Deep Learning Based on Event-Related EEG Differentiates Children with ADHD from Healthy Controls
Abstract
1. Introduction
2. Experimental Section
2.1. Participants
2.2. Task
2.3. EEG Recording and Analysis
2.4. Deep Learning
3. Results and Discussion
3.1. Behavioural Analysis
3.2. Deep Learning Results
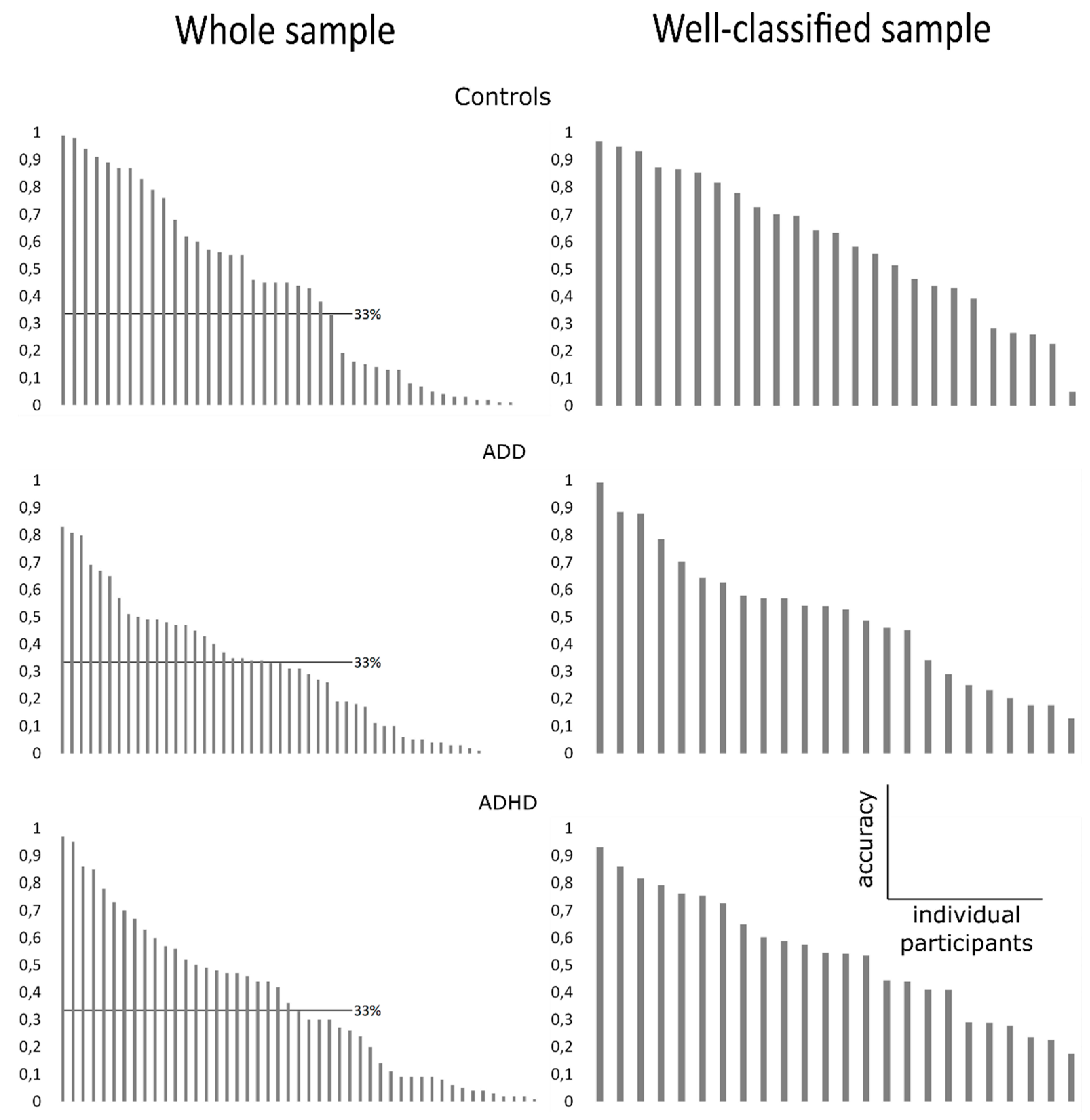
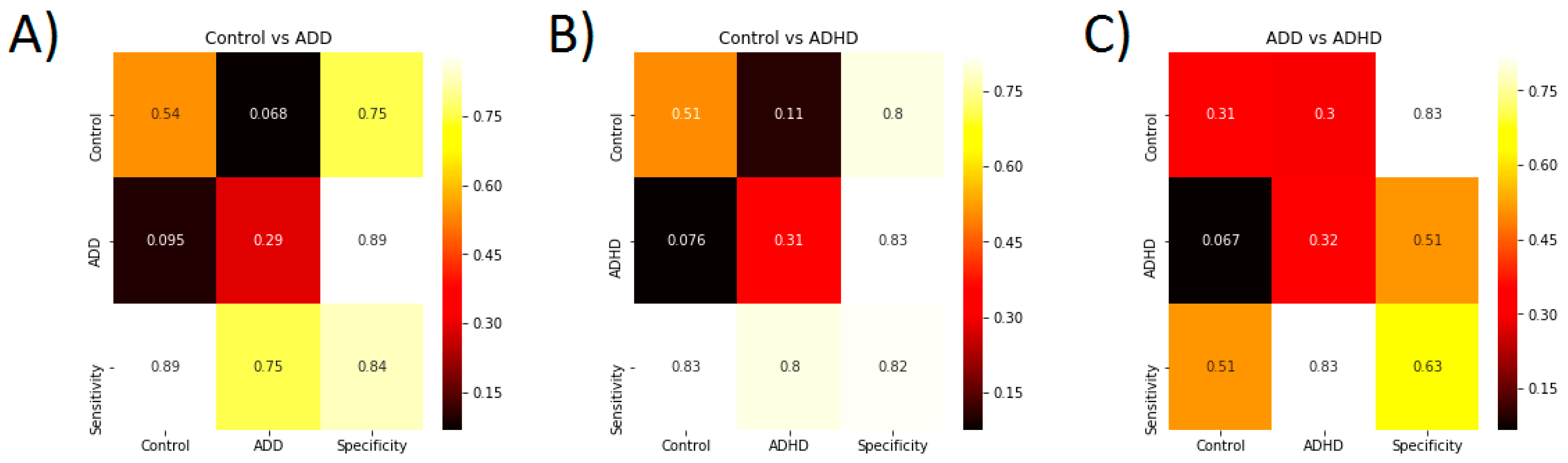
3.3. Classification Accuracy
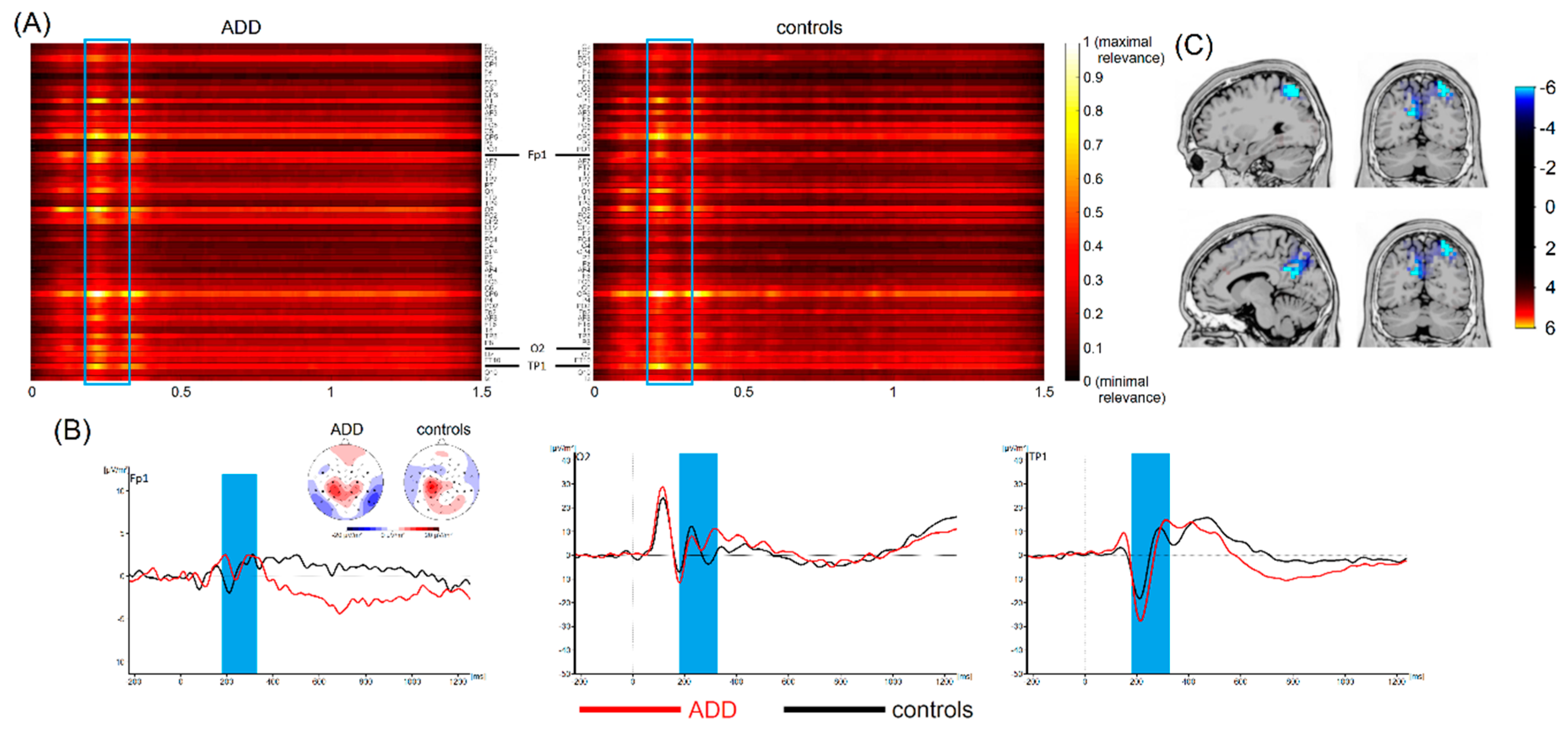
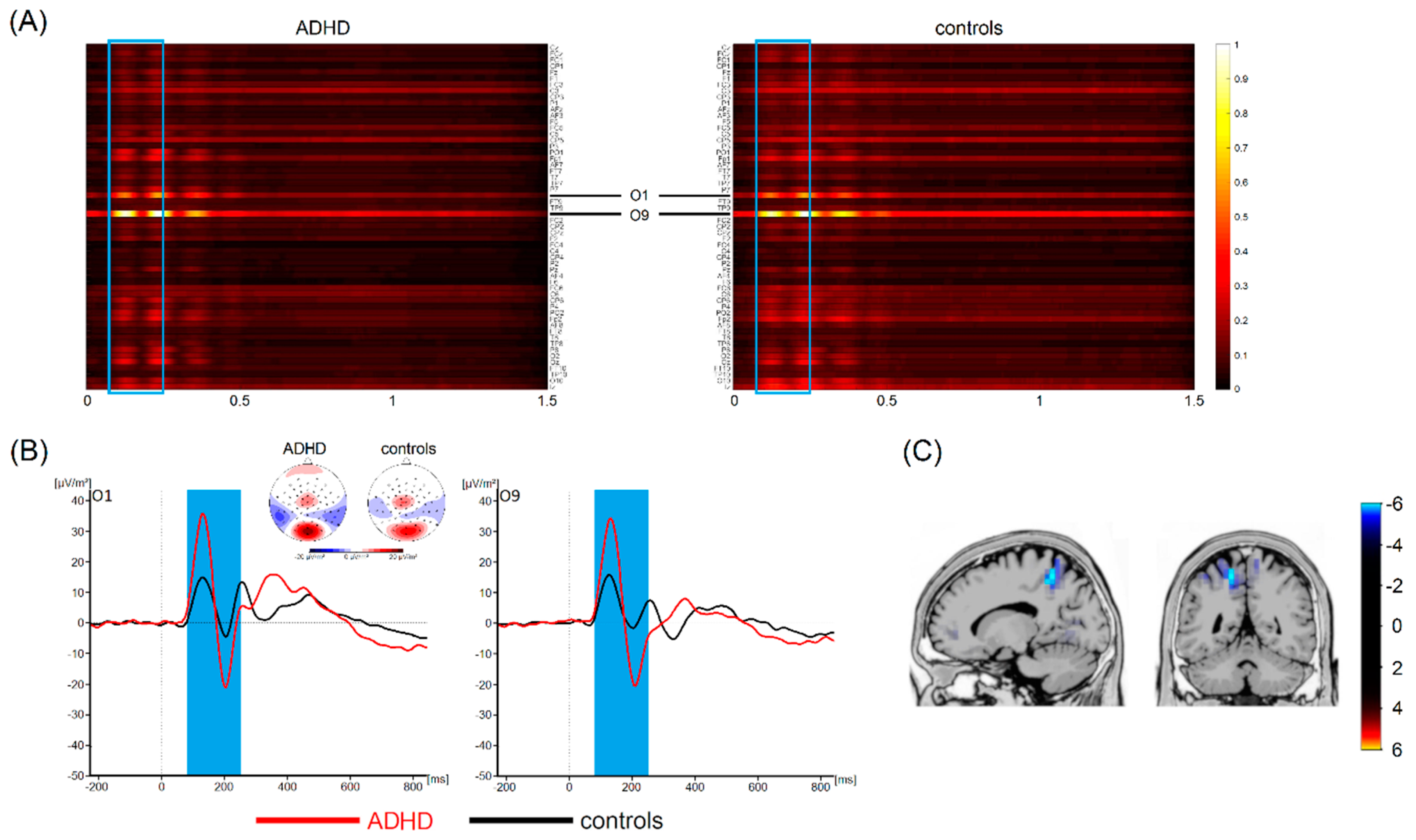
3.4. Most Important Neurophysiological Features
3.5. General Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kieling, R.; Rohde, L.A. ADHD in children and adults: diagnosis and prognosis. Curr. Top. Behav. Neurosci. 2012, 9, 1–16. [Google Scholar] [PubMed]
- Thomas, R.; Sanders, S.; Doust, J.; Beller, E.; Glasziou, P. Prevalence of attention-deficit/hyperactivity disorder: a systematic review and meta-analysis. Pediatrics 2015, 135, e994–e1001. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, N.; Mohammadi, M.R.; Araghi, S.M.; Zarafshan, H. Neurocognitive Profile of Children with Attention Deficit Hyperactivity Disorders (ADHD): A comparison between subtypes. Iran J. Psychiatry 2014, 9, 197–202. [Google Scholar] [PubMed]
