Rapid Antibody-Based COVID-19 Mass Surveillance: Relevance, Challenges, and Prospects in a Pandemic and Post-Pandemic World
Abstract
1. Introduction
2. The Relevance of Rapid Tests in a Pandemic Diagnosis and Surveillance
- (i)
- They can be accessed in rural and urban settings, developed, and developing countries alike, like a simple blood glucometer or pregnancy strip test.
- (ii)
- They can be used for the mass diagnosis utilizing rapid tests that prevent patient leakage as diagnosis and following remediation steps can happen simultaneously.
- (iii)
- They can significantly reduce the turnaround time for test results, which becomes crucial in a community transmissible disease outbreak.
- (iv)
- They can be combined with biostatistical and bioinformatics analysis, which facilitate the quick recording of an enormous number of test results, data storage for easy reference and practical use for constant patient monitoring.
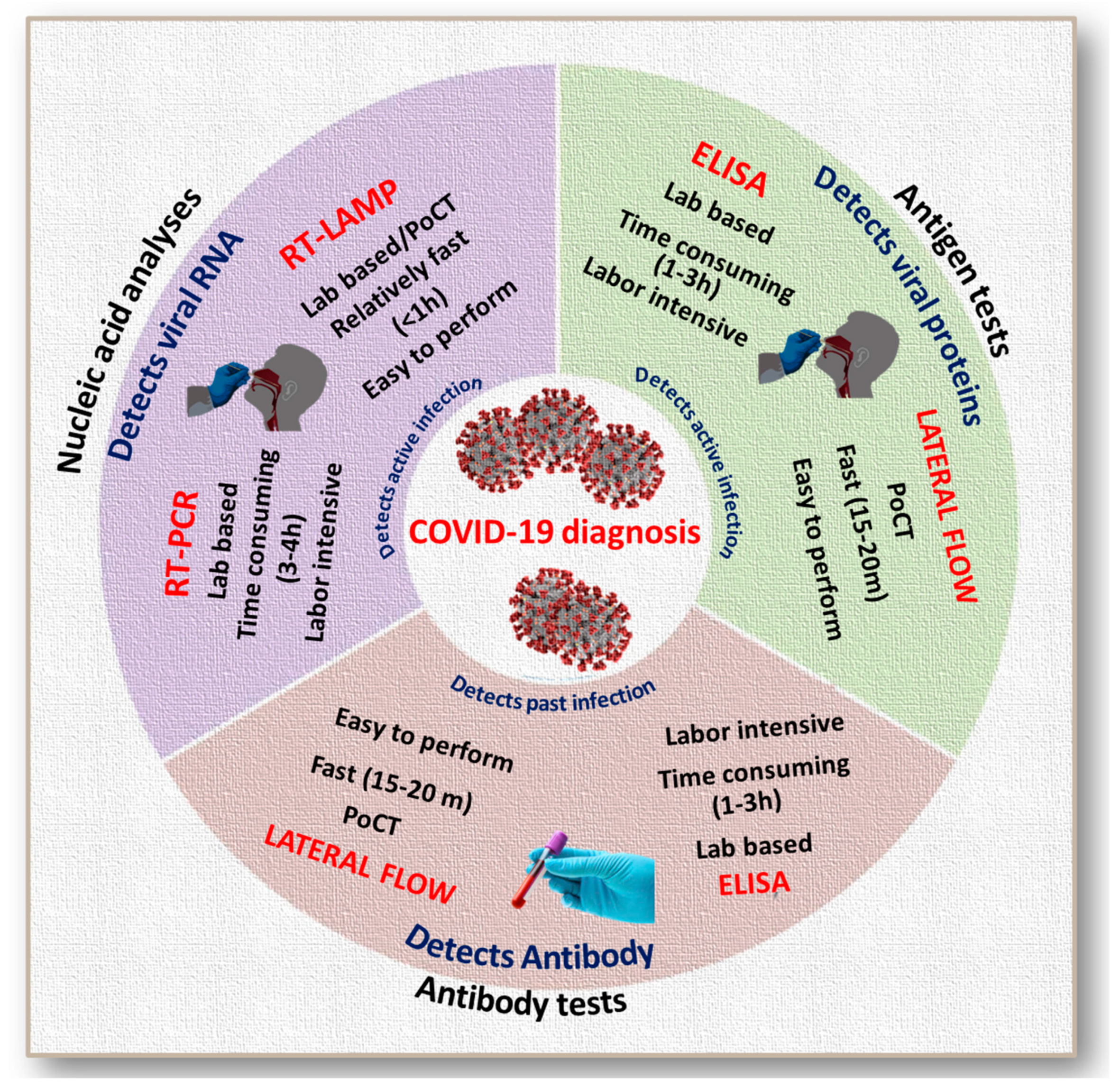
3. Evolution of Antibody-Based Mass Testing
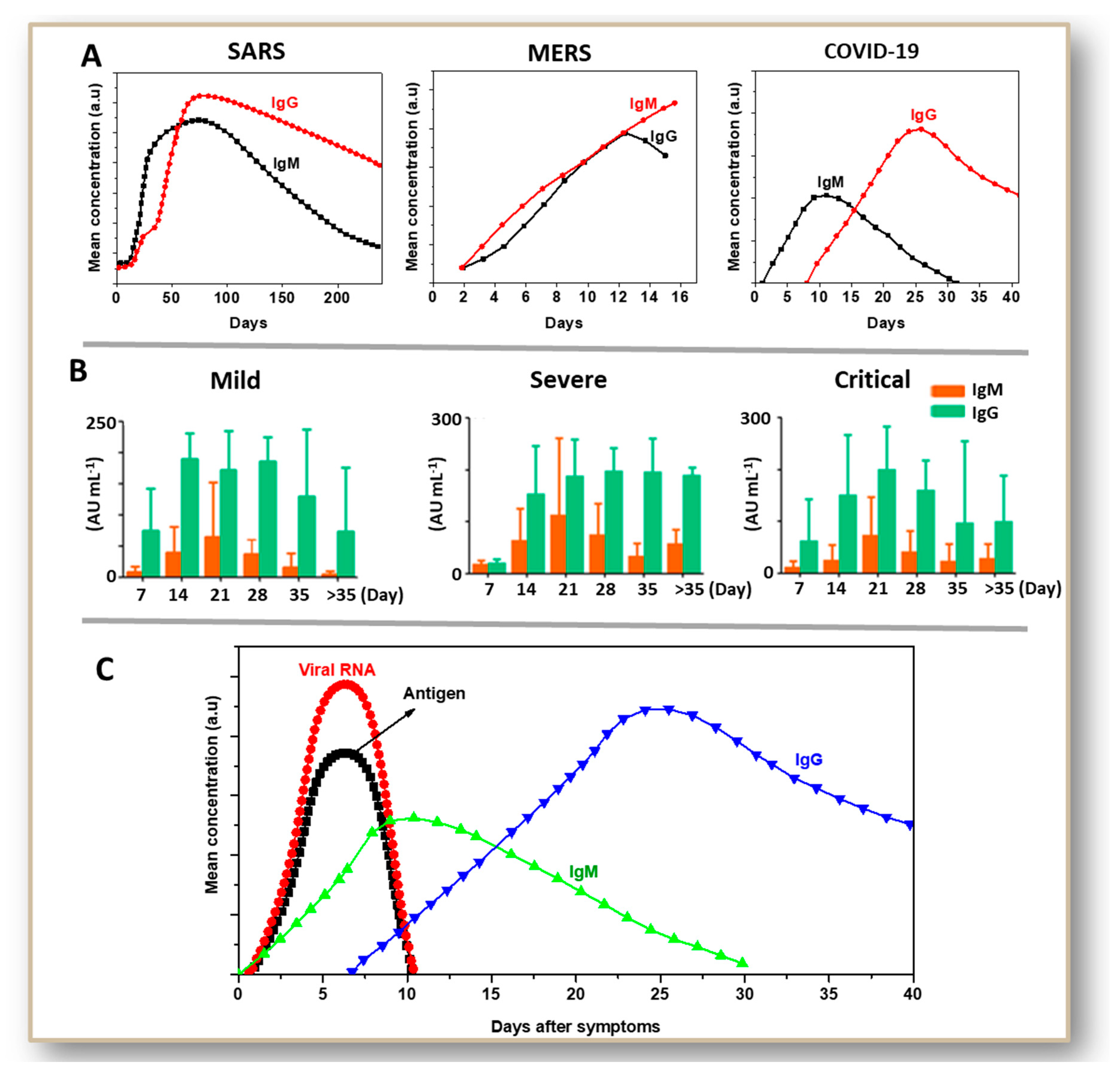
4. Antibody-Based Tests for SARS and MERS: A Basis for COVID-19 Testing
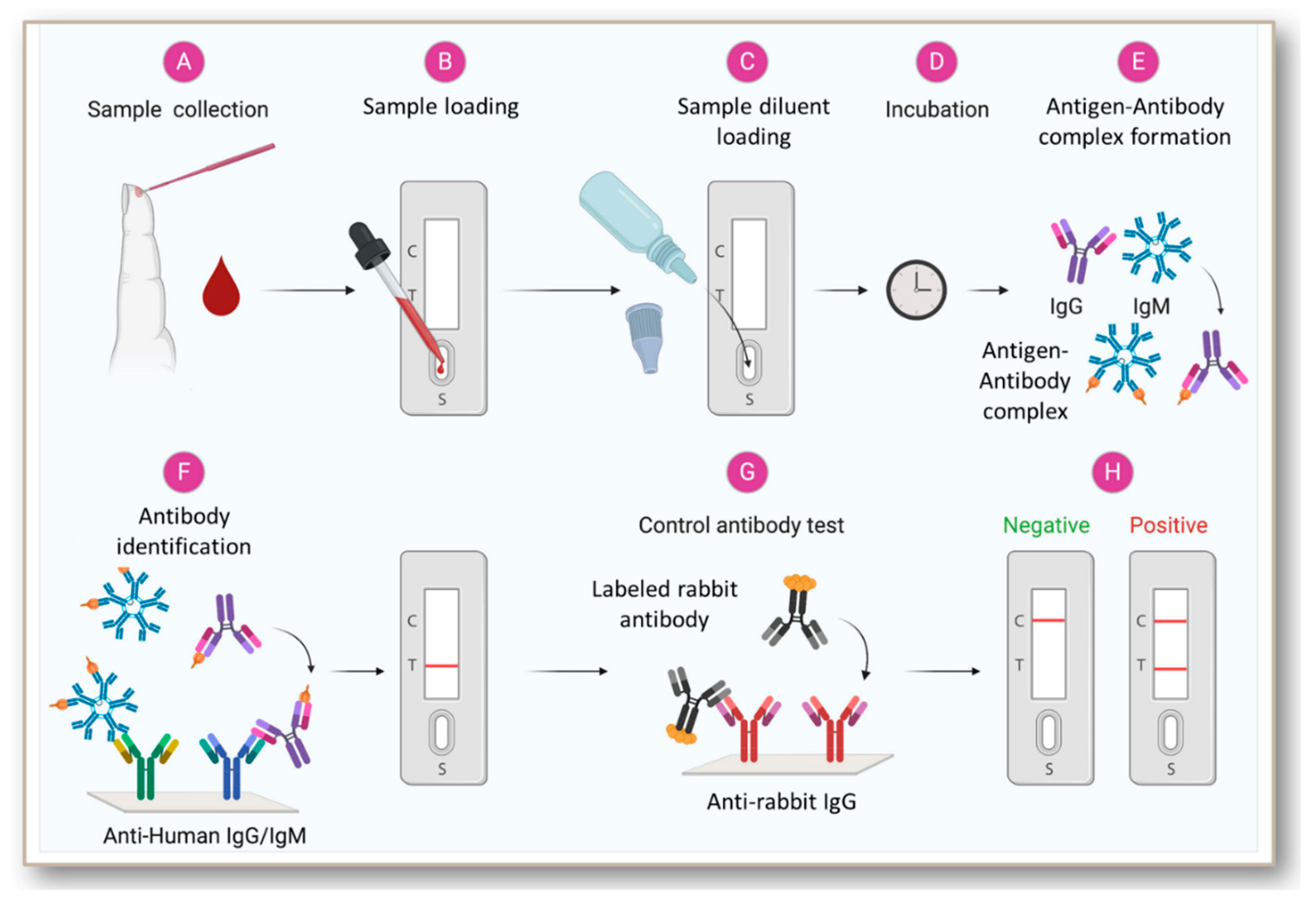
