Transcriptional Expression in Human Periodontal Ligament Cells Subjected to Orthodontic Force: An RNA-Sequencing Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Selection
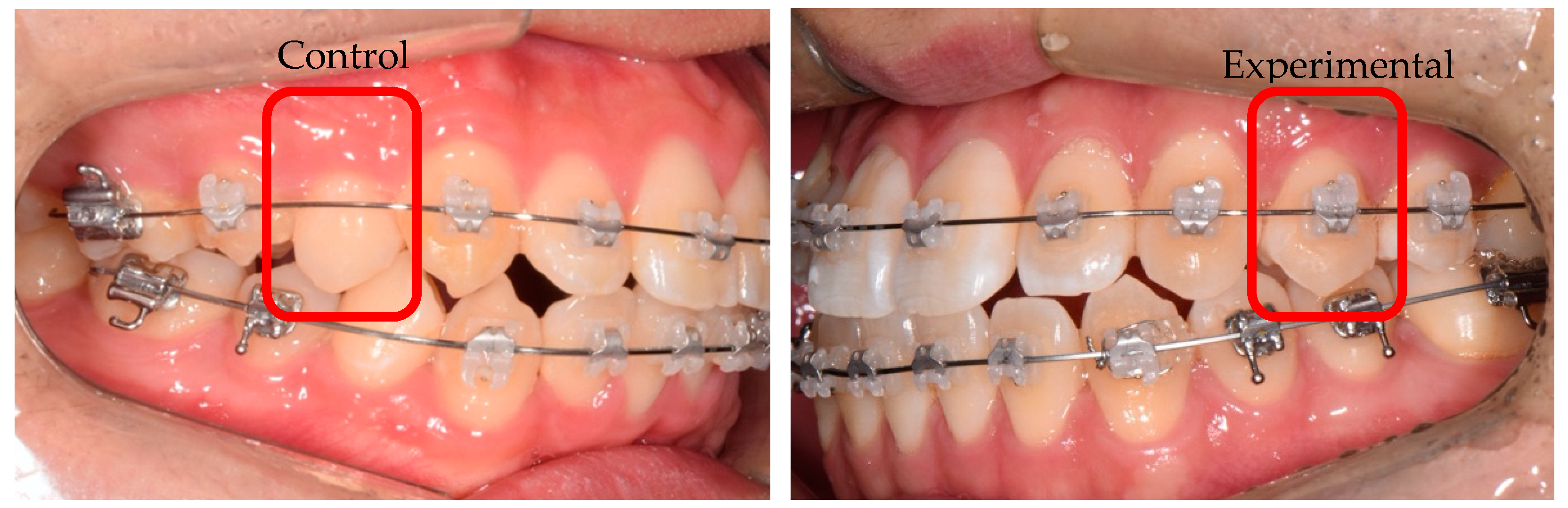
2.2. Mechanical Stimulus (Applying Orthodontic Force)
2.3. RNA Sequencing
2.4. Real-Time PCR
2.5. Statistical Analysis
3. Results
3.1. Extracted Premolar Samples
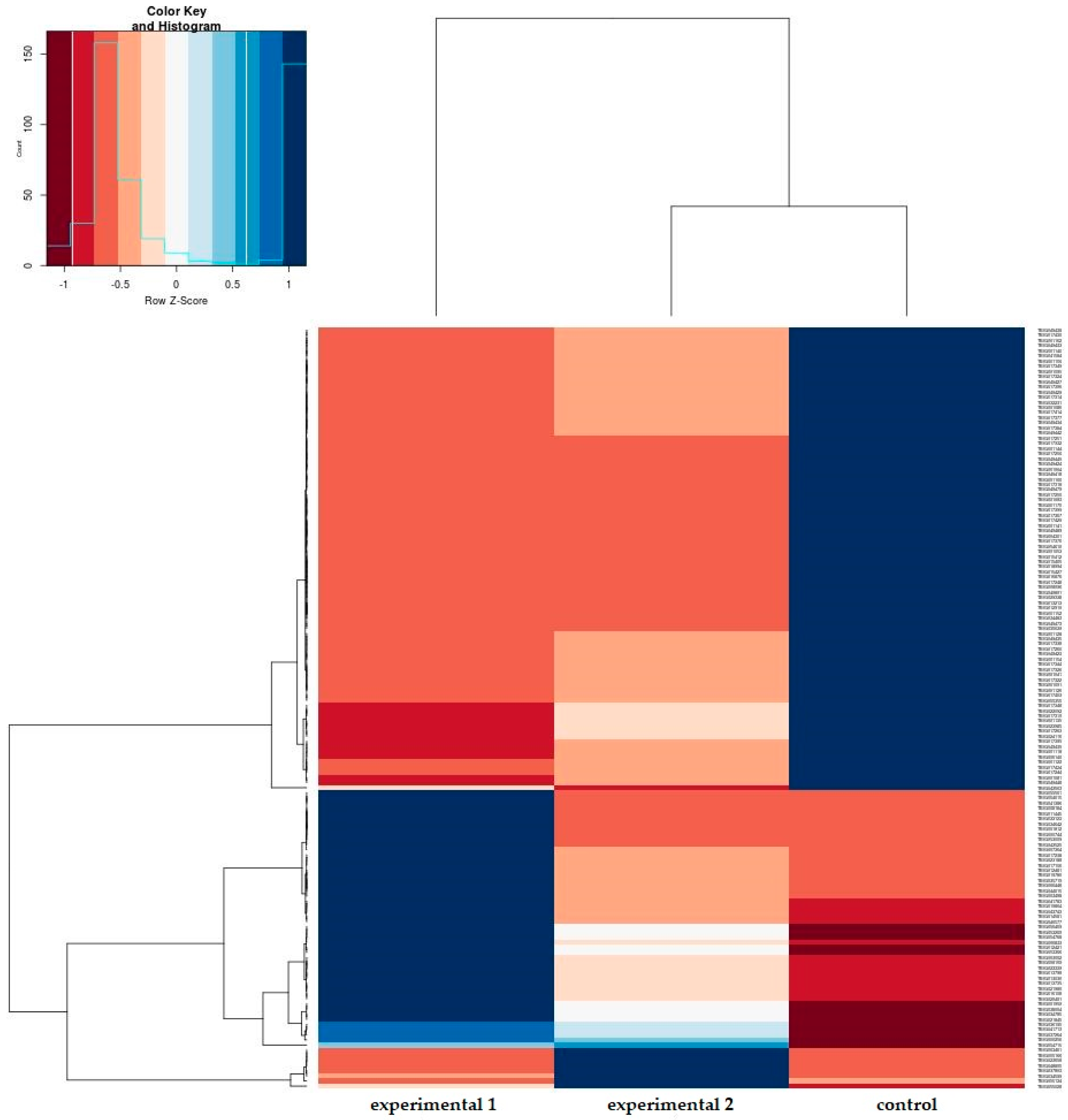
3.2. Gene Set Enrichment Analysis of Mechanically Stimulated PDL Cells
3.3. DEG Analysis of Mechanically Stimulated PDL Cells
3.4. Validation of DEGs in Mechanically Stimulated PDL Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A










| Subject No. | Sex | Age | Tooth allocation | |
|---|---|---|---|---|
| Experimental Group | Control Group | |||
| 1 | Female | 28 years 2 months | Mx Lt 2nd premolar | Mn Lt 2nd premolar |
| 2 | Male | 22 years 0 months | Mx Lt 1st premolar | Mn Lt 1st premolar |
| 3 | Male | 17 years 6 months | Mx Rt 1st premolar | Mx Lt 1st premolar |
| 4 | Male | 20 years 3 months | Mx Lt 2nd premolar, Mn Lt 2nd premolar | Mn Rt 1st premolar |
| 5 | Female | 20 years 9 months | Mn Lt 1st premolar | Mx Lt 1st premolar |
| 6 | Female | 25 years 7 months | Mn Lt 1st premolar | Mx Lt 1st premolar |
| 7 | Female | 31 years 0 months | Mx Lt 1st premolar, Mn Lt 1st premolar | Mx Rt 1st premolar, Mn Rt 1st premolar |
| 8 | Female | 20 years 8 months | Mx Rt 1st premolar, Mn Rt 1st premolar | Mx Lt 1st premolar, Mn Lt 1st premolar |
| 9 | Male | 18 years 4 months | Mx Lt 1st premolar, Mn Lt 1st premolar | Mx Rt 1st premolar, Mn Rt 1st premolar |
| 10 | Female | 18 years 6 months | Mx Lt 1st premolar | Mx Rt 1st premolar |
| 11 | Male | 25 years 1 months | Mx Rt 1st premolar | Mn Rt 1st premolar |
References
- Kirschneck, C.; Batschkus, S.; Proff, P.; Kostler, J.; Spanier, G.; Schroder, A. Valid gene expression normalization by RT-qPCR in studies on hPDL fibroblasts with focus on orthodontic tooth movement and periodontitis. Sci. Rep. 2017, 7, 14751. [Google Scholar] [CrossRef] [PubMed]
- Seo, B.M.; Miura, M.; Gronthos, S.; Bartold, P.M.; Batouli, S.; Brahim, J.; Young, M.; Robey, P.G.; Wang, C.Y.; Shi, S. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet 2004, 364, 149–155. [Google Scholar] [CrossRef]