- Randall, K.D.; Brocki, K.C.; Kerns, K.A. Cognitive control in children with ADHD-C: how efficient are they? Child Neuropsychol. 2009, 15, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, C.; González-Castro, P.; Cueli, M.; Areces, D.; González-Pienda, J.A. Attention Deficit/Hyperactivity Disorder (ADHD) Diagnosis: An Activation-Executive Model. Front. Psychol. 2016, 07. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Weibman, D.; Halperin, J.M.; Li, X. A Review of Heterogeneity in Attention Deficit/Hyperactivity Disorder (ADHD). Front. Hum. Neurosci. 2019, 13, 42. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, S.; Pedersen, M.L.; Mowinckel, A.M.; Biele, G. Modelling ADHD: A review of ADHD theories through their predictions for computational models of decision-making and reinforcement learning. Neurosci. Biobehav. Rev. 2016, 71, 633–656. [Google Scholar] [CrossRef] [PubMed]
- Barth, B.; Mayer-Carius, K.; Strehl, U.; Kelava, A.; Häußinger, F.B.; Fallgatter, A.J.; Ehlis, A.-C. Identification of neurophysiological biotypes in attention deficit hyperactivity disorder. Psychiatry Clin. Neurosci. 2018, 72, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Bluschke, A.; von der Hagen, M.; Papenhagen, K.; Roessner, V.; Beste, C. Conflict processing in juvenile patients with neurofibromatosis type 1 (NF1) and healthy controls - Two pathways to success. Neuroimage Clin. 2017, 14, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.C.; Ouyang, C.-S.; Chiang, C.-T.; Yang, R.-C.; Wu, R.-C.; Wu, H.-C.; Lin, L.-C. Novel method using Hjorth mobility analysis for diagnosing attention-deficit hyperactivity disorder in girls. Brain Dev. 2018. [Google Scholar] [CrossRef] [PubMed]
- Khoshnoud, S.; Nazari, M.A.; Shamsi, M. Functional brain dynamic analysis of ADHD and control children using nonlinear dynamical features of EEG signals. J. Integr. Neurosci. 2018, 17, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Lenartowicz, A.; Loo, S.K. Use of EEG to diagnose ADHD. Curr. Psychiatry Rep. 2014, 16, 498. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, C.; Bhat, S.; Acharya, U.R.; Adeli, H.; Bairy, G.M. Diagnosis of attention deficit hyperactivity disorder using imaging and signal processing techniques. Comput. Biol. Med. 2017, 88, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Uddin, L.Q.; Dajani, D.R.; Voorhies, W.; Bednarz, H.; Kana, R.K. Progress and roadblocks in the search for brain-based biomarkers of autism and attention-deficit/hyperactivity disorder. Transl. Psychiatry 2017, 7, e1218. [Google Scholar] [CrossRef] [PubMed]
- Wolfers, T.; Buitelaar, J.K.; Beckmann, C.F.; Franke, B.; Marquand, A.F. From estimating activation locality to predicting disorder: A review of pattern recognition for neuroimaging-based psychiatric diagnostics. Neurosci. Biobehav. Rev. 2015, 57, 328–349. [Google Scholar] [CrossRef] [PubMed]
- Bridwell, D.A.; Cavanagh, J.F.; Collins, A.G.E.; Nunez, M.D.; Srinivasan, R.; Stober, S.; Calhoun, V.D. Moving Beyond ERP Components: A Selective Review of Approaches to Integrate EEG and Behavior. Front. Hum. Neurosci. 2018, 12, 106. [Google Scholar] [CrossRef]
