Does Postoperative Oral and Intestinal Microbiota Correlate with the Weight-Loss Following Bariatric Surgery?—A Cohort Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Analysis of Endpoints
2.3. Collection of Swab and Fecal Samples
2.4. Treatment Protocol
2.5. Surgical Technique
2.6. DNA Isolation, Library Preparation and Sequencing
2.7. Statistical Analysis
2.8. Ethical Considerations
3. Results
3.1. Demographic Characteristics
3.2. Perioperative Characteristics
3.3. NGS Analysis
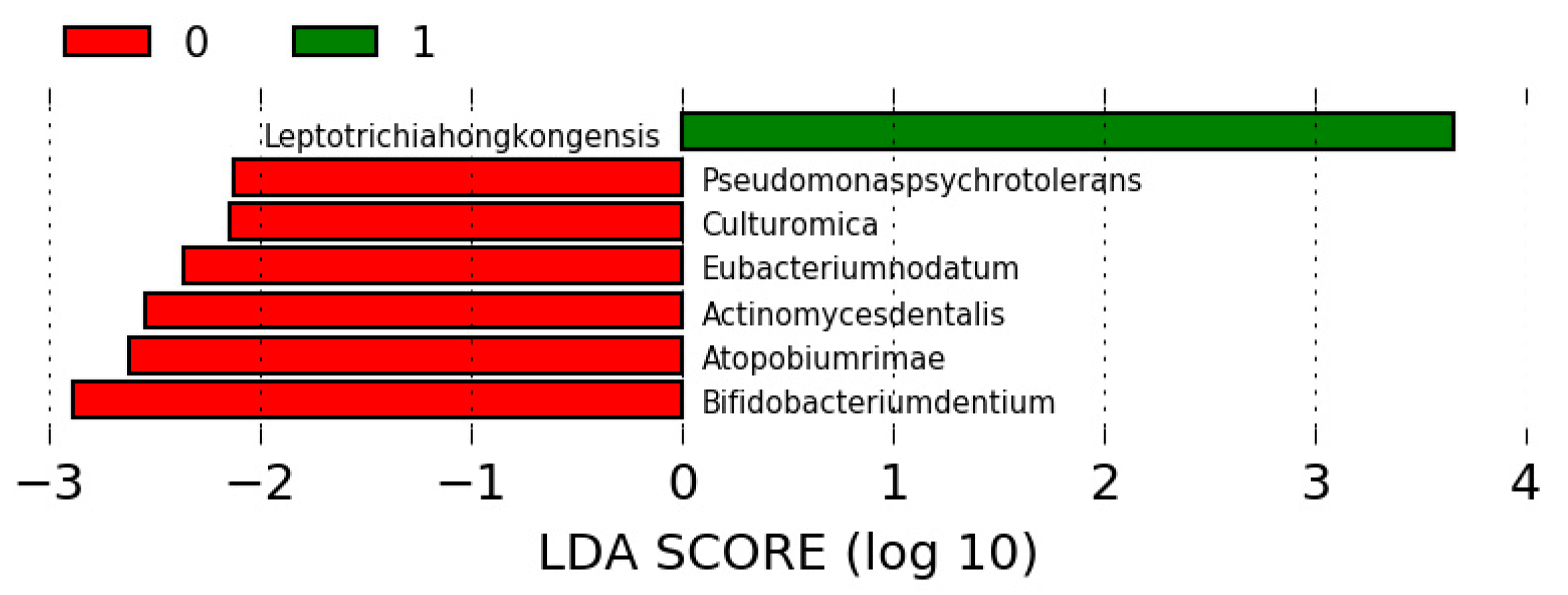
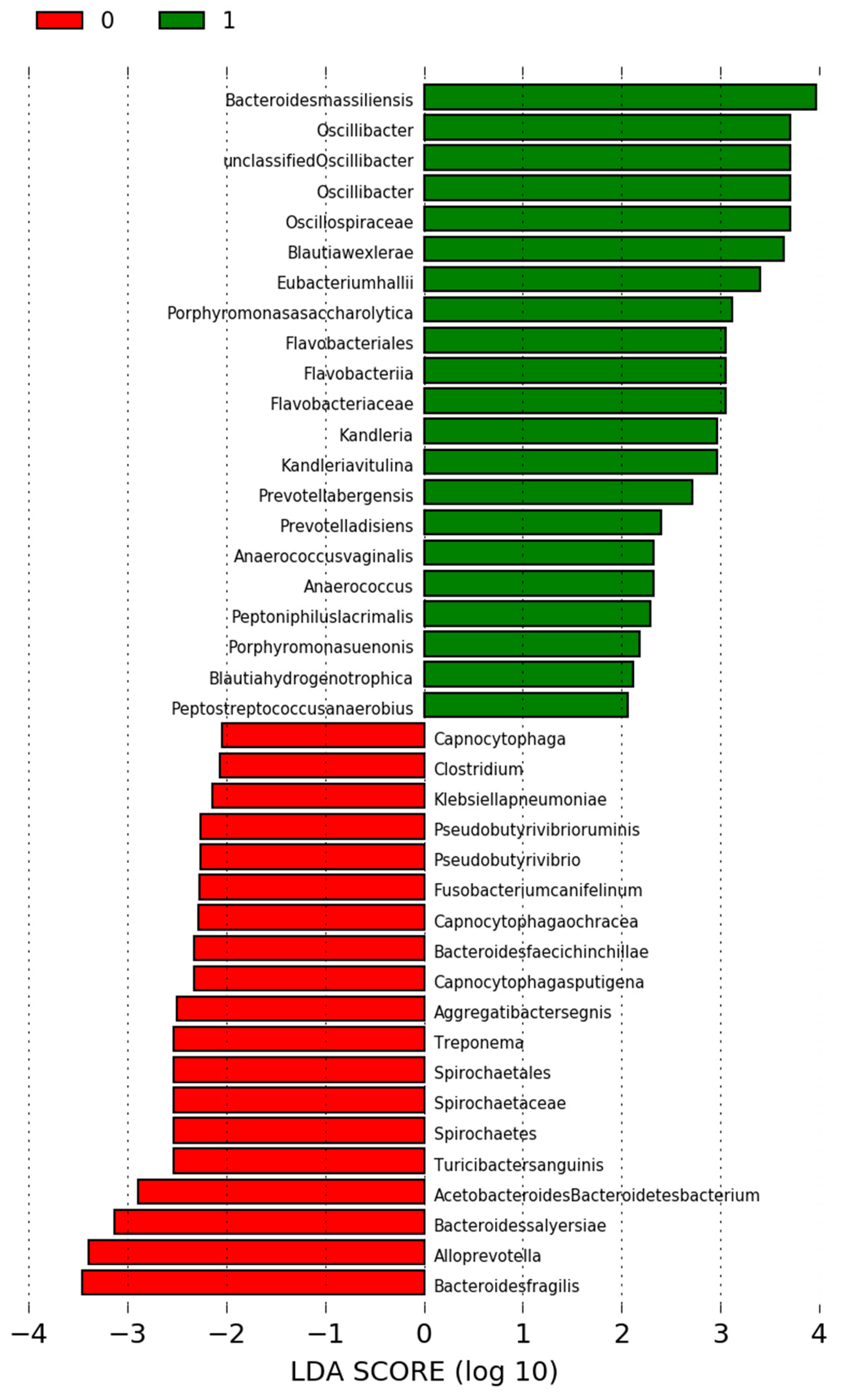
3.4. Differences in the Microbiota among the Study Groups
3.5. Differences in the Microbiota in Participants Undergoing SG
3.6. Differences in the Microbiota in Participants Who Were Undergoing RYGB
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Vu, L.; Switzer, N.J.; De Gara, C.; Karmali, S. Surgical interventions for obesity and metabolic disease. Best Pract. Res. Clin. Endocrinol. Metab. 2013, 27, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Noria, S.F.; Grantcharov, T. Biological effects of bariatric surgery on obesity-related comorbidities. Can. J. Surg. 2013, 56, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Angrisani, L.; Santonicola, A.; Iovino, P.; Formisano, G.; Buchwald, H.; Scopinaro, N. Bariatric Surgery Worldwide. Obes. Surg. 2015, 25, 1822–1832. [Google Scholar] [CrossRef]
- Waledziak, M.; Rozanska-Waledziak, A.M.; Kowalewski, P.K.; Janik, M.R.; Bragoszewski, J.; Pasnik, K.; Bednarczyk, G.; Wallner, G.; Matlok, M. Present trends in bariatric surgery in Poland. Videosurg. Other Miniinvasive Tech. 2019, 14, 86–89. [Google Scholar] [CrossRef]
- Major, P.; Stefura, T.; Malczak, P.; Wysocki, M.; Witowski, J.; Kulawik, J.; Wierdak, M.; Pisarska, M.; Pedziwiatr, M.; Budzynski, A. Postoperative Care and Functional Recovery After Laparoscopic Sleeve Gastrectomy vs. Laparoscopic Roux-en-Y Gastric Bypass Among Patients Under ERAS Protocol. Obes. Surg. 2018, 28, 1031–1039. [Google Scholar] [CrossRef]
- Chang, W.W.; Hawkins, D.N.; Brockmeyer, J.R.; Faler, B.J.; Hoppe, S.W.; Prasad, B.M. Factors influencing long-term weight loss after bariatric surgery. Surg. Obes. Relat. Dis. Off. J. Am. Soc. Bariatr. Surg. 2019, 15, 456–461. [Google Scholar] [CrossRef]
