Cell Biol. 1999, 19, 7289–7304. [Google Scholar] [CrossRef]
- Adlanmerini, M.; Solinhac, R.; Abot, A.; Fabre, A.; Raymond-Letron, I.; Guihot, A.-L.; Lenfant, F. Mutation of the palmitoylation site of estrogen receptor α In Vivo reveals tissue-specific roles for membrane versus nuclear actions. Proc. Natl. Acad. Sci. USA 2014, 111, E283–E290. [Google Scholar] [CrossRef]
- Chambliss, K.L.; Yuhanna, I.S.; Mineo, C.; Liu, P.; German, Z.; Sherman, T.S.; Shaul, P.W. Estrogen receptor alpha and endothelial nitric oxide synthase are organized into a functional signaling module in caveolae. Circ. Res. 2000, 87, E44–E52. [Google Scholar] [CrossRef] [PubMed]
- Levin, E.R. Integration of the extranuclear and nuclear actions of estrogen. Mol. Endocrinol. 2005, 19, 1951–1959. [Google Scholar] [CrossRef]
- Yao, G.; Zhang, Y.; Wang, D.; Yang, R.; Sang, H.; Han, L.; Shang, Z. GDM-Induced macrosomia is reversed by Cav-1 via AMPK-Mediated fatty acid transport and GLUT1-Mediated glucose transport in placenta. PLoS ONE 2017, 12, e0170490. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Cai, J.; Zhu, Z.; Chen, D.; Wang, J.; Wang, Q.; Chen, A.F. Hypoxic trophoblast HMGB1 induces endothelial cell hyperpermeability via the TRL-4/caveolin-1 pathway. J. Immunol. 2014, 193, 5000–5012. [Google Scholar] [CrossRef] [PubMed]
- Suemizu, H.; Aiba, K.; Yoshikawa, T.; Sharov, A.A.; Shimozawa, N.; Tamaoki, N.; Ko, M.S. Expression profiling of placentomegaly associated with nuclear transplantation of mouse ES cells. Dev. Biol. 2003, 253, 36–53. [Google Scholar] [CrossRef]
- Pawoo, N.; Heller, D.S. Placental mesenchymal dysplasia. Arch. Pathol. Lab. Med. 2014, 138, 1247–1249. [Google Scholar] [CrossRef] [PubMed]
- Koslowski, M.; Türeci, O.; Biesterfeld, S.; Seitz, G.; Huber, C.; Sahin, U. Selective activation of trophoblast-specific PLAC1 in breast cancer by CCAAT/enhancer-binding protein beta (C/EBPbeta) isoform 2. J. Biol. Chem. 2009, 284, 28607–28615. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Koslowski, M.; Paret, C.; Schmidt, M.; Türeci, O.; Sahin, U. NCOA3 is a selective co-activator of estrogen receptor α-mediated transactivation of PLAC1 in MCF-7 breast cancer cells. BMC Cancer 2013, 13, 570. [Google Scholar] [CrossRef]
- Yang, L.; Zha, T.-Q.; He, X.; Chen, L.; Zhu, Q.; Wu, W.-B.; Lu, K.-H. Placenta-Specific protein 1 promotes cell proliferation and invasion in non-small cell lung cancer. Oncol. Rep. 2018, 39, 53–60. [Google Scholar] [CrossRef]
- Chardonnens, D.; Cameo, P.; Aubert, M.L.; Pralong, F.P.; Islami, D.; Campana, A.; Bischof, P. Modulation of human cytotrophoblastic leptin secretion by interleukin-1alpha and 17beta-oestradiol and its effect on HCG secretion. Mol. Hum. Reprod. 1999, 5, 1077–1082. [Google Scholar] [CrossRef] [PubMed]