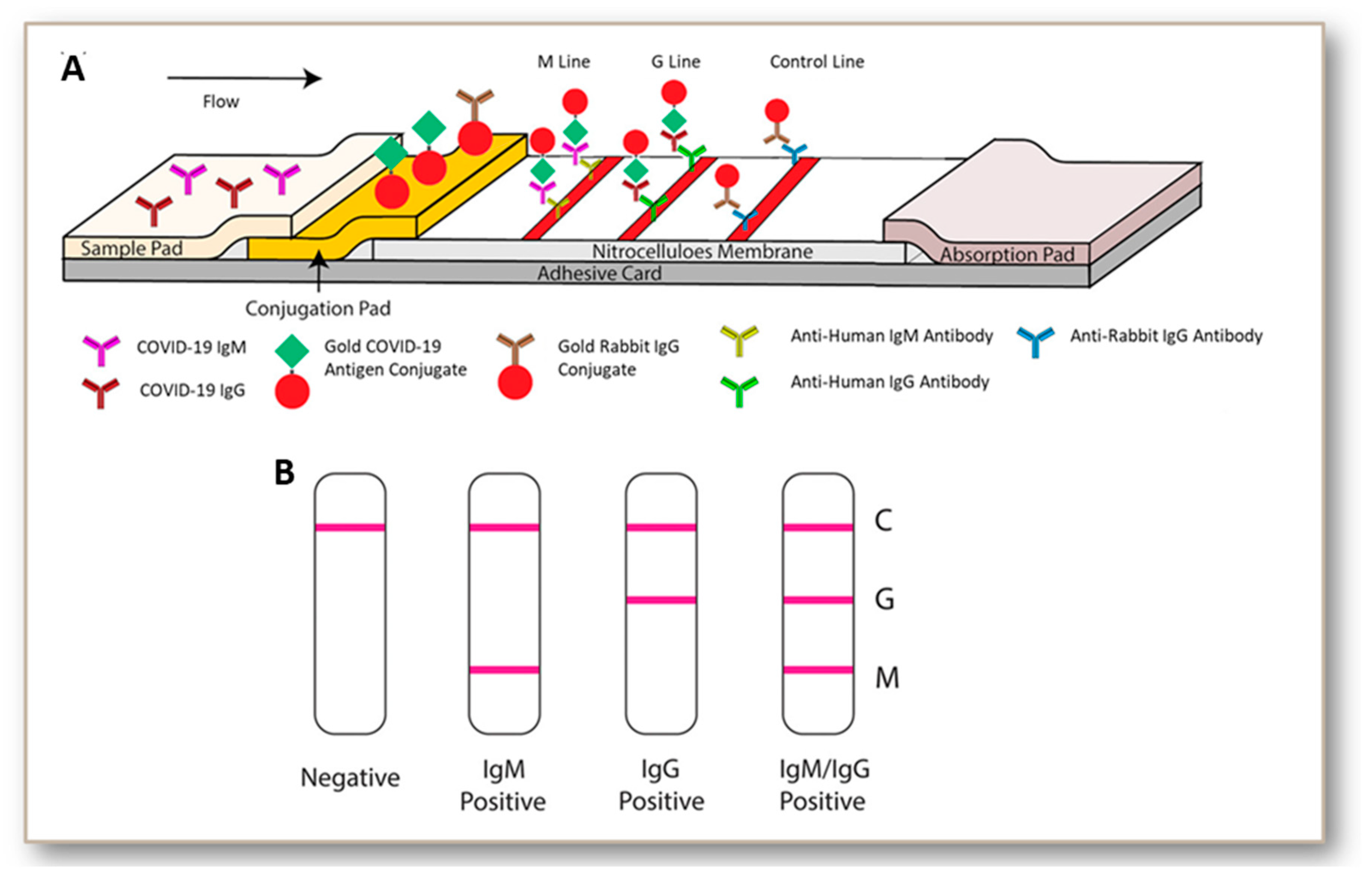
5. Development and Status of Immunoglobulin-Based Rapid Tests for COVID-19
- Binding antibody detection: these tests use specific reagents against individual isotypes such as IgM, IgG, IgA, and are tested against purified viral proteins in BSL2 laboratories.
- (a)
- The rapid tests or PoCTs utilize lateral flow devices for individual or combined antibody detection.
- (b)
- The laboratory assays are based on ELISA or chemiluminescent immunoassay (CIA).
- Neutralizing antibody assay: these tests involve infecting and incubating cultured cells with the virus isolated from patient samples, and then determining the functional ability of antibodies to prevent infections in vitro.
- (a)
- Virus neutralization tests use plaque reduction or microneutralization on clinical isolates.
- (b)
- Pseudovirus neutralization tests, on the contrary, use recombinant pseudoviruses.
6. Challenges and Pitfalls
7. Significant Advantages and Prospects
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Worldometers Covid-19 Coronavirus Pandemic. Available online: https://www.worldometers.info/coronavirus/ (accessed on 16 October 2020).