- Aasbrenn, M.; Schnurr, T.M.; Have, C.T.; Svendstrup, M.; Hansen, D.L.; Worm, D.; Balslev-Harder, M.; Hollensted, M.; Grarup, N.; Burgdorf, K.S.; et al. Genetic Determinants of Weight Loss After Bariatric Surgery. Obes. Surg. 2019, 29, 2554–2561. [Google Scholar] [CrossRef] [PubMed]
- Carden, A.; Blum, K.; Arbaugh, C.J.; Trickey, A.; Eisenberg, D. Low socioeconomic status is associated with lower weight-loss outcomes 10-years after Roux-en-Y gastric bypass. Surg. Endosc. 2019, 33, 454–459. [Google Scholar] [CrossRef]
- Orłowski, M.; Janik, M.; Franczak, P.; Frask, A.; Michalik, M. Is it possible to improve long-term results of laparoscopic adjustable gastric banding with appropriate patient selection? Videosurg. Other Miniinvasive Tech. 2020, 15, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Cadena-Obando, D.; Ramírez-Rentería, C.; Ferreira-Hermosillo, A.; Albarrán-Sanchez, A.; Sosa-Eroza, E.; Molina-Ayala, M.; Espinosa-Cárdenas, E. Are there really any predictive factors for a successful weight loss after bariatric surgery? BMC Endocr. Disord. 2020, 20, 20. [Google Scholar] [CrossRef]
- Backhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Backhed, F.; Mithieux, G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 2014, 156, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Grenham, S.; Clarke, G.; Cryan, J.F.; Dinan, T.G. Brain-gut-microbe communication in health and disease. Front. Physiol. 2011, 2, 94. [Google Scholar] [CrossRef] [PubMed]
- Lone, J.B.; Koh, W.Y.; Parray, H.A.; Paek, W.K.; Lim, J.; Rather, I.A.; Jan, A.T. Gut microbiome: Microflora association with obesity and obesity-related comorbidities. Microb. Pathog. 2018, 124, 266–271. [Google Scholar] [CrossRef]
- Szeliga, J.; Wyleżoł, M.; Major, P.; Budzyński, A.; Binda, A.; Proczko-Stepaniak, M.; Boniecka, I.; Matłok, M.; Sekuła, M.; Kaska, Ł.; et al. Metabolic and Bariatric Surgery Chapter of the Association of Polish Surgeons. Bariatric and metabolic surgery care standards. Videosurg. Other Miniinvasive Tech. 2020, 15, 391–394. [Google Scholar] [CrossRef]
- Elm, E.V.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gøtzsche, P.C.; Vandenbroucke, J.P. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement: Guidelines for reporting observational studies. Lancet 2007, 85, 867–872. [Google Scholar] [CrossRef]