- López Fontana, C.M.; Maselli, M.E.; Pérez Elizalde, R.F.; Di Milta Mónaco, N.A.; Uvilla Recupero, A.L.; López Laur, J.D. Leptin increases prostate cancer aggressiveness. J. Physiol. Biochem. 2011, 67, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Castellucci, M.; De Matteis, R.; Meisser, A.; Cancello, R.; Monsurrò, V.; Islami, D.; Bischof, P. Leptin modulates extracellular matrix molecules and metalloproteinases: Possible implications for trophoblast invasion. Mol. Hum. Reprod. 2000, 6, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Chrelias, G.; Makris, G.M.; Papanota, A.M.; Spathis, A.; Salamalekis, G.; Sergentanis, T.N.; Chrelias, C. Serum inhibin and leptin: Risk factors for pre-eclampsia? Clin. Chim. Acta. 2016, 463, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Gambino, Y.P.; Maymó, J.L.; Pérez Pérez, A.; Calvo, J.C.; Sánchez-Margalet, V.; Varone, C.L. Elsevier trophoblast research award lecture: Molecular mechanisms underlying estrogen functions in trophoblastic cells—Focus on leptin expression. Placenta 2012, 33, S63–S70. [Google Scholar] [CrossRef] [PubMed]
- Fatima, S.S.; Alam, F.; Chaudhry, B.; Khan, T.A. Elevated levels of chemerin, leptin, and interleukin-18 in gestational diabetes mellitus. J. Matern. Fetal Neonatal Med. 2017, 30, 1023–1028. [Google Scholar] [CrossRef]
- Jeon, E.J.; Hong, S.Y.; Lee, J.H. Adipokines and insulin resistance according to characteristics of pregnant women with gestational diabetes mellitus. Diabetes Metab. 2017, 41, 457–465. [Google Scholar] [CrossRef]
- Lobo, T.F.; Torloni, M.R.; Mattar, R.; Nakamura, M.U.; Alexandre, S.M.; Daher, S. Adipokine levels in overweight women with early-onset gestational diabetes mellitus. J. Endocrinol. Invest. 2018, 42, 149–156. [Google Scholar] [CrossRef]
- Popova, P.; Vasilyeva, L.; Tkachuck, A.; Puzanov, M.; Golovkin, A.; Bolotko, Y.; Grineva, E. A randomised, controlled study of different glycaemic targets during gestational diabetes treatment: Effect on the level of adipokines in cord blood and ANGPTL4. Int. J. Endocrinol. 2018, 2018, 6481658. [Google Scholar] [CrossRef]
- Sweeting, A.N.; Wong, J.; Appelblom, H.; Ross, G.P.; Kouru, H.; Williams, P.F.; Hyett, J.A. A novel early pregnancy risk prediction model for gestational diabetes mellitus. Fetal Diagn. Ther. 2019, 45, 76–84. [Google Scholar] [CrossRef]
- El Shahat, A.M.; Ahmed, A.B.; Ahmed, M.R.; Mohamed, H.S. Maternal serum leptin as a marker of preeclampsia. Arch. Gynecol. Obstet. 2013, 288, 1317–1322. [Google Scholar] [CrossRef] [PubMed]
- Güngör, Z.B.; Ekmekçi, H.; Tüten, A.; Toprak, S.; Ayaz, G.; Çalışkan, O.; Balcı Ekmekçi, Ö. Is there any relationship between adipocytokines and angiogenesis factors to address endothelial dysfunction and platelet aggregation in untreated patients with preeclampsia? Arch. Gynecol. Obstet. 2017, 296, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Gurugubelli Krishna, R.; Vishnu Bhat, B. Molecular mechanisms of intrauterine growth restriction. J. Matern. Fetal Neonatal Med. 2017, 31, 2634–2640. [Google Scholar] [CrossRef] [PubMed]
- Nezar, M.A.S.; El-Baky, M.A.; Soliman, O.A.S.; Abdel-Hady, H.A.S.; Hammad, A.M.; Al-Haggar, M.S. Endothelin-1 and leptin as markers of intrauterine growth restriction. Indian J. Pediatr. 2009, 76, 485–488. [Google Scholar] [CrossRef]
- Rönn, M.; Lind, L.; Örberg, J.; Kullberg, J.; Söderberg, S.; Larsson, A.; Lind, P.M. Bisphenol A is related to circulating levels of adiponectin, leptin and ghrelin, but not to fat mass or fat distribution in humans. Chemosphere 2014, 112, 42–48. [Google Scholar] [CrossRef]
- Ariemma, F.; D’Esposito, V.; Liguoro, D.; Oriente, F.; Cabaro, S.; Liotti, A.; Valentino, R. Low-Dose Bisphenol-A impairs adipogenesis and generates dysfunctional 3T3-L1 adipocytes. PLoS ONE 2016, 11, e0150762. [Google Scholar] [CrossRef]
- Chang, W.-L.; Liu, Y.-W.; Dang, Y.-L.; Jiang, X.-X.; Xu, H.; Huang, X.; Wang, H. PLAC8, a new marker for human interstitial extravillous trophoblast cells, promotes their invasion and migration. Development 2018, 145. [Google Scholar] [CrossRef]
- Sasaki, D.; Kotoh, J.; Watadani, R.; Matsumoto, K. New animal models reveal that coenzyme Q2 (Coq2) and placenta-specific 8 (Plac8) are candidate genes for the onset of type 2 diabetes associated with obesity in rats. Mamm. Genome 2015, 26, 619–629. [Google Scholar] [CrossRef]
- Blue, E.K.; Sheehan, B.M.; Nuss, Z.V.; Boyle, F.A.; Hocutt, C.M.; Gohn, C.R.; Haneline, L.S. Epigenetic regulation of placenta-specific 8 contributes to altered function of endothelial colony-forming cells exposed to intrauterine gestational diabetes mellitus. Diabetes 2015, 64, 2664–2675. [Google Scholar] [CrossRef]
- Jimenez-Preitner, M.; Berney, X.; Thorens, B. Plac8 is required for white adipocyte differentiation In Vitro and cell number control In Vivo. PLoS ONE 2012, 7, e48767. [Google Scholar] [CrossRef]
- Jimenez-Preitner, M.; Berney, X.; Uldry, M.; Vitali, A.; Cinti, S.; Ledford, J.G.; Thorens, B. Plac8 is an inducer of C/EBPβ required for brown fat differentiation, thermoregulation, and control of body weight. Cell Metab. 2011, 14, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Rogulski, K.; Li, Y.; Rothermund, K.; Pu, L.; Watkins, S.; Yi, F.; Prochownik, E.V. Onzin, a c-Myc-repressed target, promotes survival and transformation by modulating the Akt–Mdm2–p53 pathway. Oncogene 2005, 24, 7524–7541. [Google Scholar] [CrossRef] [PubMed]
- Berry, S.A.; Srivastava, C.H.; Rubin, L.R.; Phipps, W.R.; Pescovitz, O.H. Growth hormone-releasing hormone-like messenger ribonucleic acid and immunoreactive peptide are present in human testis and placenta. J. Clin. Endocrinol. Metab. 1992, 75, 281–284. [Google Scholar] [PubMed]
- Liu, A.X.; Zhang, D.; Zhu, Y.M.; Gao, H.J.; Jiang, J.Y.; Hu, X.L.; Lv, P.P.; Leung, P.C.; Huang, H.F. Impact of axis of GHRH and GHRH Receptor on cell viability and apoptosis of the placental choriocarcinoma cell line. Curr. Mol. Med. 2016, 16, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Perry, I.D.; Krishnan, L.; Murphy, S.P. SLC11A1 is expressed in the human placenta across multiple gestational ages. Placenta 2019, 75, 23–26. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Dutta, S.; Mukherjee, S.; Mukherjee, N.; Dutta, A.; Mukherjee, A.; Sinha, S.; Panda, C.K.; Chaudhuri, K.; Roy, A.L.; et al. Potential contribution of SIM2 and ETS2 functional polymorphisms in Down syndrome associated malignancies. BMC Med. Genet. 2013, 14, 12. [Google Scholar] [CrossRef][Green Version]
- Fan, C.-M.; Kuwana, E.; Bulfone, A.; Fletcher, C.F.; Copeland, N.G.; Jenkins, N.A.; Tessier-Lavigne, M. Expression patterns of two murine homologs of drosophila single-minded suggest possible roles in embryonic patterning and in the pathogenesis of Down Syndrome. Mol. Cell Neurosci. 1996, 7, 519. [Google Scholar] [CrossRef]
- Yang, X.; Glazebrook, P.; Ranasinghe, G.C.; Haghiac, M.; Calabuig-Navarro, V.; Minium, J.; O’Tierney-Ginn, P. Fatty acid transporter expression and regulation is impaired in placental macrovascular endothelial cells in obese women. J. Matern. Fetal Neonatal Med. 2019, 32, 971–978. [Google Scholar] [CrossRef]
- Gilmartin, A.B.H.; Ural, S.H.; Repke, J.T. Gestational diabetes mellitus. Rev. Obstet. Gynecol. 2008, 1, 129–134. [Google Scholar]
- Petry, C.J. Gestational diabetes: Risk factors and recent advances in its genetics and treatment. Br. J. Nutr. 2010, 104, 775–787. [Google Scholar] [CrossRef]
- Schmidt, M.I.; Duncan, B.B.; Reichelt, A.J.; Branchtein, L.; Matos, M.C.; Forti, A.C.E.; Yamashita, T. Gestational diabetes mellitus diagnosed with a 2-h 75-g oral glucose tolerance test and adverse pregnancy outcomes. Diabetes Care 2001, 24, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
| Gene | Sequence |
|---|---|
| CAV1 | F: 5′ACCCACTCTTTGAAGCTGTTG3′ |
| R: 5′GAACTTGAAATTGGCACCAGG3′ | |
| Leptin | F: 5′CCTGACTGGTGCTATAGGCTGGA3′ |
| R: 5′GTGAGTGCGGTTTGACCACTG3′ | |
| hPLAC8 | F: 5′GGGTGTCAAGTTGCAGCTGAT3′ |
| R: 5′TAGATCCAGGGATGCCATATCG3′ | |
| Syncytin 2 | F: 5′AGCAGCCGTAGTCCTTCAAA3′ |
| R: 5′AGGGGAAGAACCCAAGAGAA3′ | |
| ERα | F: 5′GCCCTCCCTCCCTGAAC3′ |
| R: 5′TCAACTACCATTTACCCTCATC3′ | |
| ERβ | F: 5′TCCTCCCAGCAGCAATCC3′ |
| R: 5′CCAGCAGCAGGTCATACAC3′ | |
| GPR30 | F: 5′GTTCCTCTCGTGCCTCTAC3′ |
| R: 5′ACCGCCAGGTTGATGAAG3′ | |
| TOP1 | F: 5′CCTTCCCTCTCTCCCATTTC3′ |
| R: 5′AGCCACGACTGCTTCAAGTT3′ |
| Index | Biological Pathway | p Value | Input Genes |
|---|---|---|---|
| 1 | Leptin/insulin overlap | 0.0016 | suppressor of cytokine signaling 3 (SOCS3), suppressor of cytokine signaling 1 (SOCS1), leptin (LEP), insulin receptor substrate 2 (IRS2) |
| 2 | Differentiation of white and brown adipocyte | 0.0098 | PLAC8-like 1 (PLAC8), LEP, SMAD family member 9 (SMAD9), zinc finger protein 423 (ZNF423), PPARG coactivator 1 beta (PPARGC1B) |
| 3 | Kit receptor signalling pathway | 0.0136 | ribosomal protein S6 kinase, 90 kDa, polypeptide 3 (RPS6KA3), (mitogen-activated protein kinase 8) MAPK8, SOCS1, ribosomal protein S6 kinase B1 (RPS6KB1), SHC (Src homology 2 domain containing) transforming protein 1 (SHC1), inositol polyphosphate-5-phosphatase D (INPP5D), FYN proto-oncogene, Src family tyrosine kinase (FYN), microtubule-associated protein tau (MAPT) |
| 4 | Integrin-mediated cell adhesion | 0.0205 | vasodilator-stimulated phosphoprotein (VASP), G protein-coupled receptor kinase interacting ArfGAP 2 (GIT2), (SHC1), Rho-associated, coiled-coil containing protein kinase 2 (ROCK2), CAV1, integrin, alpha 1 (ITGA1), integrin, alpha X (complement component 3 receptor 4 subunit) (ITGAX), Rap guanine nucleotide exchange factor (GEF) 1 (RAPGEF1), p21 protein (Cdc42/Rac)-activated kinase 6 (PAK6), integrin, alpha 6 (ITGA6), FYN |
| 5 | Prolactin signalling pathway | 0.0211 | SOCS3, MAPK8, SOCS1, RPS6KB1, SHC1, erb-b2 receptor tyrosine kinase 2 (ERBB2), IRS2, FYN |
| 6 | Leptin signalling pathway | 0.0211 | SOCS3, MAPK8, RPS6KB1, SOCS1, SHC1, ROCK2, LEP, ERBB2, BCL2-associated X protein (BAX), FYN |
| 7 | IL-2 signalling pathway | 0.0244 | SOCS3, RPS6KB1, SHC1, FYN, N-myc (and STAT) interactor (NMI), MAPT |
| 8 | Angiopoietin-like protein 8 regulatory pathway | 0.0265 | SHC1, solute carrier family 2 (facilitated glucose transporter), member 1 (SLC2A1), protein kinase, AMP-activated, gamma 1 non-catalytic subunit (PRKAG1), phosphoinositide-3-kinase, regulatory subunit 3 (gamma) (PIK3R3), IRS2, RPS6KA3, MAPK8, sestrin 3 (SESN3), RPS6KB1, RAPGEF1, mitogen-activated protein kinase kinase kinase 6 (MAP3K6), son of sevenless homolog 2 (SOS2), mitogen-activated protein kinase kinase kinase 5 (MAP3K5) |
| 9 | Insulin signalling | 0.0268 | syntaxin binding protein 4 (STXBP4), SHC1, SLC2A1, PIK3R3, RPS6KA3, SOCS3, MAPK8, SOCS1, RPS6KB1, RAPGEF1, tribbles pseudokinase 3 (TRIB3), MAP3K6, SOS2, MAP3K5 |
| 10 | Focal adhesion | 0.0403 | vasodilator-stimulated phosphoprotein (VASP), von Willebrand factor (VWF), SHC1, ROCK2, laminin, beta 2 (laminin S) (LAMB2), CAV1, ITGA1, PIK3R3, Rho GTPase activating protein 5 (ARHGAP5), myosin light chain kinase (MYLK), MAPK8, ERBB2, RAPGEF1, p21 protein (Cdc42/Rac)-activated kinase 6 (PAK6), filamin B, beta (FLNB), ITGA6, FYN |
| Index | Term | p Value | Input Genes |
|---|---|---|---|
| 1 | Differentiation of white and brown adipocyte | 0.002 | PLAC8, SMAD family member 1 (SMAD1), LEP, PPARGC1B |
| 2 | Methylation pathways | 0.015 | nicotinamide N-methyltransferase (NNMT), thiopurine S-methyltransferase (TPMT) |
| 3 | Mechanoregulation and pathology of YAP/TAZ via Hippo and non-Hippo mechanisms | 0.019 | integrin, beta 4 (ITGB4), integrin, beta 3 (ITGB3), macrophage stimulating 1 (MST1), actin, gamma 2 (ACTG2) |
| 4 | Photodynamic therapy-induced unfolded protein response | 0.021 | DNA-damage-inducible transcript 3 (DDIT3), TRIB3, activating transcription factor 3 (ATF3) |
| 5 | Alanine and aspartate metabolism | 0.027 | alanine-glyoxylate aminotransferase (AGXT), argininosuccinate synthase 1 (ASS1) |
| 6 | Mitochondrial gene expression WP391 | 0.064 | GA binding protein transcription factor, beta subunit 1(GABPB1), PPARGC1B |
| 7 | Hypertrophy model | 0.070 | ATF3, heparin-binding EGF-like growth factor (HBEGF) |
| 8 | Complement activation | 0.083 | Complement C 3 (C3), C15 |
| 9 | Exercise-induced circadian regulation | 0.087 | DAZ associated protein 2 (DAZAP2), cryptochrome circadian clock 2 (CRY2) |
| (10 | NRF2 pathway | 0.101 | ATP-binding cassette, sub-family C (CFTR/MRP), member 3 (ABCC3), early growth response 1 (EGR1), solute carrier family 6 member 9 (SLC6A9), glutathione S-transferase alpha 4 (GSTA4), solute carrier family 39 member 7 SLC39A7, HBEGF |
| Index | Name of Biological Pathway | p-Value | Input Genes |
|---|---|---|---|
| 1 | nuclear factor erythroid 2-related factor 2 (NRF2) pathway | 0.00464 | ATP-binding cassette, sub-family C (CFTR/MRP), member 3 (ABCC3); gamma-glutamyltransferase light chain 1 (GGTLC1); carbonyl reductase 1 (CBR1); NAD(P)H dehydrogenase, quinone 1 (NQO1); solute carrier family 6 member 15 (SLC6A15); solute carrier family 2 member 5 (SLC2A5); glutathione S-transferase mu 5 (GSTM5) |
| 2 | Role of Osx and miRNAs in tooth development | 0.01831 | HNF1 homeobox A (HNF1A); Sp7 transcription factor (SP7) |
| 3 | Oxidative damage | 0.01849 | B-cell CLL/lymphoma 2 (BCL2); cytochrome c, somatic (CYCS); tumor necrosis factor receptor superfamily, member 1B (TNFRSF1B) |