- Faust, O.; Hagiwara, Y.; Hong, T.J.; Lih, O.S.; Acharya, U.R. Deep learning for healthcare applications based on physiological signals: A review. Comput. Methods Programs Biomed. 2018, 161, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Miotto, R.; Wang, F.; Wang, S.; Jiang, X.; Dudley, J.T. Deep learning for healthcare: review, opportunities and challenges. Brief. Bioinform. 2018, 19, 1236–1246. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Bashivan, P.; Rish, I.; Yeasin, M.; Codella, N. Learning Representations from EEG with Deep Recurrent-Convolutional Neural Networks. Available online: https://arxiv.org/abs/1511.06448 (accessed on 8 February 2019).
- Schirrmeister, R.T.; Springenberg, J.T.; Fiederer, L.D.J.; Glasstetter, M.; Eggensperger, K.; Tangermann, M.; Hutter, F.; Burgard, W.; Ball, T. Deep learning with convolutional neural networks for EEG decoding and visualization. Hum. Brain Mapp. 2017, 38, 5391–5420. [Google Scholar] [CrossRef]
- Stober, S.; Sternin, A.; Owen, A.M.; Grahn, J.A. Deep Feature Learning for EEG Recordings. Available online: https://arxiv.org/abs/1511.04306 (accessed on 8 February 2019).