- Gralinski, L.E.; Menachery, V.D. Return of the coronavirus: 2019-nCoV. Viruses 2020, 12, 135. [Google Scholar] [CrossRef]
- He, X.; Lau, E.H.Y.; Wu, P.; Deng, X.; Wang, J.; Hao, X.; Lau, Y.C.; Wong, J.Y.; Guan, Y.; Tan, X.; et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020, 26, 672–675. [Google Scholar] [CrossRef]
- Wang, P.; Anderson, N.; Pan, Y.; Poon, L.; Charlton, C.; Zelyas, N.; Persing, D.; Rhoads, D.; Babcock, H. The SARS-CoV-2 Outbreak: Diagnosis, Infection Prevention, and Public Perception. Clin. Chem. 2020, 65, 644–651. [Google Scholar] [CrossRef]
- De Ridder, D.; Sandoval, J.; Vuilleumier, N.; Stringhini, S.; Spechbach, H.; Joost, S.; Kaiser, L.; Guessous, I. Geospatial digital monitoring of COVID-19 cases at high spatiotemporal resolution. Lancet Digit. Heal. 2020, 2, e393–e394. [Google Scholar] [CrossRef]
- Ghaffari, A.; Meurant, R.; Ardakani, A. COVID-19 serological tests: How well do they actually perform? Diagnostics 2020, 10, 453. [Google Scholar] [CrossRef]
- Baumgarth, N.; Tung, J.W.; Herzenberg, L.A. Inherent specificities in natural antibodies: A key to immune defense against pathogen invasion. Springer Semin. Immunopathol. 2005, 26, 347–362. [Google Scholar] [CrossRef]
- Long, B.; Brady, W.J.; Koyfman, A.; Gottlieb, M. Cardiovascular complications in COVID-19. Am. J. Emerg. Med. 2020, 5, 802–810. [Google Scholar] [CrossRef]
- Kochi, A.N.; Tagliari, A.P.; Forleo, G.B.; Fassini, G.M.; Tondo, C. Cardiac and arrhythmic complications in patients with COVID-19. J. Cardiovasc. Electrophysiol. 2020, 31, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Rhee, J.W.; Cheng, P.; Waliany, S.; Chang, A.; Witteles, R.M.; Maecker, H.; Davis, M.M.; Nguyen, P.K.; Wu, S.M. Cardiovascular Complications in Patients with COVID-19: Consequences of Viral Toxicities and Host Immune Response. Curr. Cardiol. Rep. 2020, 22, 32. [Google Scholar] [CrossRef] [PubMed]
- Lodigiani, C.; Iapichino, G.; Carenzo, L.; Cecconi, M.; Ferrazzi, P.; Sebastian, T.; Kucher, N.; Studt, J.D.; Sacco, C.; Alexia, B.; et al. Venous and arterial thromboembolic complications in COVID-19 patients admitted to an academic hospital in Milan, Italy. Thromb. Res. 2020, 191, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Filatov, A.; Sharma, P.; Hindi, F.; Espinosa, P.S. Neurological Complications of Coronavirus Disease (COVID-19): Encephalopathy. Cureus 2020, 12, e7352. [Google Scholar] [CrossRef] [PubMed]
- Needham, E.J.; Chou, S.H.Y.; Coles, A.J.; Menon, D.K. Neurological Implications of COVID-19 Infections. Neurocrit. Care 2020, 32, 667–671. [Google Scholar] [CrossRef] [PubMed]
- Varatharaj, A.; Thomas, N.; Ellul, M.A.; Davies, N.W.S.; Pollak, T.A.; Tenorio, E.L.; Sultan, M.; Easton, A.; Breen, G.; Zandi, M.; et al. Neurological and neuropsychiatric complications of COVID-19 in 153 patients: A UK-wide surveillance study. Lancet Psychiatry 2020, 7, 875–882. [Google Scholar] [CrossRef]
- Ahmad, I.; Rathore, F.A. Neurological manifestations and complications of COVID-19: A literature review. J. Clin. Neurosci. 2020, 77, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Gan, R.; Rosoman, N.P.; Henshaw, D.J.E.; Noble, E.P.; Georgius, P.; Sommerfeld, N. COVID-19 as a viral functional ACE2 deficiency disorder with ACE2 related multi-organ disease. Med. Hypotheses 2020, 144, 110024. [Google Scholar] [CrossRef]
- Wu, J.; Liu, J.; Li, S.; Peng, Z.; Xiao, Z.; Wang, X.; Yan, R.; Luo, J. Detection and analysis of nucleic acid in various biological samples of COVID-19 patients. Travel Med. Infect. Dis. 2020, 101673. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; Qian, K.; Zhang, Z.; Lin, F.; Xie, Y.; Liu, Y.; Yang, Z. CRISPR-cas systems based molecular diagnostic tool for infectious diseases and emerging 2019 novel coronavirus (COVID-19) pneumonia. J. Drug Target. 2020, 28, 727–731. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Augustine, R.; Hasan, A.; Das, S.; Ahmed, R.; Mori, Y.; Notomi, T.; Kevadiya, B.D.; Thakor, A.S. Loop-Mediated Isothermal Amplification (LAMP): A Rapid, Sensitive, Specific, and Cost-Effective Point-of-Care Test for Coronaviruses in the Context of COVID-19 Pandemic. Biology 2020, 9, 182. [Google Scholar] [CrossRef]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.H.; Choi, M.; Ku, K.B.; Lee, C.S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef]
- Wu, J.L.; Tseng, W.P.; Lin, C.H.; Lee, T.F.; Chung, M.Y.; Huang, C.H.; Chen, S.Y.; Hsueh, P.R.; Chen, S.C. Four point-of-care lateral flow immunoassays for diagnosis of COVID-19 and for assessing dynamics of antibody responses to SARS-CoV-2. J. Infect. 2020, 81, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.R.; Mirza, F.; Al-Hail, H.; Sundararaju, S.; Xaba, T.; Iqbal, M.; Alhussain, H.; Yassine, H.M.; Perez-Lopez, A.; Tang, P. Detection of SARS-CoV-2 RNA by direct RT-qPCR on nasopharyngeal specimens without extraction of viral RNA. PLoS ONE 2020, 15, e0236564. [Google Scholar] [CrossRef]
- Center for Health Security, Molecular-based Tests for COVID-19. Available online: https://www.centerforhealthsecurity.org/resources/COVID-19/molecular-based-tests/ (accessed on 16 October 2020).
- Shuren, J.E. Coronavirus (COVID-19) Update: FDA Authorizes First Antigen Test to Help in the Rapid Detection of the Virus that Causes COVID-19 in Patients|FDA. Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-antigen-test-help-rapid-detection-virus-causes (accessed on 30 July 2020).
- Mak, G.C.; Cheng, P.K.; Lau, S.S.; Wong, K.K.; Lau, C.S.; Lam, E.T.; Chan, R.C.; Tsang, D.N. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020, 129, 104500. [Google Scholar] [CrossRef] [PubMed]
- Grant, B.D.; Anderson, C.E.; Williford, J.R.; Alonzo, L.F.; Glukhova, V.A.; Boyle, D.S.; Weigl, B.H.; Nichols, K.P. SARS-CoV-2 Coronavirus Nucleocapsid Antigen-Detecting Half-Strip Lateral Flow Assay Toward the Development of Point of Care Tests Using Commercially Available Reagents. Anal. Chem. 2020, 92, 11305–11309. [Google Scholar] [CrossRef] [PubMed]
- Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W.; et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1953–1966. [Google Scholar] [CrossRef] [PubMed]
- Peiris, J.S.M.; Lai, S.T.; Poon, L.L.M.; Guan, Y.; Yam, L.Y.C.; Lim, W.; Nicholls, J.; Yee, W.K.S.; Yan, W.W.; Cheung, M.T.; et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003, 361, 1319–1325. [Google Scholar] [CrossRef]
- Louie, J.K.; Hacker, J.K.; Mark, J.; Gavali, S.S.; Yagi, S.; Espinosa, A.; Schnurr, D.P.; Cossen, C.K.; Isaacson, E.R.; Glaser, C.A.; et al. SARS and common viral infections. Emerg. Infect. Dis. 2004, 10, 1143–1146. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Wong, B.H.L.; Tsoi, H.W.; Fung, A.M.Y.; Chan, K.H.; Tam, V.K.P.; Peiris, J.S.M.; Yuen, K.Y. Detection of Specific Antibodies to Severe Acute Respiratory Syndrome (SARS) Coronavirus Nucleocapsid Protein for Serodiagnosis of SARS Coronavirus Pneumonia. J. Clin. Microbiol. 2004, 42, 2306–2309. [Google Scholar] [CrossRef]
- Younes, N.; Al-Sadeq, D.W.; AL-Jighefee, H.; Younes, S.; Al-Jamal, O.; Daas, H.I.; Yassine, H.M.; Nasrallah, G.K. Challenges in laboratory diagnosis of the novel coronavirus SARS-CoV-2. Viruses 2020, 12, 582. [Google Scholar] [CrossRef]
- Marcel, S.; Christian, A.L.; Richard, N.; Silvia, S.; Emma, H.; Jacques, F.; Marcel, Z.; Gabriela, S.; Manuel, B.; Annelies, W.S.; et al. COVID-19 epidemic in Switzerland: On the importance of testing, contact tracing and isolation. Swiss Med. Wkly. 2020, 150, w20225. [Google Scholar] [CrossRef]
- Abbasi, J. The Promise and Peril of Antibody Testing for COVID-19. JAMA 2020, 323, 2019–2021. [Google Scholar] [CrossRef] [PubMed]
- Ritzi-Lehnert, M. Development of chip-compatible sample preparation for diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 2012, 12, 189–206. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Bang, D.D.; Wolff, A. 2019 Novel coronavirus disease (COVID-19): Paving the road for rapid detection and point-of-care diagnostics. Micromachines 2020, 11, 306. [Google Scholar] [CrossRef]
- Pathak, M.; Patel, S.K.; Jigyasa, R.; Tiwari, R.; Dhama, K.; Sah, R.; Rabaan, A.A.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. Global Threat of SARS-CoV-2/COVID-19 and the Need for More and Better Diagnostic Tools. Arch. Med. Res. 2020, 51, 450. [Google Scholar] [CrossRef] [PubMed]
- Dochez, A.R.; Avery, O.T. The elaboration of specific soluble substance by pneumococcus during growth. J. Exp. Med. 1917, 26, 477–493. [Google Scholar] [CrossRef]
- Berson, S.A.; Yalow, R.S. Quantitative aspects of the reaction between insulin and insulin-binding antibody. J. Clin. Invest. 1959, 38, 1996–2016. [Google Scholar] [CrossRef]