- Bright, R.; Hynes, K.; Gronthos, S.; Bartold, P.M. Periodontal ligament-derived cells for periodontal regeneration in animal models: A systematic review. J. Periodontal. Res. 2015, 50, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Kraft, D.C.; Bindslev, D.A.; Melsen, B.; Abdallah, B.M.; Kassem, M.; Klein-Nulend, J. Mechanosensitivity of dental pulp stem cells is related to their osteogenic maturity. Eur. J. Oral Sci. 2010, 118, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Lekic, P.; McCulloch, C.A. Periodontal ligament cell population: The central role of fibroblasts in creating a unique tissue. Anat. Rec. 1996, 245, 327–341. [Google Scholar] [CrossRef]
- Lee, S.Y.; Yoo, H.I.; Kim, S.H. CCR5-CCL Axis in PDL during Orthodontic Biophysical Force Application. J. Dent. Res. 2015, 94, 1715–1723. [Google Scholar] [CrossRef]
- Zhang, C.; Li, J.; Zhang, L.; Zhou, Y.; Hou, W.; Quan, H.; Li, X.; Chen, Y.; Yu, H. Effects of mechanical vibration on proliferation and osteogenic differentiation of human periodontal ligament stem cells. Arch. Oral Biol. 2012, 57, 1395–1407. [Google Scholar] [CrossRef]
- Pavlin, D.; Gluhak-Heinrich, J. Effect of mechanical loading on periodontal cells. Crit. Rev. Oral Biol. Med. 2001, 12, 414–424. [Google Scholar] [CrossRef]
- Mine, K.; Kanno, Z.; Muramoto, T.; Soma, K. Occlusal forces promote periodontal healing of transplanted teeth and prevent dentoalveolar ankylosis: An experimental study in rats. Angle Orthod. 2005, 75, 637–644. [Google Scholar] [CrossRef]
- Xu, C.; Fan, Z.; Shan, W.; Hao, Y.; Ma, J.; Huang, Q.; Zhang, F. Cyclic stretch influenced expression of membrane connexin 43 in human periodontal ligament cell. Arch. Oral Biol. 2012, 57, 1602–1608. [Google Scholar] [CrossRef]
- Ma, J.; Zhao, D.; Wu, Y.; Xu, C.; Zhang, F. Cyclic stretch induced gene expression of extracellular matrix and adhesion molecules in human periodontal ligament cells. Arch. Oral Biol. 2015, 60, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Sprogar, S.; Vaupotic, T.; Cor, A.; Drevensek, M.; Drevensek, G. The endothelin system mediates bone modeling in the late stage of orthodontic tooth movement in rats. Bone 2008, 43, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Di Domenico, M.; D’Apuzzo, F.; Feola, A.; Cito, L.; Monsurro, A.; Pierantoni, G.M.; Berrino, L.; De Rosa, A.; Polimeni, A.; Perillo, L. Cytokines and VEGF induction in orthodontic movement in animal models. J. Biomed. Biotechnol. 2012, 2012, 201689. [Google Scholar] [CrossRef] [PubMed]
- Howard, P.S.; Kucich, U.; Taliwal, R.; Korostoff, J.M. Mechanical forces alter extracellular matrix synthesis by human periodontal ligament fibroblasts. J. Periodontal. Res. 1998, 33, 500–508. [Google Scholar] [CrossRef]
- Kokai, S.; Kanno, Z.; Koike, S.; Uesugi, S.; Takahashi, Y.; Ono, T.; Soma, K. Retrospective study of 100 autotransplanted teeth with complete root formation and subsequent orthodontic treatment. Am. J. Orthod. Dentofacial. Orthop. 2015, 148, 982–989. [Google Scholar] [CrossRef]
- Min, H.-K.Y.H.-S.S.G.L.B.-M. Effect of Mechanical Stress on the Proliferation and Expression of Cell Cycle Regulators in Human Periodontal Ligament Cells. J. Korean Acad. Periodontol. 1999, 29, 593–605. [Google Scholar]
- Kukurba, K.R.; Montgomery, S.B. RNA Sequencing and Analysis. Cold Spring Harb. Protoc. 2015, 2015, 951–969. [Google Scholar] [CrossRef]
- Qian, X.; Ba, Y.; Zhuang, Q.; Zhong, G. RNA-Seq technology and its application in fish transcriptomics. OMICS 2014, 18, 98–110. [Google Scholar] [CrossRef]
- Davanian, H.; Stranneheim, H.; Bage, T.; Lagervall, M.; Jansson, L.; Lundeberg, J.; Yucel-Lindberg, T. Gene expression profiles in paired gingival biopsies from periodontitis-affected and healthy tissues revealed by massively parallel sequencing. PLoS ONE 2012, 7, e46440. [Google Scholar] [CrossRef]
- Kurt, G.; Iseri, H.; Kisnisci, R. Rapid tooth movement and orthodontic treatment using dentoalveolar distraction (DAD). Long-term (5 years) follow-up of a Class II case. Angle Orthod. 2010, 80, 597–606. [Google Scholar] [CrossRef]
- Fu, W.J.; Hu, J.; Spencer, T.; Carroll, R.; Wu, G. Statistical models in assessing fold change of gene expression in real-time RT-PCR experiments. Comput. Biol. Chem. 2006, 30, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Nishiyama, T.; Shimizu, K.; Kadota, K. TCC: An R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics 2013, 14, 219. [Google Scholar] [CrossRef] [PubMed]
- Yee, J.A.; Kimmel, D.B.; Jee, W.S. Periodontal ligament cell kinetics following orthodontic tooth movement. Cell Tissue Kinet 1976, 9, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, N.; Schroder, A.; Jantsch, J.; Spanier, G.; Proff, P.; Kirschneck, C. The role of mechanotransduction versus hypoxia during simulated orthodontic compressive strain-an in vitro study of human periodontal ligament fibroblasts. Int. J. Oral Sci. 2019, 11, 33. [Google Scholar] [CrossRef]
- Choung, H.W.; Lee, D.S.; Park, Y.H.; Lee, Y.S.; Bai, S.; Yoo, S.H.; Lee, J.H.; You, H.K.; Park, J.C. The effect of CPNE7 on periodontal regeneration. Connect. Tissue Res. 2019, 1–12. [Google Scholar] [CrossRef]
- Billuart, P.; Bienvenu, T.; Ronce, N.; des Portes, V.; Vinet, M.C.; Zemni, R.; Roest Crollius, H.; Carrie, A.; Fauchereau, F.; Cherry, M.; et al. Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature 1998, 392, 923–926. [Google Scholar] [CrossRef]
- Govek, E.E.; Newey, S.E.; Van Aelst, L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005, 19, 1–49. [Google Scholar] [CrossRef]
- Ishida, A.; Okuno, S.; Kitani, T.; Kameshita, I.; Fujisawa, H. Regulation of multifunctional Ca2+/calmodulin-dependent protein kinases by Ca2+/calmodulin-dependent protein kinase phosphatase. Biochem. Biophys. Res. Commun. 1998, 253, 159–163. [Google Scholar] [CrossRef]
- Toms, S.R.; Lemons, J.E.; Bartolucci, A.A.; Eberhardt, A.W. Nonlinear stress-strain behavior of periodontal ligament under orthodontic loading. Am. J. Orthod. Dentofacial. Orthop. 2002, 122, 174–179. [Google Scholar] [CrossRef]
- Lin, W.L.; McCulloch, C.A.; Cho, M.I. Differentiation of periodontal ligament fibroblasts into osteoblasts during socket healing after tooth extraction in the rat. Anat. Rec. 1994, 240, 492–506. [Google Scholar] [CrossRef] [PubMed]



| Genes | Transcript ID | Forward | Reverse |
|---|---|---|---|
| CC2D1B | NM_032449.2 | GAGTCGCAGCTAGCCTCTGT | TCTGTCTCAGGGCTCCTGTT |
| TMEM253 | NM_001146683.1 | CTTGCTGAGCCAGAGGAAAC | CAAACCAGGAACCTCTTCCA |
| TENM4 | NM_001098816.2 | CCGTCTTCCTTTCTGACAGC | ATCAGCCCAAACTTGTCCAC |
| CPNE3 | NM_003909.5 | TCGACCACTGGTGATGAAAA | CCGATGAACCATTAGCCAGT |
| MYOM2 | NM_003970.4 | CGGTGAATACAAGGCAACCT | TCACATATCTGCAGCCAAGC |
| PPM1F | NM_014634.4 | GTACAGCAGGGACAGGTGGT | ACAGGCAAGCAGCAGGTAGT |
| PCDP1 | NM_001271049.2 | TCAACAAGTAGCACGCAAGG | ATCCGCCTCAGGAAGAATTT |
| ATP2A1 | NM_173201.3 | TGGCTCTTCTTCCGCTACAT | GCCTCGAAGACCTCACAGTC |
| GPR171 | NM_013308.3 | CAACCGTTGTGTGGCTAATG | TATGATGTAGCCCGTGGTCA |
| OPHN1 | NM_002547.3 | GTCCCCAAGCAGGCCTAT | GTCCATTGGTGGCCTTTG |