- Lawhern, V.J.; Solon, A.J.; Waytowich, N.R.; Gordon, S.M.; Hung, C.P.; Lance, B.J. EEGNet: A Compact Convolutional Network for EEG-based Brain-Computer Interfaces. J. Neural Eng. 2018, 15, 056013. [Google Scholar] [CrossRef] [PubMed]
- Coull, J.T.; Cheng, R.-K.; Meck, W.H. Neuroanatomical and neurochemical substrates of timing. Neuropsychopharmacology 2011, 36, 3–25. [Google Scholar] [CrossRef] [PubMed]
- Merchant, H.; de Lafuente, V. Introduction to the neurobiology of interval timing. Adv. Exp. Med. Biol. 2014, 829, 1–13. [Google Scholar] [PubMed]
- Petter, E.A.; Lusk, N.A.; Hesslow, G.; Meck, W.H. Interactive roles of the cerebellum and striatum in sub-second and supra-second timing: Support for an initiation, continuation, adjustment, and termination (ICAT) model of temporal processing. Neurosci. Biobehav. Rev. 2016, 71, 739–755. [Google Scholar] [CrossRef] [PubMed]
- Doehnert, M.; Brandeis, D.; Schneider, G.; Drechsler, R.; Steinhausen, H.-C. A neurophysiological marker of impaired preparation in an 11-year follow-up study of attention-deficit/hyperactivity disorder (ADHD). J. Child Psychol. Psychiatry 2013, 54, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.-L.; Gau, S.S.-F.; Hsu, W.-Y.; Wu, Y.-Y. Deficits in interval timing measured by the dual-task paradigm among children and adolescents with attention-deficit/hyperactivity disorder. J. Child. Psychol. Psychiatry 2010, 51, 223–232. [Google Scholar] [CrossRef]
- Pretus, C.; Picado, M.; Ramos-Quiroga, A.; Carmona, S.; Richarte, V.; Fauquet, J.; Vilarroya, Ó. Presence of Distractor Improves Time Estimation Performance in an Adult ADHD Sample. J. Atten. Disord. 2016. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.; Cubillo, A.; Barrett, N.; Giampietro, V.; Simmons, A.; Brammer, M.; Rubia, K. Neurofunctional effects of methylphenidate and atomoxetine in boys with attention-deficit/hyperactivity disorder during time discrimination. Biol. Psychiatry 2013, 74, 615–622. [Google Scholar] [CrossRef]
- Smith, A.; Taylor, E.; Rogers, J.W.; Newman, S.; Rubia, K. Evidence for a pure time perception deficit in children with ADHD. J. Child. Psychol. Psychiatry 2002, 43, 529–542. [Google Scholar] [CrossRef]
- Walg, M.; Oepen, J.; Prior, H. Adjustment of Time Perception in the Range of Seconds and Milliseconds: The Nature of Time-Processing Alterations in Children With ADHD. J. Atten. Disord. 2015, 19, 755–763. [Google Scholar] [CrossRef]
- Wilson, T.W.; Heinrichs-Graham, E.; White, M.L.; Knott, N.L.; Wetzel, M.W. Estimating the passage of minutes: deviant oscillatory frontal activity in medicated and unmedicated ADHD. Neuropsychology 2013, 27, 654–665. [Google Scholar] [CrossRef]
- Walg, M.; Hapfelmeier, G.; El-Wahsch, D.; Prior, H. The faster internal clock in ADHD is related to lower processing speed: WISC-IV profile analyses and time estimation tasks facilitate the distinction between real ADHD and pseudo-ADHD. Eur. Child. Adolesc. Psychiatry 2017, 26, 1177–1186. [Google Scholar] [CrossRef]
- Bluschke, A.; Schuster, J.; Roessner, V.; Beste, C. Neurophysiological mechanisms of interval timing dissociate inattentive and combined ADHD subtypes. Sci. Rep. 2018, 8, 2033. [Google Scholar] [CrossRef]
- Döpfner, M.; Görtz-Dorten, A.; Lehmkuhl, G. Diagnostik-System für Psychische Störungen im Kindes- und Jugendalter nach ICD-10 und DSM-IV, DISYPS-II; Huber: Bern, Switzerland, 2008. [Google Scholar]
- Beste, C.; Saft, C.; Andrich, J.; Müller, T.; Gold, R.; Falkenstein, M. Time processing in Huntington’s disease: a group-control study. PLoS ONE 2007, 2, e1263. [Google Scholar] [CrossRef]
- Wild-Wall, N.; Willemssen, R.; Falkenstein, M.; Beste, C. Time estimation in healthy ageing and neurodegenerative basal ganglia disorders. Neurosci. Lett. 2008, 442, 34–38. [Google Scholar] [CrossRef]
- Nunez, P.L.; Pilgreen, K.L. The spline-Laplacian in clinical neurophysiology: a method to improve EEG spatial resolution. J. Clin. Neurophysiol. 1991, 8, 397–413. [Google Scholar] [CrossRef]
- Marco-Pallarés, J.; Grau, C.; Ruffini, G. Combined ICA-LORETA analysis of mismatch negativity. Neuroimage 2005, 25, 471–477. [Google Scholar] [CrossRef]
- Pascual-Marqui, R.D. Standardized low-resolution brain electromagnetic tomography (sLORETA): technical details. Methods Find. Exp. Clin. Pharmacol. 2002, 24, 5–12. [Google Scholar]
- Sekihara, K.; Sahani, M.; Nagarajan, S.S. Localization bias and spatial resolution of adaptive and non-adaptive spatial filters for MEG source reconstruction. Neuroimage 2005, 25, 1056–1067. [Google Scholar] [CrossRef]
- Dippel, G.; Beste, C. A causal role of the right inferior frontal cortex in implementing strategies for multi-component behaviour. Nat. Commun. 2015, 6, 6587. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. Available online: https://arxiv.org/abs/1412.6980 (accessed on 8 February 2019).