- Dindo, D.; Demartines, N.; Clavien, P.-A. Classification of Surgical Complications. Ann. Surg. 2004, 240, 205–213. [Google Scholar] [CrossRef]
- Grover, B.T.; Morell, M.C.; Kothari, S.N.; Borgert, A.J.; Kallies, K.J.; Baker, M.T. Defining Weight Loss After Bariatric Surgery: A Call for Standardization. Obes. Surg. 2019, 29, 3493–3499. [Google Scholar] [CrossRef]
- Fouladi, F.; Brooks, A.E.; Fodor, A.A.; Carroll, I.M.; Bulik-Sullivan, E.C.; Tsilimigras, M.C.B.; Sioda, M.; Steffen, K.J. The Role of the Gut Microbiota in Sustained Weight Loss Following Roux-en-Y Gastric Bypass Surgery. Obes. Surg. 2019, 29, 1259–1267. [Google Scholar] [CrossRef]
- Sroka-Oleksiak, A.; Gosiewski, T.; Pabian, W.; Gurgul, A. Next-Generation Sequencing as a Tool to Detect Vaginal Microbiota Disturbances during Pregnancy. Microorganisms 2020, 8, 1813. [Google Scholar] [CrossRef] [PubMed]
- Brzychczy-Włoch, M.; Pabian, W.; Majewska, E.; Zuk, M.G.; Kielbik, J.; Gosiewski, T.; Bulanda, M.G. Dynamics of colonization with group B streptococci in relation to normal flora in women during subsequent trimesters of pregnancy. New Microbiol. 2014, 37, 307–319. [Google Scholar] [PubMed]
- Sroka-Oleksiak, A.; Młodzińska, A.; Bulanda, M.; Salamon, D.; Major, P.; Stanek, M.; Gosiewski, T. Metagenomic Analysis of Duodenal Microbiota Reveals a Potential Biomarker of Dysbiosis in the Course of Obesity and Type 2 Diabetes: A Pilot Study. J. Clin. Med. 2020, 9, 369. [Google Scholar] [CrossRef]
- Malczak, P.; Pisarska, M.; Piotr, M.; Wysocki, M.; Budzynski, A.; Pedziwiatr, M. Enhanced Recovery after Bariatric Surgery: Systematic Review and Meta-Analysis. Obes. Surg. 2017, 27, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Major, P.; Wysocki, M.; Janik, M.; Stefura, T.; Walędziak, M.; Pędziwiatr, M.; Kowalewski, P.; Paśnik, K.; Budzyński, A. Impact of age on postoperative outcomes in bariatric surgery. Acta Chir. Belg. 2018, 118, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Malla, M.A.; Dubey, A.; Kumar, A.; Yadav, S.; Hashem, A.; Abd Allah, E.F. Exploring the Human Microbiome: The Potential Future Role of Next-Generation Sequencing in Disease Diagnosis and Treatment. Front. Immunol. 2018, 9, 2868. [Google Scholar] [CrossRef]
- Luijten, J.C.H.B.M.; Vugts, G.; Nieuwenhuijzen, G.A.P.; Luyer, M.D.P. The Importance of the Microbiome in Bariatric Surgery: A Systematic Review. Obes. Surg. 2019, 29, 2338–2349. [Google Scholar] [CrossRef]
- Dror, T.; Dickstein, Y.; Dubourg, G.; Paul, M. Microbiota manipulation for weight change. Microb. Pathog. 2017, 106, 146–161. [Google Scholar] [CrossRef]
- Oduro-Donkor, D.; Turner, M.C.; Farnaud, S.; Renshaw, D.; Kyrou, I.; Hanson, P.; Hattersley, J.; Weickert, M.O.; Menon, V.; Randeva, H.S.; et al. Modification of fecal microbiota as a mediator of effective weight loss and metabolic benefits following bariatric surgery. Expert Rev. Endocrinol. Metab. 2020, 15, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Xuan, S.; Wang, Z. Oral microbiota: A new view of body health. Food Sci. Hum. Wellness 2019, 8, 8–15. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Deo, P.N.; Deshmukh, R. Oral microbiome: Unveiling the fundamentals. J. Oral Maxillofac. Pathol. 2019, 23, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; Paul, S.; Dutta, C. Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Front. Microbiol. 2017, 8, 1162. [Google Scholar] [CrossRef] [PubMed]
- Hasan, N.; Yang, H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ 2019, 7, e7502. [Google Scholar] [CrossRef]
- Grembi, J.A.; Nguyen, L.H.; Haggerty, T.D.; Gardner, C.D.; Holmes, S.P.; Parsonnet, J. Gut microbiota plasticity is correlated with sustained weight loss on a low-carb or low-fat dietary intervention. Sci. Rep. 2020, 10, 1405. [Google Scholar] [CrossRef]
- Anhê, F.F.; Varin, T.V.; Schertzer, J.D.; Marette, A. The Gut Microbiota as a Mediator of Metabolic Benefits after Bariatric Surgery. Can. J. Diabetes 2017, 41, 439–447. [Google Scholar] [CrossRef]
- Makaronidis, J.M.; Batterham, R.L. Potential Mechanisms Mediating Sustained Weight Loss Following Roux-en-Y Gastric Bypass and Sleeve Gastrectomy. Endocrinol. Metab. Clin. N. Am. 2016, 45, 539–552. [Google Scholar] [CrossRef]
- Davies, N.K.; O’Sullivan, J.M.; Plank, L.D.; Murphy, R. Altered gut microbiome after bariatric surgery and its association with metabolic benefits: A systematic review. Surg. Obes. Relat. Dis. 2019, 15, 656–665. [Google Scholar] [CrossRef]
- Pucci, A.; Batterham, R.L. Mechanisms underlying the weight loss effects of RYGB and SG: Similar, yet different. J. Endocrinol. Investig. 2019, 42, 117–128. [Google Scholar] [CrossRef] [PubMed]

| Parameter | Total | Group 1 | Group 2 | p |
|---|---|---|---|---|
| Total, n (%) | 31 (100) | 20 (64.5) | 11 (35.5) | - |
| Mean age, years ± SD | 43.5 ± 12.3 | 41.95 ± 11.01 | 46.46 ± 14.85 | 0.340 |
| Sex (female), n (%) | 20 (64.5) | 13 (65) | 7 (63.6) | 0.940 |
| Median maximal weight, kg (IQR) | 132 (122.5–151) | 129.5 (125.5–140.3) | 132 (121.8–153.8) | 0.999 |
| Median maximal BMI, kg/m2 (IQR) | 47.3 (43.6–52.5) | 48.2 (45–50.4) | 47.2 (43.1–55.1) | 0.563 |
| Median preoperative BMI, kg/m2 (IQR) | 44.1 (40.7–48.3) | 43.3 (42.7–48.7) | 45.7 (40.2–47.6) | 0.367 |
| Type 2 Diabetes, n (%) | 5 (16.1) | 2 (10) | 3 (27.3) | 0.211 |
| Diabetes complications, n (%) | 2 (6.5) | 1 (5) | 1 (9.1) | 0.657 |
| Hyperlipidemia, n (%) | 5 (16.1) | 4 (20) | 1 (9.1) | 0.429 |
| Steatohepatitis, n (%) | 6 (19.4) | 4 (20) | 2 (18.2) | 0.902 |
| Hypertension, n (%) | 22 (71) | 13 (65) | 9 (81.8) | 0.324 |
| Cardiovascular disorders, n (%) | 5 (16.1) | 4 (20) | 1 (9.1) | 0.429 |
| Respiratory disorders, n (%) | 4 (12.9) | 4 (20) | 0 | 0.112 |
| Varicose veins, n (%) | 5 (16.1) | 3 (15) | 2 (18.2) | 0.818 |
| Smoking, n (%) | 5 (16.1) | 3 (15) | 2 (18.2) | 0.818 |
| Parameter | Total | Group 1 | Group 2 | p |
|---|---|---|---|---|
| Total, n (%) | 31 (100) | 20 (64.5) | 11 (35.5) | - |
| Operation, n (%) | 0.902 | |||
| -SG | 25 (80.6) | 16 (80) | 9 (81.8) | |
| -RYGB | 6 (19.4) | 4 (20) | 2 (18.2) | |
| ASA class, n (%) | 0.818 | |||
| -II | 26 (83.9) | 17 (85) | 9 (81.8) | |
| -III | 5 (16.1) | 3 (15) | 2 (18.2) | |
| Median SG operative time, min. ± SD | 100 (67.5–110) | 102.5 (75–102.5) | 90 (67.5–111.25) | 0.623 |
| Median RYGB operative time, min. ± SD | 125 (120–137.5) | 125 (120–160) | 140 (120–132.5) | |
| Postoperative complications, n (%) | 1 (3.2) | 1 (5) | 0 | 0.451 |
| %TBWL ± SD | 30.4 ± 11.4 | 36.3 ± 9.7 | 19.7 ± 3.7 | <0.001 |
| %EWL ± SD | 55.2 ± 17.6 | 65.3 ± 12.7 | 36.9 ± 7 | <0.001 |
| %EBMIL ± SD | 61.3 ± 20.1 | 71.6 ± 16.8 | 42.6 ± 8.6 | <0.001 |
| Swab Samples | ||||
|---|---|---|---|---|
| Taxonomic Level | Abundance | Reads PF Classified to Taxonomic Level | % Reads PF Classified to Taxonomic Level | |
| Kingdom | 2 | 53.20702439 | 99.85% | |
| Phylum | 48 | 53.06839024 | 99.59% | |
| Class | 85 | 52.95868293 | 99.38% | |
| Order | 120 | 52.90856098 | 99.29% | |
| Family | 297 | 52.72834146 | 98.5% | |
| Genus | 1230 | 52.20009756 | 97.95% | |
| Species | 2091 | 39.75509756 | 74.65% | |
| Shanon-Wienner Index of diversity | 3.03 | |||
| Feacal Samples | ||||
| Taxonomic Level | Abundance | Reads PF Classified to Taxonomic Level | % Reads PF Classified to Taxonomic Level | |
| Kingdom | 2 | 55.96700000 | 99.85% | |
| Phylum | 45 | 55.79125806 | 99.54% | |
| Class | 84 | 55.57280645 | 99.16% | |
| Order | 117 | 55.54338710 | 99.11% | |
| Family | 285 | 55.09909677 | 98.32% | |
| Genus | 1075 | 53.84638710 | 96.10% | |
| Species | 1892 | 46.19551613 | 82.54% | |
| Shanon-Wienner Index of diversity | 3.54 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stefura, T.; Zapała, B.; Stój, A.; Gosiewski, T.; Skomarovska, O.; Krzysztofik, M.; Pędziwiatr, M.; Major, P. Does Postoperative Oral and Intestinal Microbiota Correlate with the Weight-Loss Following Bariatric Surgery?—A Cohort Study. J. Clin. Med. 2020, 9, 3863. https://doi.org/10.3390/jcm9123863
Stefura T, Zapała B, Stój A, Gosiewski T, Skomarovska O, Krzysztofik M, Pędziwiatr M, Major P. Does Postoperative Oral and Intestinal Microbiota Correlate with the Weight-Loss Following Bariatric Surgery?—A Cohort Study. Journal of Clinical Medicine. 2020; 9(12):3863. https://doi.org/10.3390/jcm9123863
Chicago/Turabian StyleStefura, Tomasz, Barbara Zapała, Anastazja Stój, Tomasz Gosiewski, Oksana Skomarovska, Marta Krzysztofik, Michał Pędziwiatr, and Piotr Major. 2020. "Does Postoperative Oral and Intestinal Microbiota Correlate with the Weight-Loss Following Bariatric Surgery?—A Cohort Study" Journal of Clinical Medicine 9, no. 12: 3863. https://doi.org/10.3390/jcm9123863
APA StyleStefura, T., Zapała, B., Stój, A., Gosiewski, T., Skomarovska, O., Krzysztofik, M., Pędziwiatr, M., & Major, P. (2020). Does Postoperative Oral and Intestinal Microbiota Correlate with the Weight-Loss Following Bariatric Surgery?—A Cohort Study. Journal of Clinical Medicine, 9(12), 3863. https://doi.org/10.3390/jcm9123863