| TANGO2 | NM_152906.6 | TCCCTGGAGGAAGCTGTG | GCTGCGCCTCTTCATTGT |
| UAP1 | NM_001324116.1 | TCCAAAGCTGGGCAAGAG | GGTTCCATTCGTGCATCC |
| RP1-34H18.1 | ENST00000550042.1 | GCGGAGGAGGGAAGAAAG | AAAACCAACCGAGGCACA |
| GAPDH | TCCGCGGCTATATGAAAACAG | TCGTAGTGGGCTTGCTG AA |
| Gene Name | Description | Control | Experimental | log2fc | p Value | q Value |
|---|---|---|---|---|---|---|
| OPHN1 | oligophrenin1 | 5.31 | 781 | 7.2 | 5.00E-05 | 0.0149 |
| TANGO2 | transport and golgi organization 2 homolog (Drosophila) | 6.66 | 86.1 | 3.69 | 5.00E-05 | 0.0149 |
| CC2D1B | coiled-coil and C2 domaincontaining 1B | 12 | 150 | 3.58 | 5.00E-05 | 0.0149 |
| UAP1 | UDP-N-acteylglucosaminepyrophosphorylase1 | 43 | 468 | 3.45 | 5.00E-05 | 0.0149 |
| TMEM253 | transmembrane protein 253 | 0.836 | 7.66 | 3.2 | 5.00E-05 | 0.0149 |
| TENM4 | teneurin transmembrane protein 4 | 13.5 | 93.6 | 2.8 | 5.00E-05 | 0.014911 |
| ABHD4 | abhydrolase domain containing 4 | 20.9 | 137 | 2.71 | 5.00E-05 | 0.0149 |
| CPNE3 | copine III | 23.7 | 134 | 2.5 | 5.00E-05 | 0.0149 |
| RP11-820L6.1 | - | 1.76 | 9.19 | 2.38 | 5.00E-05 | 0.0149 |
| SPATA22 | spermatogenesis associated 22 | 0.379 | 1.89 | 2.31 | 5.00E-05 | 0.0149 |
| SCUBE1 | signalpeptide, CUB domain, EGF-like1 | 0.217 | 0.88 | 2.02 | 0.0001 | 0.0277 |
| ARMC8 | armadillo repeat containing 8 | 13.5 | 54 | 2 | 5.00E-05 | 0.0149 |
| ERI2 | ERI1 exoribonuclease family member 2 | 6.69 | 26.5 | 1.99 | 5.00E-05 | 0.0149 |
| Gene Name | Description | Control | Experimental | log2fc | p Value | q Value |
|---|---|---|---|---|---|---|
| ITCH | itchy E3 ubiquitin protein ligase | 41.6 | 14.4 | −1.53 | 5.00E-05 | 0.0149 |
| ARL14EP | ADP-ribosylation factor-like 14 effector protein | 71.8 | 22.8 | −1.65 | 5.00E-05 | 0.0149 |
| RFX1 | regulatory factor X, 1 (influences HLA class II expression) | 17.1 | 5.47 | −1.65 | 5.00E-05 | 0.0149 |
| PLEKHH1 | pleckstrin homology domain containing, family H (with MyTH4 domain) member1 | 6.91 | 2.18 | −1.66 | 5.00E-05 | 0.0149 |
| EHBP1L1 | EH domain binding protein1-like 1 | 162 | 46 | −1.81 | 5.00E-05 | 0.0149 |
| PIGQ | phosphatidylinositol glycan anchor biosynthesis, class Q | 286 | 81.7 | −1.81 | 5.00E-05 | 0.0149 |
| ASPG | asparaginase homolog (S.cerevisiae) | 56.8 | 15.2 | −1.9 | 5.00E-05 | 0.0149 |
| BOD1L1 | biorientation of chromosomes in cell division 1-like 1 | 92.5 | 22.5 | −2.04 | 5.00E-05 | 0.0149 |
| PTPN12 | protein tyrosine phosphatase, non-receptortype12 | 268 | 55.9 | −2.26 | 5.00E-05 | 0.0149 |
| USH1C | Usher syndrome 1C(autosomal recessive,severe) | 4.9 | 0.927 | −2.4 | 5.00E-05 | 0.0149 |
| MYOM2 | myomesin 2 | 18.2 | 3.03 | −2.58 | 5.00E-05 | 0.0149 |
| PRR11 | proline rich11 | 16.7 | 2.72 | −2.62 | 5.00E-05 | 0.0149 |
| CCDC91 | coiled-coil domain containing 91 | 166 | 26.8 | −2.63 | 5.00E-05 | 0.0149 |
| PPM1F | protein phosphatase,Mg2+/Mn2+ dependent, 1F | 165 | 24.5 | −2.75 | 5.00E-05 | 0.0149 |
| PCDP1 | Homo sapiens primary ciliary dyskinesia protein1 (PCDP1),transcript variant1, mRNA. | 2.35 | 0.254 | −3.21 | 5.00E-05 | 0.0149 |
| TSPAN8 | tetraspanin 8 | 34.8 | 3.1 | −3.49 | 5.00E-05 | 0.0149 |
| RCCD1 | RCC1 domain containing1 | 114 | 7.76 | −3.87 | 5.00E-05 | 0.0149 |
| ATP2A1 | ATPase, Ca++ transporting,cardiac muscle,fast twitch 1 | 13.7 | 0.916 | −3.91 | 5.00E-05 | 0.0149 |
| GPR171 | G protein-coupled receptor 171 | 49 | 0.782 | −5.97 | 5.00E-05 | 0.0149 |
| RP1-34H18.1 | - | 4.14E+03 | 0.774 | −12.4 | 5.00E-05 | 0.0149 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.; Kang, H.E.; Yook, J.I.; Yu, H.-S.; Kim, E.; Cha, J.-Y.; Choi, Y.J. Transcriptional Expression in Human Periodontal Ligament Cells Subjected to Orthodontic Force: An RNA-Sequencing Study. J. Clin. Med. 2020, 9, 358. https://doi.org/10.3390/jcm9020358
Kim K, Kang HE, Yook JI, Yu H-S, Kim E, Cha J-Y, Choi YJ. Transcriptional Expression in Human Periodontal Ligament Cells Subjected to Orthodontic Force: An RNA-Sequencing Study. Journal of Clinical Medicine. 2020; 9(2):358. https://doi.org/10.3390/jcm9020358
Chicago/Turabian StyleKim, Kyunam, Hee Eun Kang, Jong In Yook, Hyung-Seog Yu, Euiseong Kim, Jung-Yul Cha, and Yoon Jeong Choi. 2020. "Transcriptional Expression in Human Periodontal Ligament Cells Subjected to Orthodontic Force: An RNA-Sequencing Study" Journal of Clinical Medicine 9, no. 2: 358. https://doi.org/10.3390/jcm9020358
APA StyleKim, K., Kang, H. E., Yook, J. I., Yu, H.-S., Kim, E., Cha, J.-Y., & Choi, Y. J. (2020). Transcriptional Expression in Human Periodontal Ligament Cells Subjected to Orthodontic Force: An RNA-Sequencing Study. Journal of Clinical Medicine, 9(2), 358. https://doi.org/10.3390/jcm9020358