- Varoquaux, G.; Raamana, P.R.; Engemann, D.A.; Hoyos-Idrobo, A.; Schwartz, Y.; Thirion, B. Assessing and tuning brain decoders: Cross-validation, caveats, and guidelines. NeuroImage 2017, 145, 166–179. [Google Scholar] [CrossRef]
- Combrisson, E.; Jerbi, K. Exceeding chance level by chance: The caveat of theoretical chance levels in brain signal classification and statistical assessment of decoding accuracy. J. Neurosci. Methods 2015, 250, 126–136. [Google Scholar] [CrossRef]
- Beste, C.; Baune, B.T.; Falkenstein, M.; Konrad, C. Variations in the TNF-α gene (TNF-α -308G→A) affect attention and action selection mechanisms in a dissociated fashion. J. Neurophysiol. 2010, 104, 2523–2531. [Google Scholar] [CrossRef]
- Herrmann, C.S.; Knight, R.T. Mechanisms of human attention: event-related potentials and oscillations. Neurosci. Biobehav. Rev. 2001, 25, 465–476. [Google Scholar] [CrossRef]
- Luck, S.J.; Kappenman, E.S. The Oxford Handbook of Event-Related Potential Components; Oxford University Press: Oxford, New York, NY, USA, 2013; ISBN 978-0-19-932804-8. [Google Scholar]
- Schneider, D.; Beste, C.; Wascher, E. On the time course of bottom-up and top-down processes in selective visual attention: an EEG study. Psychophysiology 2012, 49, 1492–1503. [Google Scholar] [CrossRef]
- Gohil, K.; Bluschke, A.; Roessner, V.; Stock, A.-K.; Beste, C. ADHD patients fail to maintain task goals in face of subliminally and consciously induced cognitive conflicts. Psychol. Med. 2017, 47, 1771–1783. [Google Scholar] [CrossRef]
- Arnsten, A.F.T.; Rubia, K. Neurobiological circuits regulating attention, cognitive control, motivation, and emotion: disruptions in neurodevelopmental psychiatric disorders. J. Am. Acad. Child. Adolesc. Psychiatry 2012, 51, 356–367. [Google Scholar] [CrossRef]
- Kasparek, T.; Theiner, P.; Filova, A. Neurobiology of ADHD From Childhood to Adulthood: Findings of Imaging Methods. J. Atten. Disord. 2015, 19, 931–943. [Google Scholar] [CrossRef]
- Ptak, R. The frontoparietal attention network of the human brain: action, saliency, and a priority map of the environment. Neuroscientist 2012, 18, 502–515. [Google Scholar] [CrossRef]
- Kompatsiari, K.; Candrian, G.; Mueller, A. Test-retest reliability of ERP components: A short-term replication of a visual Go/NoGo task in ADHD subjects. Neurosci. Lett. 2016, 617, 166–172. [Google Scholar] [CrossRef]
- Bluschke, A.; Gohil, K.; Petzold, M.; Roessner, V.; Beste, C. Neural mechanisms underlying successful and deficient multi-component behavior in early adolescent ADHD. Neuroimage Clin. 2018, 18, 533–542. [Google Scholar] [CrossRef]
- Bluschke, A.; Broschwitz, F.; