Clustering Heatmap for Visualizing and Exploring Complex and High-dimensional Data Related to Chronic Kidney Disease
Abstract
1. Introduction
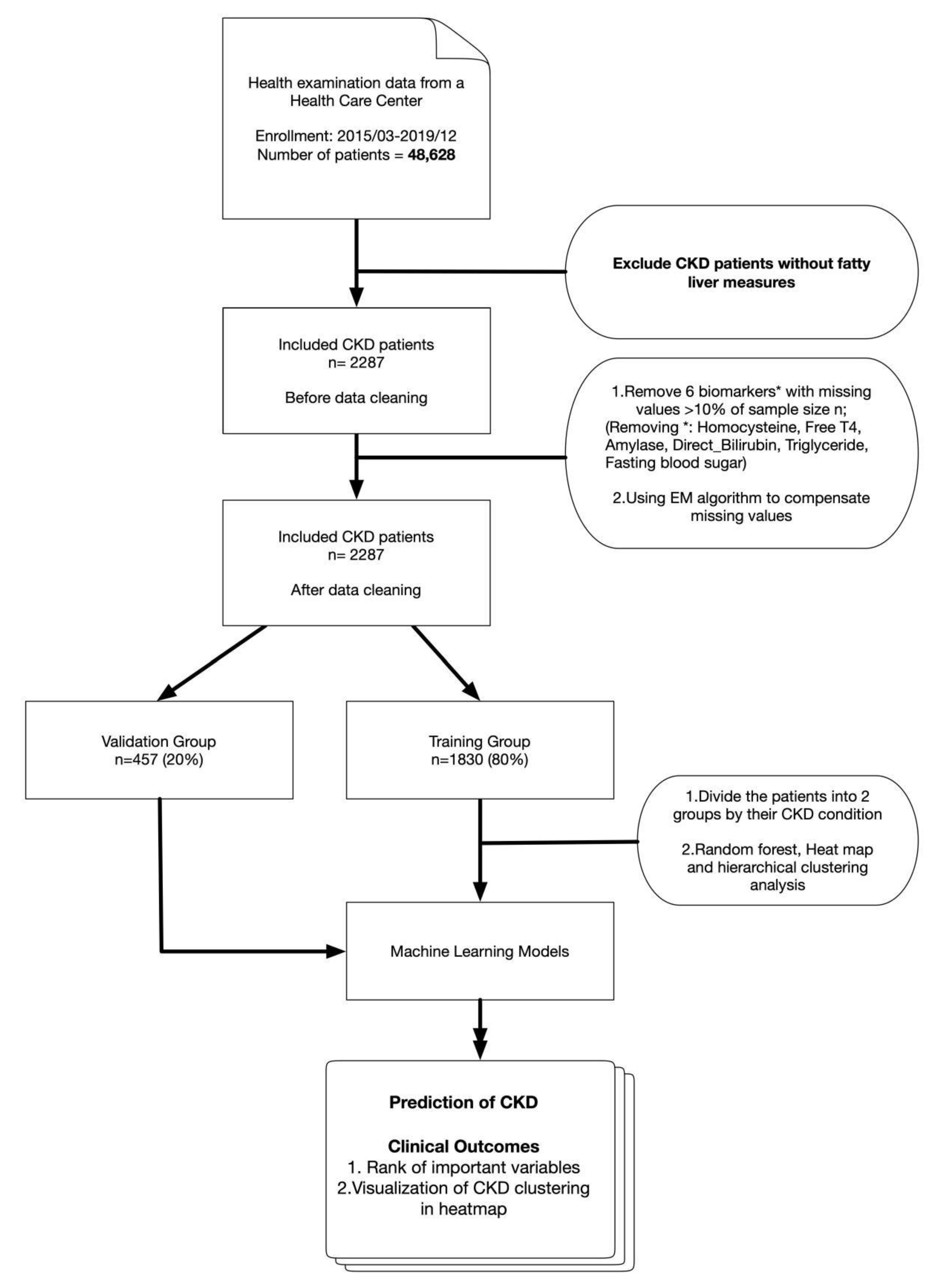
2. Experimental Section
2.1. Study Design
2.2. Setting
2.3. Inclusion and Exclusion Criteria
2.4. Data
2.5. Definitions of Measurement Cutoffs and Calculations
2.6. Statistical Analysis
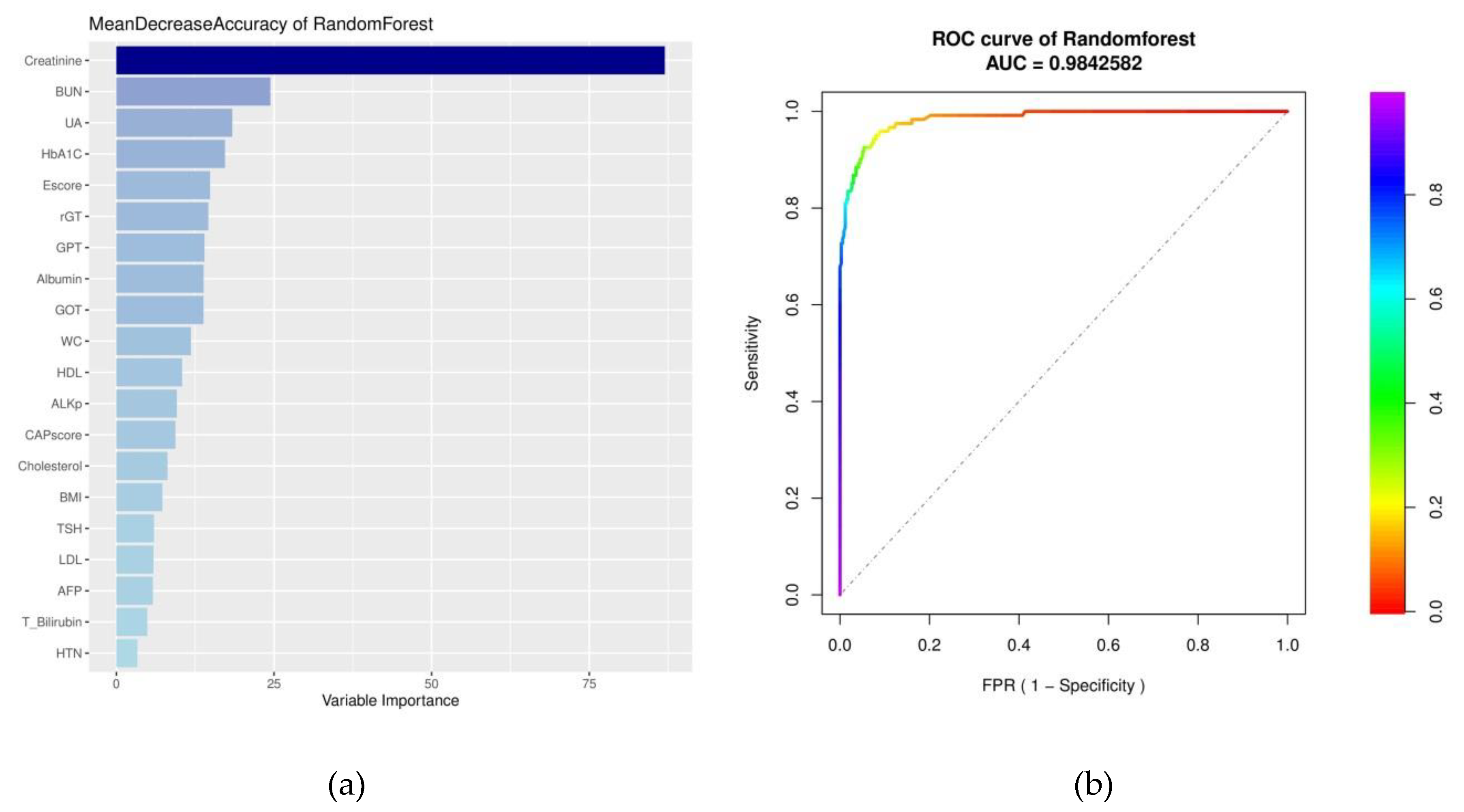
2.6.1. Receiver Operating Characteristic Curve
2.6.2. Random Forest
2.6.3. Missing Values
2.6.4. Multivariate Analysis
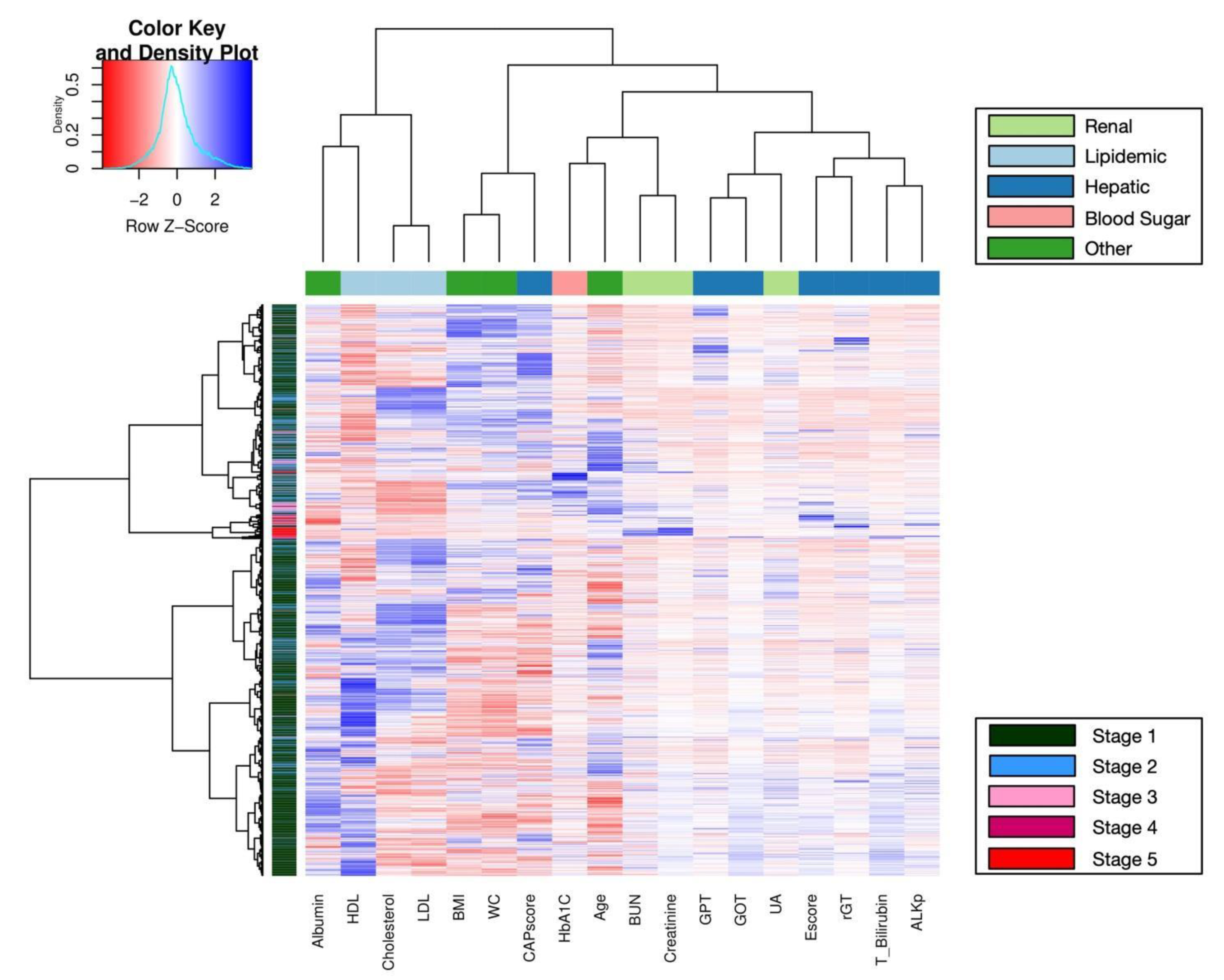
2.6.5. Heatmap and Clustering
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nugent, R.A.; Fathima, S.F.; Feigl, A.B.; Chyung, D. The burden of chronic kidney disease on developing nations: A 21st century challenge in global health. Nephron Clin. Pract. 2011, 118, C269–C276. [Google Scholar] [CrossRef] [PubMed]
- Hill, N.R.; Fatoba, S.T.; Oke, J.L.; Hirst, J.A.; O’Callaghan, C.A.; Lasserson, D.S.; Hobbs, F.D.R. Global prevalence of chronic kidney disease—A systematic review and meta-analysis. PLoS ONE 2016, 11, e158765. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.P.; Cheng, T.Y.D.; Tsai, M.K.; Chang, Y.C.; Chan, H.T.; Tsai, S.P.; Chiang, P.H.; Hsu, C.C.; Sung, P.K.; Hsu, Y.H.; et al. All-cause mortality attributable to chronic kidney disease: A prospective cohort study based on 462 293 adults in taiwan. Lancet 2008, 371, 2173–2182. [Google Scholar] [CrossRef]
- Al-Aly, Z.; Zeringue, A.; Fu, J.; Rauchman, M.I.; McDonald, J.R.; El-Achkar, T.M.; Balasubramanian, S.; Nurutdinova, D.; Xian, H.; Stroupe, K.; et al. Rate of kidney function decline associates with mortality. J. Am. Soc. Nephrol.: JASN 2010, 21, 1961–1969. [Google Scholar] [CrossRef]
- Coresh, J.; Turin, T.C.; Matsushita, K.; Sang, Y.; Ballew, S.H.; Appel, L.J.; Arima, H.; Chadban, S.J.; Cirillo, M.; Djurdjev, O.; et al. Decline in estimated glomerular filtration rate and subsequent risk of end-stage renal disease and mortality. Jama 2014, 311, 2518–2531. [Google Scholar] [CrossRef]
- Matsushita, K.; Selvin, E.; Bash, L.D.; Franceschini, N.; Astor, B.C.; Coresh, J. Change in estimated gfr associates with coronary heart disease and mortality. J. Am. Soc. Nephrol.: JASN 2009, 20, 2617–2624. [Google Scholar] [CrossRef]
- O’Hare, A.M.; Batten, A.; Burrows, N.R.; Pavkov, M.E.; Taylor, L.; Gupta, I.; Todd-Stenberg, J.; Maynard, C.; Rodriguez, R.A.; Murtagh, F.E.M.; et al. Trajectories of kidney function decline in the 2 years before initiation of long-term dialysis. Am. J. Kidney Dis. 2012, 59, 513–522. [Google Scholar]
- Rosansky, S.J. Renal function trajectory is more important than chronic kidney disease stage for managing patients with chronic kidney disease. Am. J. Nephrol. 2012, 36, 1–10. [Google Scholar] [CrossRef]
- Charleonnan, A.; Fufaung, T.; Niyomwong, T.; Chokchueypattanakit, W.; Suwannawach, S.; Ninchawee, N. Predictive analytics for chronic kidney disease using machine learning techniques. In Proceedings of the 2016 Management and Innovation Technology International Conference (MITicon), Bang-San, Chonburi, Thailand, 12–14 October 2016; pp. MIT-80–MIT-83. [Google Scholar]
- Oeda, S.; Takahashi, H.; Imajo, K.; Seko, Y.; Ogawa, Y.; Moriguchi, M.; Yoneda, M.; Anzai, K.; Aishima, S.; Kage, M.; et al. Accuracy of liver stiffness measurement and controlled attenuation parameter using fibroscan® m/xl probes to diagnose liver fibrosis and steatosis in patients with nonalcoholic fatty liver disease: A multicenter prospective study. J. Gastroenterol. 2019. [Google Scholar] [CrossRef]
- Lin, Y.-J.; Lin, C.-H.; Wang, S.-T.; Lin, S.-Y.; Chang, S.-S. Noninvasive and convenient screening of metabolic syndrome using the controlled attenuation parameter technology: An evaluation based on self-paid health examination participants. J. Clin. Med. 2019, 8, 1775. [Google Scholar] [CrossRef]
- Lee, J.I.; Lee, H.W.; Lee, K.S. Value of controlled attenuation parameter in fibrosis prediction in nonalcoholic steatohepatitis. World J. Gastroenterol. 2019, 25, 4959–4969. [Google Scholar] [CrossRef] [PubMed]
- Ben Yakov, G.; Sharma, D.; Alao, H.; Surana, P.; Kapuria, D.; Etzion, O.; Rivera, E.; Huang, A.; Koh, C.; Heller, T.; et al. Vibration controlled transient elastography (fibroscan®) in sickle cell liver disease—Could we strike while the liver is hard? Br. J. Haematol. 2019, 187, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Vuppalanchi, R.; Chalasani, N. Nonalcoholic fatty liver disease and nonalcoholic steatohepatitis: Selected practical issues in their evaluation and management. Hepatology 2009, 49, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Alberti, K.; Zimmet, P.; Shaw, J. Metabolic syndrome—A new world-wide definition. A consensus statement from the international diabetes federation. Diabetic Med. 2006, 23, 469–480. [Google Scholar] [CrossRef]
- Ma, Y.-C.; Zuo, L.; Chen, J.-H.; Luo, Q.; Yu, X.-Q.; Li, Y.; Xu, J.-S.; Huang, S.-M.; Wang, L.-N.; Huang, W.; et al. Modified glomerular filtration rate estimating equation for chinese patients with chronic kidney disease. J. Am. Soc. Nephrol.: JASN 2006, 17, 2937–2944. [Google Scholar] [CrossRef]
- National Kidney Foundation. K/doqi clinical practice guidelines for chronic kidney disease: Evaluation, classification, and stratification. Am. J. Kidney Dis. Off. J. Natl. Kidney Found. 2002, 39, S1–S266. [Google Scholar]
- Fox, J.; Monette, G. Generalized collinearity diagnostics. J. Am. Stat. Assoc. 1992, 87, 178–183. [Google Scholar] [CrossRef]
- Fox, J.; Bates, D.; Firth, D.; Friendly, M.; Gorjanc, G.; Graves, S.; Heiberger, R.; Monette, G.; Nilsson, H.; Ripley, B.; et al. The car package. R Found. Stat. Comput. 2007. [Google Scholar]
- Fawcett, T. An introduction to roc analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar]
- Breiman, L.; Stone, C.J.; Friedman, J.H.; Olshen, R.A. Classification and Regression Trees; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Therneau, T.M.; Atkinson, B.; Ripley, B. The RPART Package. 2010. Available online: https://cran.r-project.org/web/packages/rpart/index.html (accessed on 27 September 2019).
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- von Hippel, P.T. Biases in spss 12.0 missing value analysis. Am. Stat. 2004, 58, 160–164. [Google Scholar] [CrossRef]
- Anderson, T.W. An Introduction to Multivariate Statistical Analysis; Wiley: New York, NY, USA, 1962. [Google Scholar]
- Rokach, L.; Maimon, O. Clustering methods. In Data mining and knowledge discovery handbook; Springer US: Boston, MA, USA, 2005; pp. 321–352. [Google Scholar]
- Ward, J.H. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Cormack, R.M. A review of classification. J. R. Stat. Soc. Ser. A 1971, 134, 321–353. [Google Scholar] [CrossRef]
- Wilkinson, L.; Friendly, M. The history of the cluster heat map. Am. Stat. 2009, 63, 179–184. [Google Scholar] [CrossRef]
- Perrot, A.; Bourqui, R.; Hanusse, N.; Lalanne, F.; Auber, D. Large interactive visualization of density functions on big data infrastructure. In Proceedings of the 2015 IEEE 5th Symposium on large Data Analysis and Visualization (lDAV), Chicago, IL, USA, 25–26 October 2015; pp. 99–106. [Google Scholar]
- Warnes, G.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Mächler, M.; Magnusson, A.; Möller, S. Gplots: Various r Programming Tools for Plotting Data. 2005. Available online: https://cran.r-project.org/web/packages/gplots/index.html (accessed on 27 September 2019).
- Eddowes, P.J.; Sasso, M.; Allison, M.; Tsochatzis, E.; Anstee, Q.M.; Sheridan, D.; Guha, I.N.; Cobbold, J.F.; Deeks, J.J.; Paradis, V.; et al. Accuracy of fibroscan controlled attenuation parameter and liver stiffness measurement in assessing steatosis and fibrosis in patients with nonalcoholic fatty liver disease. Gastroenterology 2019, 156, 1717–1730. [Google Scholar] [CrossRef]
- Mantovani, A.; Zaza, G.; Byrne, C.D.; Lonardo, A.; Zoppini, G.; Bonora, E.; Targher, G. Nonalcoholic fatty liver disease increases risk of incident chronic kidney disease: A systematic review and meta-analysis. Metab. Clin. Exp. 2018, 79, 64–76. [Google Scholar] [CrossRef]
- Musso, G.; Cassader, M.; Cohney, S.; De Michieli, F.; Pinach, S.; Saba, F.; Gambino, R. Fatty liver and chronic kidney disease: Novel mechanistic insights and therapeutic opportunities. Diabetes Care 2016, 39, 1830–1845. [Google Scholar] [CrossRef]
- Kuo, C.F.; Luo, S.F.; See, L.C.; Ko, Y.S.; Chen, Y.M.; Hwang, J.S.; Chou, I.J.; Chang, H.C.; Chen, H.W.; Yu, K.H. Hyperuricaemia and accelerated reduction in renal function. Scand. J. Rheumatol. 2011, 40, 116–121. [Google Scholar] [CrossRef]
- Satirapoj, B.; Supasyndh, O.; Chaiprasert, A.; Ruangkanchanasetr, P.; Kanjanakul, I.; Phulsuksombuti, D.; Utainam, D.; Choovichian, P. Relationship between serum uric acid levels with chronic kidney disease in a southeast asian population. Nephrology 2010, 15, 253–258. [Google Scholar] [CrossRef]
- Kang, D.-H.; Nakagawa, T. Uric acid and chronic renal disease: Possible implication of hyperuricemia on progression of renal disease. Semin. Nephrol. 2005, 25, 43–49. [Google Scholar] [CrossRef]
- Reiss, A.B.; Voloshyna, I.; De Leon, J.; Miyawaki, N.; Mattana, J. Cholesterol metabolism in ckd. Am. J. Kidney Dis. 2015, 66, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Castelao, A.; Navarro-Gonzalez, J.F.; Luis Gorriz, J.; de Alvaro, F. The concept and the epidemiology of diabetic nephropathy have changed in recent years. J. Clin. Med. 2015, 4, 1207–1216. [Google Scholar] [CrossRef] [PubMed]
- Satirapoj, B.; Adler, S.G. Prevalence and management of diabetic nephropathy in western countries. Kidney Dis. 2015, 1, 61. [Google Scholar] [CrossRef] [PubMed]
- Michishita, R.; Matsuda, T.; Kawakami, S.; Tanaka, S.; Kiyonaga, A.; Tanaka, H.; Morito, N.; Higaki, Y. Hypertension and hyperglycemia and the combination thereof enhances the incidence of chronic kidney disease (ckd) in middle-aged and older males. Clin. Exp. Hypertens. (New York, N.Y.: 1993) 2017, 39, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Zelnick, L.R.; Weiss, N.S.; Kestenbaum, B.R.; Robinson-Cohen, C.; Heagerty, P.J.; Tuttle, K.; Hall, Y.N.; Hirsch, I.B.; de Boer, I.H. Diabetes and ckd in the united states population, 2009-2014. Clin. J. Am. Soc. Nephrol.: CJASN 2017, 12, 1984–1990. [Google Scholar] [CrossRef]
- Maciejczyk, M.; Szulimowska, J.; Taranta-Janusz, K.; Werbel, K.; Wasilewska, A.; Zalewska, A. Salivary frap as a marker of chronic kidney disease progression in children. Antioxidants 2019, 8, 409. [Google Scholar] [CrossRef]
| CKD Stage 1 | CKD Stage 2 | CKD Stage 3–5 | ||
|---|---|---|---|---|
| n1 = 1715 | n2 = 370 | n3 = 202 | ||
| Factors | No. (%) | No. (%) | No. (%) | p-Value |
| Sex | ||||
| Female | 1017 (59.3%) | 96(25.9%) | 63(31.2%) | <0.001 |
| Male | 698 (40.7%) | 274(74.1%) | 139(68.8%) | |
| Hypertension | ||||
| Normal | 1299 (75.7%) | 214(57.8%) | 112(55.4%) | <0.001 |
| High | 416 (24.3%) | 156(42.2%) | 90(44.6%) | |
| Median (IQR) | ||||
| Age, years | 42 (35–51) | 53 (46–60.75) | 63.5 (55.25–73) | <0.001 |
| Albumin, g/dL | 4.6 (4.4–4.8) | 4.6 (4.5–4.8) | 4.3 (3.8–4.5) | <0.001 |
| BMI, kg/m2 | 23.4 (21.1–25.99) | 24.7 (22.7–27.02) | 25.05 (22.15–27.98) | <0.001 |
| WC, cm | 80.5 (73.5–88) | 86 (80–92) | 87.58 (82–95.75) | <0.001 |
| AFP, ng/mL | 2.35 (1.62–3.27) | 2.62 (1.98–3.62) | 2.69 (2.15–4.48) | <0.001 |
| ALKp, IU/L | 60 (50–72) | 67 (55–79) | 79.69 (65.68–96) | <0.001 |
| GOT, IU/L | 20 (17–24) | 23 (20–28) | 23.1 (19–34) | <0.001 |
| GPT, IU/L | 18 (13–28) | 23 (17–33) | 22 (15–35) | <0.001 |
| T_Bilirubin, mg/dL | 0.6 (0.4–0.8) | 0.6 (0.5–0.8) | 0.6 (0.4–0.9) | 0.002 |
| γGT, U/L | 17 (12–27) | 23 (17–34) | 34.63 (21–70.90) | <0.001 |
| CAPscore, dB/m | 241 (208–281) | 260 (227–301.8) | 246 (203–296.5) | <0.001 |
| Escore, kPa | 4.3 (3.5–5.1) | 4.6 (3.7–5.6) | 7.9 (4.925–13.525) | <0.001 |
| BUN, mg/dL | 12 (10–14.34) | 15 (13–18) | 23.5 (16.78–38.82) | <0.001 |
| Creatinine, mg/dL | 0.7 (0.6–0.8) | 1 (1–1.1) | 1.7 (1.425–3.9) | <0.001 |
| UA, mg/dL | 5.1 (4.3–6.3) | 6.3 (5.5–7.3) | 6.6 (5.7–7.975) | <0.001 |
| Cholesterol, mg/dL | 187 (165–209) | 191.5 (165–217) | 169 (130.2–195) | <0.001 |
| HbA1C, % | 5.4 (5.2–5.6) | 5.525 (5.3–5.9) | 5.7 (5.3–6.3) | <0.001 |
| HDL, mg/dL | 54 (45–65) | 49 (41–58) | 46 (37–57.9) | <0.001 |
| LDL, mg/dL | 121 (102–143) | 131 (102.2–152) | 107 (80.25–128.75) | <0.001 |
| TSH, μIU/mL | 1.81(1.2–2.545) | 2.075 (1.465–2.835) | 2.009 (1.54–2.565) | <0.001 |
| Factors | Odds Ratio | 95% CI OR | VIF | ΔVIF | p-Value |
|---|---|---|---|---|---|
| BMI, kg/m2 | 0.904 | (0.854, 0.958) | 3.977 | 3.975 | 0.001* |
| WC, cm | 1.046 | (1.023, 1.069) | 4.170 | 4.169 | <0.001* |
| Cholesterol, mg/dL | 0.983 | (0.973, 0.994) | 10.598 | Δ | 0.002* |
| Hypertension | 0.213 | (0.654, 1.099) | 1.123 | 1.122 | 0.213 |
| HbA1C, % | 1.248 | (1.087, 1.434) | 1.223 | 1.178 | 0.002* |
| HDL, mg/dL | 1.005 | (0.994, 1.016) | 1.945 | 1.334 | 0.363 |
| LDL, mg/dL | 1.016 | (1.005, 1.028) | 10.286 | 1.077 | 0.004* |
| Albumin, g/dL | 0.793 | (0.536, 1.174) | 1.355 | 1.364 | 0.247 |
| ALKp, IU/L | 1.001 | (0.995, 1.006) | 1.202 | 1.209 | 0.761 |
| GOT, IU/L | 1.030 | (1.012, 1.050) | 3.832 | 3.801 | 0.001* |
| GPT, IU/L | 0.977 | (0.966, 0.988) | 3.847 | 3.803 | <0.001* |
| γGT, U/L | 1.002 | (0.997, 1.006) | 1.452 | 1.448 | 0.425 |
| T_Bilirubin, mg/dL | 1.088 | (0.930, 1.273) | 1.182 | 1.180 | 0.291 |
| CAPscore, dB/m | 1.003 | (1.000, 1.006) | 1.646 | 1.647 | 0.044† |
| Escore, kPa | 1.012 | (0.985, 1.040) | 1.622 | 1.595 | 0.393 |
| AFP, ng/mL | 1.000 | (1.000, 1.000) | 1.141 | 1.129 | 0.528 |
| BUN, mg/dL | 1.229 | (1.190, 1.270) | 1.063 | 1.062 | <0.001* |
| UA, mg/dL | 1.478 | (1.349, 1.619) | 1.346 | 1.333 | <0.001* |
| TSH, μIU/mL | 1.047 | (1.004, 1.091) | 1.016 | 1.015 | 0.031 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, C.-S.; Lin, C.-H.; Lin, Y.-J.; Lin, S.-Y.; Wang, S.-T.; L Wu, J.; Tsai, M.-H.; Chang, S.-S. Clustering Heatmap for Visualizing and Exploring Complex and High-dimensional Data Related to Chronic Kidney Disease. J. Clin. Med. 2020, 9, 403. https://doi.org/10.3390/jcm9020403
Yu C-S, Lin C-H, Lin Y-J, Lin S-Y, Wang S-T, L Wu J, Tsai M-H, Chang S-S. Clustering Heatmap for Visualizing and Exploring Complex and High-dimensional Data Related to Chronic Kidney Disease. Journal of Clinical Medicine. 2020; 9(2):403. https://doi.org/10.3390/jcm9020403
Chicago/Turabian StyleYu, Cheng-Sheng, Chang-Hsien Lin, Yu-Jiun Lin, Shiyng-Yu Lin, Sen-Te Wang, Jenny L Wu, Ming-Hui Tsai, and Shy-Shin Chang. 2020. "Clustering Heatmap for Visualizing and Exploring Complex and High-dimensional Data Related to Chronic Kidney Disease" Journal of Clinical Medicine 9, no. 2: 403. https://doi.org/10.3390/jcm9020403
APA StyleYu, C.-S., Lin, C.-H., Lin, Y.-J., Lin, S.-Y., Wang, S.-T., L Wu, J., Tsai, M.-H., & Chang, S.-S. (2020). Clustering Heatmap for Visualizing and Exploring Complex and High-dimensional Data Related to Chronic Kidney Disease. Journal of Clinical Medicine, 9(2), 403. https://doi.org/10.3390/jcm9020403