- Engvall, E.; Jonsson, K.; Perlmann, P. Enzyme-linked immunosorbent assay. II. Quantitative assay of protein antigen, immunoglobulin g, by means of enzyme-labelled antigen and antibody-coated tubes. BBA-Protein Struct. 1971, 251, 427–434. [Google Scholar] [CrossRef]
- Busso, N.; Collart, M.; Vassalli, J.-D.; Belin, D. Antagonist effect of RU 486 on transcription of glucocorticoid-regulated genes. Exp. Cell Res. 1987, 173, 425–430. [Google Scholar] [CrossRef]
- Leuvering, J.H.W.; Thai, P.J.H.M.; Van der Waart, M.V.; Schuurs, A.H.W.M. Sol particle immunoassay (spia). J. Immunoass. 1980, 1, 77–91. [Google Scholar] [CrossRef]
- Talman, E.A.; Boughner, D.R. Glutaraldehyde fixation alters the internal shear properties of porcine aortic heart valve tissue. Ann. Thorac. Surg. 1995, 60. [Google Scholar] [CrossRef]
- Posthuma-Trumpie, G.A.; Korf, J.; van Amerongen, A. Lateral flow (immuno) assay: Its strengths, weaknesses, opportunities and threats A literature survey. Anal. Bioanal. Chem. 2009, 393, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Rosenstein, R.W.; Bloomster, T.G. Solid Phase Assay Employing Capillary Flow. U.S. Patent 4,855,240, 8 August 1989. [Google Scholar]
- May, K.; Prior, M.E.; Richards, I. Capillary Immunoassay and Device Therefor Comprising Mobilizable Particulate Labelled Reagents. U.S. Patent 6,818,455, 16 November 2004. [Google Scholar]
- Xiang, T.; Jiang, Z.; Zheng, J.; Lo, C.; Tsou, H.; Ren, G.; Zhang, J.; Huang, A.; Lai, G. A novel double antibody sandwich-lateral flow immunoassay for the rapid and simple detection of hepatitis C virus. Int. J. Mol. Med. 2012, 30, 1041–1047. [Google Scholar] [CrossRef]
- Shi, L.; Wu, F.; Wen, Y.; Zhao, F.; Xiang, J.; Ma, L. A novel method to detect Listeria monocytogenes via superparamagnetic lateral flow immunoassay. Anal. Bioanal. Chem. 2015, 407, 529–535. [Google Scholar] [CrossRef]
- Wiriyachaiporn, N.; Sirikett, H.; Maneeprakorn, W.; Dharakul, T. Carbon nanotag based visual detection of influenza A virus by a lateral flow immunoassay. Microchim. Acta 2017, 184, 1827–1835. [Google Scholar] [CrossRef]
- Wang, R.; Kim, K.; Choi, N.; Wang, X.; Lee, J.; Jeon, J.H.; Rhie, G.; Choo, J. Highly sensitive detection of high-risk bacterial pathogens using SERS-based lateral flow assay strips. Sens. Actuators B Chem. 2018, 270, 72–79. [Google Scholar] [CrossRef]
- Azar, M.M.; Landry, L. Detection of Influenza A and B Viruses and Respiratory Syncytial Virus by Use of Clinical Laboratory Improvement Amendments of 1988 (CLIA)-Waived Point-of-Care Assays: A Paradigm Shift to Molecular Tests. J. Clin. Microbiol. 2018, 56, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kozel, T.R.; Burnham-Marusich, A.R. Point-of-care testing for infectious diseases: Past, present, and future. J. Clin. Microbiol. 2017, 55, 2313–2320. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.; Kaplan, S.E.; Lopez, J.C.; Stiles, J.; Lu, X.; Tang, Y.W. Parallel validation of three molecular devices for simultaneous detection and identification of influenza A and B and respiratory syncytial viruses. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef] [PubMed]
- Loeffelholz, M.J.; Tang, Y.W. Laboratory diagnosis of emerging human coronavirus infections–the state of the art. Emerg. Microbes Infect. 2020, 9, 747–756. [Google Scholar] [CrossRef]
- Wood, C.S.; Thomas, M.R.; Budd, J.; Mashamba-Thompson, T.P.; Herbst, K.; Pillay, D.; Peeling, R.W.; Johnson, A.M.; McKendry, R.A.; Stevens, M.M. Taking connected mobile-health diagnostics of infectious diseases to the field. Nature 2019, 566, 467–474. [Google Scholar] [CrossRef]
- Borucki, M.K.; Lao, V.; Hwang, M.; Gardner, S.; Adney, D.; Munster, V.; Bowen, R.; Allen, J.E. Middle East Respiratory Syndrome Coronavirus Intra-Host Populations Are Characterized by Numerous High Frequency Variants. PLoS ONE 2016, 11, e0146251. [Google Scholar] [CrossRef] [PubMed]
- Rafael, M.E.; Taylor, T.; Magill, A.; Lim, Y.W.; Girosi, F.; Allan, R. Reducing the burden of childhood malaria in Africa: The role of improved. Nature 2006, 444, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Shulman, S.T.; Bisno, A.L.; Clegg, H.W.; Gerber, M.A.; Kaplan, E.L.; Lee, G.; Martin, J.M.; Van Beneden, C. Clinical practice guideline for the diagnosis and management of group a streptococcal pharyngitis: 2012 update by the infectious diseases society of America. Clin. Infect. Dis. 2012, 55, e86–e102. [Google Scholar] [CrossRef] [PubMed]
- Baptista, P.V. Nanodiagnostics: Leaving the research lab to enter the clinics? Diagnosis 2014, 1, 305–309. [Google Scholar] [CrossRef]
- Tallury, P.; Malhotra, A.; Byrne, L.M.; Santra, S. Nanobioimaging and sensing of infectious diseases. Adv. Drug Deliv. Rev. 2010, 62, 424–437. [Google Scholar] [CrossRef]
- Paul, A.; Hasan, A.; Rodes, L.; Sangaralingam, M.; Prakash, S. Bioengineered baculoviruses as new class of therapeutics using micro and nanotechnologies: Principles, prospects and challenges. Adv. Drug Deliv. Rev. 2014, 71, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Kaittanis, C.; Santra, S.; Perez, J.M. Emerging nanotechnology-based strategies for the identification of microbial pathogenesis. Adv. Drug Deliv. Rev. 2010, 62, 408–423. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, L.; Kong, X.; Sun, L. Application of nanodiagnostics in point-of-care tests for infectious diseases. Int. J. Nanomed. 2017, 12, 4789–4803. [Google Scholar] [CrossRef]
- Sun, B.; Straubinger, R.M.; Lovell, J.F. Current taxane formulations and emerging cabazitaxel delivery systems. Nano Res. 2018, 11, 5193–5218. [Google Scholar] [CrossRef]
- Vidrih, J.A.; Walensky, R.P.; Sax, P.E.; Freedberg, K.A. Positive Epstein-Barr virus heterophile antibody tests in patients with primary human immunodeficiency virus infection. Am. J. Med. 2001, 111, 192–194. [Google Scholar] [CrossRef]
- Rabe, I.B.; Staples, J.E.; Villanueva, J.; Hummel, K.B.; Johnson, J.A.; Rose, L.; Hills, S.; Wasley, A.; Fischer, M.; Powers, A.M. Interim guidance for interpretation of Zika virus antibody test results. Morb. Mortal. Wkly. Rep. 2016, 65, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Zumla, A.; Chan, J.F.W.; Azhar, E.I.; Hui, D.S.C.; Yuen, K.Y. Coronaviruses-drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016, 15, 327–347. [Google Scholar] [CrossRef] [PubMed]
- Kirchdoerfer, R.N.; Cottrell, C.A.; Wang, N.; Pallesen, J.; Yassine, H.M.; Turner, H.L.; Corbett, K.S.; Graham, B.S.; McLellan, J.S.; Ward, A.B. Pre-fusion structure of a human coronavirus spike protein. Nature 2016, 531, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Van Der Hoek, L. Human coronaviruses: What do they cause? Antivir. Ther. 2007, 12, 651–658. [Google Scholar] [PubMed]
- Memish, Z.A.; Perlman, S.; Van Kerkhove, M.D.; Zumla, A. Middle East respiratory syndrome. Lancet 2020, 395, 1063–1077. [Google Scholar] [CrossRef]
- Babadaei, M.M.N.; Hasan, A.; Bloukh, S.H.; Edis, Z.; Sharifi, M.; Kachooei, E.; Falahati, M. The expression level of angiotensin-converting enzyme 2 determines the severity of COVID-19: Lung and heart tissue as targets. J. Biomol. Struct. Dyn. 2020, 1–7. [Google Scholar] [CrossRef]
- Song, Z.; Xu, Y.; Bao, L.; Zhang, L.; Yu, P.; Qu, Y.; Zhu, H.; Zhao, W.; Han, Y.; Qin, C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses 2019, 11, 59. [Google Scholar] [CrossRef]
- Columbus, C.; Brust, K.B.; Arroliga, A.C. 2019 novel coronavirus: An emerging global threat. Baylor Univ. Med. Cent. Proc. 2020, 33, 209–212. [Google Scholar] [CrossRef]
- Prompetchara, E.; Ketloy, C.; Palaga, T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pacific J. Allergy Immunol. 2020, 38, 1–9. [Google Scholar]
- Chen, X.; Zhou, B.; Li, M.; Liang, X.; Wang, H.; Yang, G.; Wang, H.; Le, X. Serology of Severe Acute Respiratory Syndrome: Implications for Surveillance and Outcome. J. Infect. Dis. 2004, 189, 1158–1163. [Google Scholar] [CrossRef]
- Hsueh, P.R.; Hsiao, C.H.; Yeh, S.H.; Wang, W.K.; Chen, P.J.; Wang, J.T.; Chang, S.C.; Kao, C.L.; Yang, P.C.; Chen, D.S.; et al. Microbiologic characteristics, serologic responses, and clinical manifestations in severe acute respiratory syndrome, Taiwan. Emerg. Infect. Dis. 2003, 9, 1163–1167. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, S.; Li, Q.; Li, Y.; Wei, M.; Gao, H.; Donovan, C.; Wang, P.P. Determining SARS sub-clinical infection: A longitudinal seroepidemiological study in recovered SARS patients and controls after an outbreak in a general hospital. Scand. J. Infect. Dis. 2009, 41, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Callow, K.A.; Parry, H.F.; Sergeant, M.; Tyrrell, D.A.J. The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect. 1990, 105, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, P.R.; Kao, C.L.; Lee, C.N.; Chen, L.K.; Ho, M.S.; Sia, C.; De Fang, X.; Lynn, S.; Chang, T.Y.; Liu, S.K.; et al. SARS antibody test for serosurveillance. Emerg. Infect. Dis. 2004, 10, 1558–1562. [Google Scholar] [CrossRef] [PubMed]
- Spanakis, N.; Tsiodras, S.; Haagmans, B.L.; Raj, V.S.; Pontikis, K.; Koutsoukou, A.; Koulouris, N.G.; Osterhaus, A.D.M.E.; Koopmans, M.P.G.; Tsakris, A. Virological and serological analysis of a recent Middle East respiratory syndrome coronavirus infection case on a triple combination antiviral regimen. Int. J. Antimicrob. Agents 2014, 44, 528–532. [Google Scholar] [CrossRef]
- Ko, J.H.; Müller, M.A.; Seok, H.; Park, G.E.; Lee, J.Y.; Cho, S.Y.; Ha, Y.E.; Baek, J.Y.; Kim, S.H.; Kang, J.M.; et al. Serologic responses of 42 MERS-coronavirus-infected patients according to the disease severity. Diagn. Microbiol. Infect. Dis. 2017, 89, 106–111. [Google Scholar] [CrossRef]
- Zhang, G.; Nie, S.; Zhang, Z.; Zhang, Z. Longitudinal Change of Severe Acute Respiratory Syndrome Coronavirus 2 Antibodies in Patients with Coronavirus Disease 2019. J. Infect. Dis. 2020, 222, 183–188. [Google Scholar] [CrossRef]
- Mo, H.; Zeng, G.; Ren, X.; Li, H.; Ke, C.; Tan, Y.; Cai, C.; Lai, K.; Chen, R.; Chan-Yeung, M.; et al. Longitudinal profile of antibodies against SARS-coronavirus in SARS patients and their clinical significance. Respirology 2006, 11, 49–53. [Google Scholar] [CrossRef]
- Wu, L.P.; Wang, N.C.; Chang, Y.H.; Tian, X.Y.; Na, D.Y.; Zhang, L.Y.; Zheng, L.; Lan, T.; Wang, L.F.; Liang, G.D. Duration of antibody responses after severe acute respiratory syndrome. Emerg. Infect. Dis. 2007, 13, 1562–1564. [Google Scholar] [CrossRef]
- Wang, W.; Wang, H.; Deng, Y.; Song, T.; Lan, J.; Wu, G.; Ke, C.; Tan, W. Characterization of anti-MERS-CoV antibodies against various recombinant structural antigens of MERS-CoV in an imported case in China. Emerg. Microbes Infect. 2016, 5, e113. [Google Scholar] [CrossRef]
- Park, W.B.; Perera, R.A.P.M.; Choe, P.G.; Lau, E.H.Y.; Choi, S.J.; Chun, J.Y.; Oh, H.S.; Song, K.H.; Bang, J.H.; Kim, E.S.; et al. Kinetics of serologic responses to mers coronavirus infection in humans, South Korea. Emerg. Infect. Dis. 2015, 21, 2186–2189. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Albarrak, A.M.; Omrani, A.S.; Albarrak, M.M.; Farah, M.E.; Almasri, M.; Muth, D.; Sieberg, A.; Meyer, B.; Assiri, A.M.; et al. Viral Shedding and Antibody Response in 37 Patients with Middle East Respiratory Syndrome Coronavirus Infection. Clin. Infect. Dis. 2015, 62, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Long, Q.; Deng, H.; Chen, J.; Hu, J.; Liu, B.; Liao, P.; Lin, Y.; Yu, L.; Mo, Z.; Xu, Y.; et al. Antibody responses to SARS-CoV-2 in COVID-19 patients: The perspective application of serological tests in clinical practice. medRxiv 2020. [Google Scholar] [CrossRef]
- Xiao, A.T.; Gao, C.; Zhang, S. Profile of specific antibodies to SARS-CoV-2: The first report. J. Infect. 2020, 81, 147–178. [Google Scholar] [CrossRef] [PubMed]
- Tuaillon, E.; Bolloré, K.; Pisoni, A.; Debiesse, S.; Renault, C.; Marie, S.; Groc, S.; Niels, C.; Pansu, N.; Dupuy, A.M.; et al. Detection of SARS-CoV-2 antibodies using commercial assays and seroconversion patterns in hospitalized patients. J. Infect. 2020, 81, e39–e45. [Google Scholar] [CrossRef]
- Lee, C.Y.P.; Lin, R.T.P.; Renia, L.; Ng, L.F.P. Serological Approaches for COVID-19: Epidemiologic Perspective on Surveillance and Control. Front. Immunol. 2020, 11, 879. [Google Scholar] [CrossRef]
- Long, Q.X.; Tang, X.J.; Shi, Q.L.; Li, Q.; Deng, H.J.; Yuan, J.; Hu, J.L.; Xu, W.; Zhang, Y.; Lv, F.J.; et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020, 26, 1200–1204. [Google Scholar] [CrossRef]
- Huang, A.T.; Garcia-Carreras, B.; Hitchings, M.D.T.; Yang, B.; Katzelnick, L.; Rattigan, S.M.; Borgert, B.; Moreno, C.; Solomon, B.D.; Rodriguez-Barraquer, I.; et al. A systematic review of antibody mediated immunity to coronaviruses: Antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. medRxiv 2020. [Google Scholar] [CrossRef]
- Hou, H.; Wang, T.; Zhang, B.; Luo, Y.; Mao, L.; Wang, F.; Wu, S.; Sun, Z. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin. Transl. Immunol. 2020, 9, e1136. [Google Scholar] [CrossRef]
- Wu, H.S.; Chiu, S.C.; Tseng, T.C.; Lin, S.F.; Lin, J.H.; Hsu, Y.F.; Wang, M.C.; Lin, T.L.; Yang, W.Z.; Ferng, T.L.; et al. Serologic and Molecular Biologic Methods for SARS-associated Coronavirus Infection, Taiwan. Emerg. Infect. Dis. 2004, 10, 304–310. [Google Scholar] [CrossRef]
- Hsueh, P.R.; Huang, L.M.; Chen, P.J.; Kao, C.L.; Yang, P.C. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin. Microbiol. Infect. 2004, 10, 1062–1066. [Google Scholar] [CrossRef] [PubMed]
- Manopo, I.; Lu, L.; He, Q.; Chee, L.L.; Chan, S.W.; Kwang, J. Evaluation of a safe and sensitive Spike protein-based immunofluorescence assay for the detection of antibody responses to SARS-CoV. J. Immunol. Methods 2005, 296, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, K.; Chan, K.H.; Takeda, K.; Lo, K.F.; Leung, R.H.K.; Okamoto, T. Sensitive and specific enzyme-linked immunosorbent assay using chemiluminescence for detection of severe acute respiratory syndrome viral infection. J. Clin. Microbiol. 2008, 46, 302–310. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, J.S.; Chen, J.T.; Liu, Y.X.; Zhang, Z.S.; Gao, H.; Liu, Y.; Wang, X.; Ning, Y.; Liu, Y.F.; Gao, Q.; et al. A serological survey on neutralizing antibody titer of SARS convalescent sera. J. Med. Virol. 2005, 77, 147–150. [Google Scholar] [CrossRef]
- Perera, R.A.; Wang, P.; Gomaa, M.R.; El-Shesheny, R.; Kandeil, A.; Bagato, O.; Siu, L.Y.; Shehata, M.M.; Kayed, A.S.; Moatasim, Y.; et al. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, june 2013. Eurosurveillance 2013, 18, 20574. [Google Scholar] [CrossRef]
- Richardson, S.E.; Tellier, R.; Mahony, J. The laboratory diagnosis of severe acute respiratory syndrome: Emerging laboratory tests for an emerging pathogen. Clin. Biochem. Rev. 2004, 25, 133–141. [Google Scholar]
- Park, S.W.; Perera, R.A.P.M.; Choe, P.G.; Lau, E.H.Y.; Choi, S.J.; Chun, J.Y.; Oh, H.S.; Song, K.; Bang, J.H.; Kim, E.S.; et al. Comparison of serological assays in human middle east respiratory syndrome (MERS)-coronavirus infection. Eurosurveillance 2015, 20, 30042. [Google Scholar] [CrossRef]
- Algaissi, A.; Hashem, A.M. Evaluation of MERS-CoV neutralizing antibodies in sera using live virus microneutralization assay. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2020; Volume 2099, pp. 107–116. [Google Scholar]
- Arwady, M.A.; Alraddadi, B.; Basler, C.; Azhar, E.I.; Abuelzein, E.; Sindy, A.I.; Bin Sadiq, B.M.; Althaqafi, A.O.; Shabouni, O.; Banjar, A.; et al. Middle east respiratory syndrome coronavirus transmission in extended family, Saudi Arabia, 2014. Emerg. Infect. Dis. 2016, 22, 1395–1402. [Google Scholar] [CrossRef]
- Lu, S.; Lin, J.; Zhang, Z.; Xiao, L.; Jiang, Z.; Chen, J.; Hu, C.; Luo, S. Alert for non-respiratory symptoms of Coronavirus Disease 2019 (COVID-19) patients in epidemic period: A case report of familial cluster with three asymptomatic COVID-19 patients. J. Med. Virol. 2020. [Google Scholar] [CrossRef]
- Mason, R.J. Pathogenesis of COVID-19 from a cell biology perspective. Eur. Respir. J. 2020, 55, 2000607. [Google Scholar] [CrossRef]
- Kubina, R.; Dziedzic, A. Molecular and serological tests for COVID-19 a comparative review of SARS-CoV-2 coronavirus laboratory and point-of-care diagnostics. Diagnostics. 2020, 10, 434. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Ding, C.; Li, J.; Wang, Y.; Guo, H.; Lu, Z.; Wang, J.; Zheng, C.; Jin, T.; Gao, Y.; et al. Characteristics of patients with coronavirus disease (COVID-19) confirmed using an IgM-IgG antibody test. J. Med. Virol. 2020, 92, 2004–2010. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Wunderink, R.G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology 2018, 23, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Feng, Y.; Mo, X.; Zheng, P.; Wang, Q.; Li, P.; Peng, P.; Liu, X.; Chen, Z.; Huang, H.; et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microbes Infect. 2020, 9, 940–948. [Google Scholar] [CrossRef]