| 4 | Nanomaterial-induced apoptosis | 0.03164 | BCL2; CYCS |
| 5 | Nuclear receptors meta-pathway | 0.03703 | ABCC3; gamma-glutamyltransferase light chain 1 (GGTLC1); carbonyl reductase 1 (CBR1); nuclear receptor coactivator 2 (NCOA2); NQO1; SC6A15; DnaJ (Hsp40) homolog, subfamily C, member 15 (DNAJC15); SLC2A5; GSTM5 |
| 6 | Apoptosis modulation and signalling | 0.03943 | BCL2; CYCS; Bcl2 modifying factor (BMF); tumor necrosis factor receptor superfamily, member 1B (TNFRSF1B) |
| 7 | Photodynamic therapy-induced NFE2L2 (NRF2) survival signalling | 0.04431 | ABCC3; NQO1 |
| 8 | Gastric cancer network 2 | 0.07007 | collagen, type IX, alpha 1(COL9A1); family with sequence similarity 91, member A1 (FAM91A1) |
| 9 | Constitutive androstane receptor pathway | 0.07406 | ABCC3; NCOA2 |
| 10 | Oxidative stress | 0.07814 | NQO1; NADPH oxidase 1 (NOX1) |
| Index | Name of Biological Pathway | p-Value | Input Genes |
|---|---|---|---|
| 1 | mir-124 predicted interactions with cell cycle and differentiation | 0.0395 | STE20-related kinase adaptor beta (STRADB) |
| 2 | LncRNA-mediated mechanisms of therapeutic resistance | 0.0395 | hypoxia inducible factor 1, alpha subunit (HIF1A) |
| 3 | MicroRNA for targeting cancer growth and vascularization | 0.0460 | HIF1A |
| 4 | HIF1A and Peroxisome Proliferator Activated Receptor Gamma (PPARG) regulation of glycolysis | 0.0524 | HIF1A |
| 5 | TGIF disruption of SHH signalling (Hedgehog signaling pathway) | 0.0587 | TGFB-induced factor homeobox 1 (TGIF1) |
| 6 | Notch signalling pathway | 0.0631 | melanoma antigen family A,1 (MAGEA1); HIF1A |
| 7 | Steroid biosynthesis | 0.0650 | hydroxysteroid (17-beta) dehydrogenase 4 (HSD17B4) |
| 8 | Ectoderm differentiation | 0.0657 | frizzled class receptor 5 (FZD5); ribosome binding protein 1 (RRBP1); WD repeat domain 44 (WDR44) |
| 9 | G1 to S cell cycle control | 0.0686 | cyclin G2 (CCNG2); polymerase (DNA directed), epsilon, catalytic subunit (POLE) |
| 10 | NAD metabolism, sirtuins, and aging | 0.0713 | HIF1A |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Aguiar Greca, S.-C.; Kyrou, I.; Pink, R.; Randeva, H.; Grammatopoulos, D.; Silva, E.; Karteris, E. Involvement of the Endocrine-Disrupting Chemical Bisphenol A (BPA) in Human Placentation. J. Clin. Med. 2020, 9, 405. https://doi.org/10.3390/jcm9020405
de Aguiar Greca S-C, Kyrou I, Pink R, Randeva H, Grammatopoulos D, Silva E, Karteris E. Involvement of the Endocrine-Disrupting Chemical Bisphenol A (BPA) in Human Placentation. Journal of Clinical Medicine. 2020; 9(2):405. https://doi.org/10.3390/jcm9020405
Chicago/Turabian Stylede Aguiar Greca, Sophie-Christine, Ioannis Kyrou, Ryan Pink, Harpal Randeva, Dimitris Grammatopoulos, Elisabete Silva, and Emmanouil Karteris. 2020. "Involvement of the Endocrine-Disrupting Chemical Bisphenol A (BPA) in Human Placentation" Journal of Clinical Medicine 9, no. 2: 405. https://doi.org/10.3390/jcm9020405
APA Stylede Aguiar Greca, S.-C., Kyrou, I., Pink, R., Randeva, H., Grammatopoulos, D., Silva, E., & Karteris, E. (2020). Involvement of the Endocrine-Disrupting Chemical Bisphenol A (BPA) in Human Placentation. Journal of Clinical Medicine, 9(2), 405. https://doi.org/10.3390/jcm9020405