- Chen, Y.; Tong, X.; Li, Y.; Gu, B.; Yan, J.; Liu, Y.; Shen, H.; Huang, R.; Wu, C. A comprehensive, longitudinal analysis of humoral responses specific to four recombinant antigens of SARS-CoV-2 in severe and non-severe COVID-19 patients. PLoS Pathog. 2020, 16, e1008796. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.; et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Okba, N.M.A.; Muller, M.A.; Li, W.; Wang, C.; GeurtsvanKessel, C.H.; Corman, V.M.; Lamers, M.M.; Sikkema, R.S.; de Bruin, E.; Chandler, F.D.; et al. SARS-CoV-2 specific antibody responses in COVID-19 patients. medRxiv 2020. [Google Scholar] [CrossRef]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020, 92, 1524–14518. [Google Scholar] [CrossRef]
- Thevarajan, I.; Nguyen, T.H.O.; Koutsakos, M.; Druce, J.; Caly, L.; van de Sandt, C.E.; Jia, X.; Nicholson, S.; Catton, M.; Cowie, B.; et al. Breadth of concomitant immune responses prior to patient recovery: A case report of non-severe COVID-19. Nat. Med. 2020, 26, 453–455. [Google Scholar] [CrossRef]
- McElroy, A.K.; Akondy, R.S.; Davis, C.W.; Ellebedy, A.H.; Mehta, A.K.; Kraft, C.S.; Lyon, G.M.; Ribner, B.S.; Varkey, J.; Sidney, J.; et al. Human Ebola virus infection results in substantial immune activation. Proc. Natl. Acad. Sci. USA 2015, 112, 4719–4724. [Google Scholar] [CrossRef]
- Koutsakos, M.; Wheatley, A.K.; Loh, L.; Clemens, E.B.; Sant, S.; Nüssing, S.; Fox, A.; Chung, A.W.; Laurie, K.L.; Hurt, A.C.; et al. Circulating TFH cells, serological memory, and tissue compartmentalization shape human influenza-specific B cell immunity. Sci. Transl. Med. 2018, 10, eaan8405. [Google Scholar] [CrossRef] [PubMed]
- Haveri, A.; Smura, T.; Kuivanen, S.; Österlund, P.; Hepojoki, J.; Ikonen, N.; Pitkäpaasi, M.; Blomqvist, S.; Rönkkö, E.; Kantele, A.; et al. Serological and molecular findings during SARS-CoV-2 infection: The first case study in Finland, January to February 2020. Eurosurveillance 2020, 25, 2000266. [Google Scholar] [CrossRef] [PubMed]
- La Marca, A.; Capuzzo, M.; Paglia, T.; Roli, L.; Trenti, T.; Nelson, S.M. Testing for SARS-CoV-2 (COVID-19): A systematic review and clinical guide to molecular and serological in-vitro diagnostic assays. Reprod. Biomed. Online 2020, 41, 483–499. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Ren, L.; Yang, S.; Xiao, M.; Chang, D.; Yang, F.; Dela Cruz, C.S.; Wang, Y.; Wu, C.; Xiao, Y.; et al. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19). Clin. Infect. Dis. 2020, 71, 778–785. [Google Scholar] [CrossRef]
- Xiang, F.; Wang, X.; He, X.; Peng, Z.; Yang, B.; Zhang, J.; Zhou, Q.; Ye, H.; Ma, Y.; Li, H.; et al. Antibody Detection and Dynamic Characteristics in Patients with COVID-19. Clin. Infect. Dis. 2020, 1–23. [Google Scholar] [CrossRef]
- Infantino, M.; Grossi, V.; Lari, B.; Bambi, R.; Perri, A.; Manneschi, M.; Terenzi, G.; Liotti, I.; Ciotta, G.; Taddei, C.; et al. Diagnostic accuracy of an automated chemiluminescent immunoassay for anti-SARS-CoV-2 IgM and IgG antibodies: An Italian experience. J. Med. Virol. 2020, 92, 1671–1675. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, M.; Zuo, Z.; Fan, C.; Ye, F.; Cai, Z.; Wang, Y.; Cui, H.; Pan, K.; Xu, A. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int. J. Infect. Dis. 2020, 94, 49–52. [Google Scholar] [CrossRef]
- Soleimani, R.; Khourssaji, M.; Gruson, D.; Rodriguez-Villalobos, H.; Berghmans, M.; Belkhir, L.; Yombi, J.C.; Kabamba-Mukadi, B. Clinical usefulness of fully automated chemiluminescent immunoassay for quantitative antibody measurements in COVID-19 patients. J. Med. Virol. 2020. [Google Scholar] [CrossRef]
- Long, Q.X.; Liu, B.Z.; Deng, H.J.; Wu, G.C.; Deng, K.; Chen, Y.K.; Liao, P.; Qiu, J.F.; Lin, Y.; Cai, X.F.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef]
- Cai, X.F.; Chen, J.; Li Hu, J.; Long, Q.X.; Deng, H.J.; Liu, P.; Fan, K.; Liao, P.; Liu, B.Z.; Wu, G.C.; et al. A Peptide-Based Magnetic Chemiluminescence Enzyme Immunoassay for Serological Diagnosis of Coronavirus Disease 2019. J. Infect. Dis. 2020, 222, 189–193. [Google Scholar] [CrossRef]
- Perera, R.A.P.M.; Mok, C.K.P.; Tsang, O.T.Y.; Lv, H.; Ko, R.L.W.; Wu, N.C.; Yuan, M.; Leung, W.S.; Chan, J.M.C.; Chik, T.S.H.; et al. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Eurosurveillance 2020, 25, 2000421. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhu, F.; Guo, F.; Yang, B.; Wang, T. Detection of antibodies against SARS-CoV-2 in patients with COVID-19. J. Med. Virol. 2020, 92, 1735–1738. [Google Scholar] [CrossRef] [PubMed]
- Jääskeläinen, A.J.; Kekäläinen, E.; Kallio-Kokko, H.; Mannonen, L.; Kortela, E.; Vapalahti, O.; Kurkela, S.; Lappalainen, M. Evaluation of commercial and automated SARS-CoV-2 IgG and IgA ELISAs using coronavirus disease (COVID-19) patient samples. Eurosurveillance 2020, 25, 2000603. [Google Scholar] [CrossRef] [PubMed]
- Cassaniti, I.; Novazzi, F.; Giardina, F.; Salinaro, F.; Sachs, M.; Perlini, S.; Bruno, R.; Mojoli, F.; Baldanti, F. Performance of VivaDiag COVID-19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J. Med. Virol. 2020, 1–4. [Google Scholar] [CrossRef]
- Pan, Y.; Li, X.; Yang, G.; Fan, J.; Tang, Y.; Zhao, J.; Long, X.; Guo, S.; Zhao, Z.; Liu, Y.; et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J. Infect. 2020, 81, e28–e32. [Google Scholar] [CrossRef]
- Wang, X.; Guo, X.; Prof, A.; Xin, Q.; Pan, Y.; Li, J.; Chu, Y.; Feng, Y.; Wang, Q.; Youan, B. Neutralizing Antibodies Responses to SARS-CoV-2 in COVID-19 Inpatients and Convalescent Patients. medRxiv 2020. [Google Scholar] [CrossRef]
- Jia, X.; Zhang, P.; Tian, Y.; Wang, J.; Zeng, H.; Wang, J.; Jiao, L.; Chen, Z.; Zhang, L.; He, H.; et al. Clinical significance of IgM and IgG test for diagnosis of highly suspected COVID-19 infection. medRxiv 2020. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Y.; Diao, B.; Ren, F.; Wang, Y.; Ding, J.; Huang, Q. Diagnostic Indexes of a Rapid IgG/IgM Combined Antibody Test for SARS-CoV-2. medRxiv 2020. [Google Scholar] [CrossRef]
- Pan, Y.; Zhang, D.; Yang, P.; Poon, L.L.M.; Wang, Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020, 20, 411–412. [Google Scholar] [CrossRef]
- Norman, M.; Gilboa, T.; Ogata, A.F.; Maley, A.M.; Cohen, L.; Cai, Y.; Zhang, J.; Feldman, J.E.; Hauser, B.M.; Caradonna, T.M.; et al. Ultra-Sensitive High-Resolution Profiling of Anti-SARS-CoV-2 Antibodies for Detecting Early Seroconversion in COVID-19 Patients. medRxiv Prepr. Serv. Heal. Sci. 2020. [Google Scholar] [CrossRef]
- To, K.K.W.; Tsang, O.T.Y.; Leung, W.S.; Tam, A.R.; Wu, T.C.; Lung, D.C.; Yip, C.C.Y.; Cai, J.P.; Chan, J.M.C.; Chik, T.S.H.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef]
- Amanat, F.; Stadlbauer, D.; Strohmeier, S.; Nguyen, T.H.O.; Chromikova, V.; McMahon, M.; Jiang, K.; Arunkumar, G.A.; Jurczyszak, D.; Polanco, J.; et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020, 26, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Hasan, A.; Nejadi Babadaei, M.M.; Bloukh, S.H.; Chowdhury, M.E.H.; Sharifi, M.; Haghighat, S.; Falahati, M. Targeting SARS-CoV2 Spike Protein Receptor Binding Domain by Therapeutic Antibodies. Biomed. Pharmacother. 2020, 130, 110559. [Google Scholar] [CrossRef]
- Hasan, A.; Paray, B.A.; Hussain, A.; Qadir, F.A.; Attar, F.; Aziz, F.M.; Sharifi, M.; Derakhshankhah, H.; Rasti, B.; Mehrabi, M.; et al. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. J. Biomol. Struct. Dyn. 2020, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Al-Muharraqi, M.A. Testing recommendation for COVID-19 (SARS-CoV-2) in patients planned for surgery - continuing the service and ‘suppressing’ the pandemic. Br. J. Oral Maxillofac. Surg. 2020, 58, 503–505. [Google Scholar] [CrossRef] [PubMed]
- Lassaunière, R.; Frische, A.; Harboe, Z.; Nielsen, A.C.; Fomsgaard, A.; Krogfelt, K.; Jørgensen, C. Evaluation of nine commercial SARS-CoV-2 immunoassays. medRxiv 2020. [Google Scholar] [CrossRef]
- GeurtsvanKessel, C.H.; Okba, N.M.A.; Igloi, Z.; Bogers, S.; Embregts, C.W.E.; Laksono, B.M.; Leijten, L.; Rokx, C.; Rijnders, B.; Rahamat-Langendoen, J.; et al. An evaluation of COVID-19 serological assays informs future diagnostics and exposure assessment. Nat. Commun. 2020, 11, 3436. [Google Scholar] [CrossRef]
- Andrey, D.O.; Cohen, P.; Meyer, B.; Torriani, G.; Yerly, S.; Mazza, L.; Calame, A.; Arm-Vernez, I.; Guessous, I.; Stringhini, S.; et al. Diagnostic accuracy of Augurix COVID-19 IgG serology rapid test. Eur. J. Clin. Investig. 2020, 50, e13357. [Google Scholar] [CrossRef]
- Andrey, D.O.; Cohen, P.; Meyer, B.; Torriani, G.; Yerly, S.; Mazza, L.; Calame, A.; Arm-Vernez, I.; Guessous, I.; Stringhini, S.; et al. Head-to-Head Accuracy Comparison of Three Commercial COVID-19 IgM/IgG Serology Rapid Tests. J. Clin. Med. 2020, 9, 2369. [Google Scholar] [CrossRef]
- ANCY CROTTI BREAKING: BD Stops Selling COVID-19 Antibody Test it Touted in March. Available online: https://www.massdevice.com/breaking-bd-stops-selling-covid-19-antibody-test-it-touted-in-march/ (accessed on 26 September 2020).
- Kobokovich, A.; West, R.; Grovall, G. Serology-Based Tests for COVID-19. Available online: https://www.centerforhealthsecurity.org/resources/COVID-19/serology/Serology-based-tests-for-COVID-19.html (accessed on 24 July 2020).
- Salminen, T.; Juntunen, E.; Talha, S.M.; Pettersson, K. High-sensitivity lateral flow immunoassay with a fluorescent lanthanide nanoparticle label. J. Immunol. Methods 2019, 465, 39–44. [Google Scholar] [CrossRef]
- Edridge, A.W.D.; Kaczorowska, J.M.; Hoste, A.C.R.; Bakker, M.; Klein, M.; Jebbink, M.F.; Matser, A.; Kinsella, C.M.; Rueda, P.; Prins, M.; et al. Human coronavirus reinfection dynamics: Lessons for SARS-CoV-2. Medrxiv 2020, 1–10. [Google Scholar] [CrossRef]
- Kellam, P.; Barclay, W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J. Gen. Virol. 2020, 101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Du, R.H.; Li, B.; Zheng, X.S.; Yang, X.-L.; Hu, B.; Wang, Y.Y.; Xiao, G.F.; Yan, B.; Shi, Z.L.; et al. Molecular and serological investigation of 2019-nCoV infected patients: Implication of multiple shedding routes. Emerg. Microbes Infect. 2020, 9, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Demey, B.; Daher, N.; François, C.; Lanoix, J.P.; Duverlie, G.; Castelain, S.; Brochot, E. Dynamic profile for the detection of anti-SARS-CoV-2 antibodies using four immunochromatographic assays. J. Infect. 2020, 81, e6–e10. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Joyner, M.J.; Pirofski, L.-A. SARS-CoV-2 viral load and antibody responses: The case for convalescent plasma therapy. J. Clin. Investig. 2020, 130. [Google Scholar] [CrossRef]
- Du, D.; Fu, H.J.; Ren, W.-W.; Li, X.L.; Guo, L.H. PSA targeted dual-modality manganese oxide–mesoporous silica nanoparticles for prostate cancer imaging. Biomed. Pharmacother. 2020, 121, 109614. [Google Scholar] [CrossRef]
- Che, X.; Qiu, L.; Liao, Z.; Wang, Y.; Wen, K.; Pan, Y.; Hao, W.; Mei, Y.; Cheng, V.C.C.; Yuen, K. Antigenic Cross-Reactivity between Severe Acute Respiratory Syndrome–Associated Coronavirus and Human Coronaviruses 229E and OC43. J. Infect. Dis. 2005, 191, 2033–2037. [Google Scholar] [CrossRef][Green Version]
- Wang, N.; Li, S.Y.; Yang, X.; Huang, H.M.; Zhang, Y.J.; Guo, H.; Luo, C.M.; Miller, M.; Zhu, G.; Chmura, A.A.; et al. Serological Evidence of Bat SARS-Related Coronavirus Infection in Humans, China. Virol. Sin. 2018, 33, 104–107. [Google Scholar] [CrossRef]
- Jacofsky, D.; Jacofsky, E.M.; Jacofsky, M. Understanding Antibody Testing for COVID-19. J. Arthroplasty 2020, 35, S74–S81. [Google Scholar] [CrossRef]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef]
- Carbone, M.; Green, J.B.; Bucci, E.M.; Lednicky, J.A. Coronaviruses: Facts, Myths, and Hypotheses. J. Thorac. Oncol. 2020, 15, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Petherick, A. Developing antibody tests for SARS-CoV-2. Lancet 2020, 395, 1101–1102. [Google Scholar] [CrossRef]
- Grady, D. Coronavirus Test Kits Sent to States Are Flawed, C.D.C. Says. Available online: https://www.nytimes.com/2020/02/12/health/coronavirus-test-kits-cdc.html (accessed on 1 June 2020).
- Xiang, J.; Yan, M.; Li, H.; Liu, T.; Lin, C.; Huang, S.; Shen, C. Evaluation of Enzyme-Linked Immunoassay and Colloidal Gold- Immunochromatographic Assay Kit for Detection of Novel Coronavirus (SARS-Cov-2) Causing an Outbreak of Pneumonia (COVID-19). medRxiv 2020. [Google Scholar] [CrossRef]
- Pollán, M.; Pérez-Gómez, B.; Pastor-Barriuso, R.; Oteo, J.; Hernán, M.A.; Pérez-Olmeda, M.; Sanmartín, J.L.; Fernández-García, A.; Cruz, I.; Fernández de Larrea, N.; et al. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): A nationwide, population-based seroepidemiological study. Lancet 2020, 396, 535–544. [Google Scholar] [CrossRef]
- Stringhini, S.; Wisniak, A.; Piumatti, G.; Azman, A.S.; Lauer, S.A.; Baysson, H.; De Ridder, D.; Petrovic, D.; Schrempft, S.; Marcus, K.; et al. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): A population-based study. Lancet 2020, 396, 313–319. [Google Scholar] [CrossRef]
- Stubblefield, W.B.; Talbot, H.K.; Feldstein, L.; Tenforde, M.W.; Rasheed, M.A.U.; Mills, L.; Lester, S.N.; Freeman, B.; Thornburg, N.J.; Jones, I.D.; et al. Seroprevalence of SARS-CoV-2 Among Frontline Healthcare Personnel During the First Month of Caring for COVID-19 Patients—Nashville, Tennessee. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Batisse, D.; Benech, N.; Botelho-Nevers, E.; Bouiller, K.; Collarino, R.; Conrad, A.; Gallay, L.; Goehringer, F.; Gousseff, M.; Joseph, D.C.; et al. Clinical recurrences of COVID-19 symptoms after recovery: Viral relapse, reinfection or inflammatory rebound? J. Infect. 2020. [Google Scholar] [CrossRef]
- Dufort, E.M.; Koumans, E.H.; Chow, E.J.; Rosenthal, E.M.; Muse, A.; Rowlands, J.; Barranco, M.A.; Maxted, A.M.; Rosenberg, E.S.; Easton, D.; et al. Multisystem inflammatory syndrome in children in New York state. N. Engl. J. Med. 2020, 383, 347–358. [Google Scholar] [CrossRef]
- Feldstein, L.R.; Rose, E.B.; Horwitz, S.M.; Collins, J.P.; Newhams, M.M.; Son, M.B.F.; Newburger, J.W.; Kleinman, L.C.; Heidemann, S.M.; Martin, A.A.; et al. Multisystem inflammatory syndrome in U.S. Children and adolescents. N. Engl. J. Med. 2020, 383, 334–346. [Google Scholar] [CrossRef]
- Consiglio, C.R.; Cotugno, N.; Sardh, F.; Pou, C.; Amodio, D.; Rodriguez, L.; Tan, Z.; Zicari, S.; Ruggiero, A.; Pascucci, G.R.; et al. The Immunology of Multisystem Inflammatory Syndrome in Children with COVID-19. Cell 2020. [Google Scholar] [CrossRef]
- Nakra, N.A.; Blumberg, D.A.; Herrera-Guerra, A.; Lakshminrusimha, S. Multi-System Inflammatory Syndrome in Children (MIS-C) Following SARS-CoV-2 Infection: Review of Clinical Presentation, Hypothetical Pathogenesis, and Proposed Management. Children 2020, 7, 69. [Google Scholar] [CrossRef]
- Ahmed, M.; Advani, S.; Moreira, A.; Zoretic, S.; Martinez, J.; Chorath, K.; Acosta, S.; Naqvi, R.; Burmeister-Morton, F.; Burmeister, F.; et al. Multisystem inflammatory syndrome in children: A systematic review. EClinicalMedicine 2020, 26, 100527. [Google Scholar] [CrossRef] [PubMed]
- Greene, A.G.; Saleh, M.; Roseman, E.; Sinert, R. Toxic shock-like syndrome and COVID-19: A case report of multisystem inflammatory syndrome in children (MIS-C). Am. J. Emerg. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, E.; Bamford, A.; Kenny, J.; Kaforou, M.; Jones, C.E.; Shah, P.; Ramnarayan, P.; Fraisse, A.; Miller, O.; Davies, P.; et al. Clinical Characteristics of 58 Children with a Pediatric Inflammatory Multisystem Syndrome Temporally Associated with SARS-CoV-2. JAMA 2020, 324, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Dolinger, M.T.; Person, H.; Smith, R.; Jarchin, L.; Pittman, N.; Dubinsky, M.C.; Lai, J. Pediatric Crohn Disease and Multisystem Inflammatory Syndrome in Children (MIS-C) and COVID-19 Treated With Infliximab. J. Pediatr. Gastroenterol. Nutr. 2020. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Aydin, S.I.; Derespina, K.R.; Bansal, P.B.; Kowalsky, S.; Trachtman, R.; Gillen, J.K.; Perez, M.M.; Soshnick, S.H.; Conway, E.E.; et al. Multisystem Inflammatory Syndrome in Children Associated with Severe Acute Respiratory Syndrome Coronavirus 2 Infection (MIS-C): A Multi-institutional Study from New York City. J. Pediatr. 2020, 224, 24–29. [Google Scholar] [CrossRef]
- Carter, M.J.; Fish, M.; Jennings, A.; Doores, K.J.; Wellman, P.; Seow, J.; Acors, S.; Graham, C.; Timms, E.; Kenny, J.; et al. Peripheral immunophenotypes in children with multisystem inflammatory syndrome associated with SARS-CoV-2 infection. Nat. Med. 2020. [Google Scholar] [CrossRef]
- Nancy Fliesler COVID-19 and a Serious Inflammatory Syndrome in Children: Unpacking Recent Warnings. Available online: https://discoveries.childrenshospital.org/covid-19-inflammatory-syndrome-children/ (accessed on 8 October 2020).
- Jiang, L.; Tang, K.; Levin, M.; Irfan, O.; Morris, S.K.; Wilson, K.; Klein, J.D.; Bhutta, Z.A. COVID-19 and multisystem inflammatory syndrome in children and adolescents. Lancet Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Gupta, A.; Madhavan, M.V.; Sehgal, K.; Nair, N.; Mahajan, S.; Sehrawat, T.S.; Bikdeli, B.; Ahluwalia, N.; Ausiello, J.C.; Wan, E.Y.; et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020, 26, 1017–1032. [Google Scholar] [CrossRef]
- Case Series of Multisystem Inflammatory Syndrome in Adults Associated with SARS-CoV-2 Infection—United Kingdom and United States. March–August 2020. Available online: https://www.cdc.gov/mmwr/volumes/69/wr/mm6940e1.htm (accessed on 8 October 2020).
- Gbesemete, D.; Barker, M.; Lawrence, W.T.; Watson, D.; De Graaf, H.; Read, R.C. Exploring the acceptability of controlled human infection with SARSCoV2—A public consultation. BMC Med. 2020, 18, 209. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention COVID-19 Serology Surveillance Strategy. Available online: https://www.cdc.gov/coronavirus/2019-ncov/covid-data/serology-surveillance/index.html (accessed on 24 July 2020).
- Wang, L.; Shi, W.; Joyce, M.G.; Modjarrad, K.; Zhang, Y.; Leung, K.; Lees, C.R.; Zhou, T.; Yassine, H.M.; Kanekiyo, M.; et al. Evaluation of candidate vaccine approaches for MERS-CoV. Nat. Commun. 2015, 6, 7712. [Google Scholar] [CrossRef] [PubMed]
| Methodology Used for the Test | Main Findings | Number of Subjects | Categorization of Subjects | Sensitivity (%) | Specificity (%) | Positive Predictive Value | Negative Predictive Value | Reference |
|---|---|---|---|---|---|---|---|---|
| Lateral flow immunoassay (LFIA) | The IgM–IgG combined assay showed better sensitivity than a single IgM or IgG test. | 525 | 397 were PCR positive and remaining 128 were negative | 88.66 | 90.63 | NA | NA | [114] |
| Chemiluminescence assay | Showed higher efficiency for the detection of IgM and IgG anti-SARS CoV-2 antibodies | 125 | 61 were PCR positive patients and 64 were negative | 73.3 (IgM) 76.7 (IgG) | 92.2 100 | 81.5 NA | 88.1 90.1 | [122] |
| Chemiluminescence assay | IgG showed higher titre value than that of IgM | 76 | 43 were PCR positive and the remaining 33 were probable cases | 48.1 IgM 88.9 IgG | 100 IgM 90.9 IgG | NA | NA | [123] |
| Chemiluminescence assay | Automated CLIA analyzer | 176 | 125 PCR confirmed cases | 95–95.5 | 100 | 100 | NA | [124] |
| Chemiluminescence assay | Positive results after two weeks of symptom onset | 34 | All the subjects were PCR confirmed patients | 94.1 | NA | NA | NA | [89] |
| Chemiluminescence assay | Highest seroconversion was observed around 20–22 days after symptom onset | 285 | All the 285 were PCR positive patients | 94 (IgM) 100 (IgG) | NA | NA | NA | [125] |
| Chemiluminescence assay | Rapid | 476 | 276 PCR positive patients, and 200 negative volunteers | 57.2 (IgM) 71.4 (IgG) | NA | NA | NA | [126] |
| ELISA | IgM and IgG were reliably positive after one month of symptom onset | 24 | All were PCR positive patients | 74 | 100 | NA | NA | [127] |
| ELISA | All patients tested positive for IgG and some patients tested negative for IgM | 60 | Samples collected from convalescent patients | 78 IgM 100 IgG | NA | NA | NA | [128] |
| ELISA | The IgM reached 100% about one month of symptom onset | 173 | All the subjects were PCR positive patients | 100 (>15 days) | NA | NA | NA | [112] |
| ELISA | IgA, IgM, and IgG were detected from 5 days from the onset of symptoms | 140 | 82 PCR positive and 58 suspected cases | 75.6 (IgM in confirmed cases) 93.1 (IgM in probable cases) | NA | NA | NA | [120] |
| ELISA | IgG and IgA ELISAs in along with EUROLabworkstation (Euroimmu) | 39 PCR positive cases, 13 were IgG and IgA positive and 11 IgA only positive | NA | 91.9% (IgG)73.0% (IgA) | NA | NA | [129] | |
| IgM /IgG immunoassay | The rapid test | 110 | 30 were PCR positive cases, 50 were persons with respiratory symptoms and 30 were PCR negatives | 18.4 | 91.7 | 87.5 | 26.2 | [130] |
| Immunochromatography | The IgM-positive rate showed an elevation from 11.1% in early-stage to 74.2% in late-stage disease. The positive rate of IgG in patients was 3.6% in early-stage and 96.8% in late-stage disease | 105 | 105 patients | 68.6 | NA | NA | NA | [131] |
| Modified cytopathogenic assay | Patients with severe clinical manifestations showed higher antibody titre | 70 | The study comprises inpatients and Convalescent patients | 100 | NA | NA | NA | [132] |
| Immunofluorescence assay | Rapid and easy to use | 59 | 59 suspected patients; 24 PCR positive cases | 87.5 | NA | NA | NA | [133] |
| Rapid immunoassay | The accuracy was 40% in the first week and 93.9% after 2 | 179 | Patients included PCR positive (n = 90) and PCR negative (n = 89) | 85.6 | 91 | 95.1 | 82.7 | [134] |
| Rapid lateral flow assay | Fast, point of care test | 67 | 67 were PCR positive patients | 11 (<7 days) 92 (7–14 days) 96 (>14 days) | NA | NA | NA | [135] |
| Single Molecular Array (SIMOA) | Indicates disease severity | 81 | All were SARS-CoV-2 confirmed patients | 86 | 100 | NA | NA | [136] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Augustine, R.; Das, S.; Hasan, A.; S, A.; Abdul Salam, S.; Augustine, P.; Dalvi, Y.B.; Varghese, R.; Primavera, R.; Yassine, H.M.; et al. Rapid Antibody-Based COVID-19 Mass Surveillance: Relevance, Challenges, and Prospects in a Pandemic and Post-Pandemic World. J. Clin. Med. 2020, 9, 3372. https://doi.org/10.3390/jcm9103372
Augustine R, Das S, Hasan A, S A, Abdul Salam S, Augustine P, Dalvi YB, Varghese R, Primavera R, Yassine HM, et al. Rapid Antibody-Based COVID-19 Mass Surveillance: Relevance, Challenges, and Prospects in a Pandemic and Post-Pandemic World. Journal of Clinical Medicine. 2020; 9(10):3372. https://doi.org/10.3390/jcm9103372
Chicago/Turabian StyleAugustine, Robin, Suvarthi Das, Anwarul Hasan, Abhilash S, Shaheen Abdul Salam, Priya Augustine, Yogesh Bharat Dalvi, Ruby Varghese, Rosita Primavera, Hadi Mohamad Yassine, and et al. 2020. "Rapid Antibody-Based COVID-19 Mass Surveillance: Relevance, Challenges, and Prospects in a Pandemic and Post-Pandemic World" Journal of Clinical Medicine 9, no. 10: 3372. https://doi.org/10.3390/jcm9103372
APA StyleAugustine, R., Das, S., Hasan, A., S, A., Abdul Salam, S., Augustine, P., Dalvi, Y. B., Varghese, R., Primavera, R., Yassine, H. M., Thakor, A. S., & Kevadiya, B. D. (2020). Rapid Antibody-Based COVID-19 Mass Surveillance: Relevance, Challenges, and Prospects in a Pandemic and Post-Pandemic World. Journal of Clinical Medicine, 9(10), 3372. https://doi.org/10.3390/jcm9103372







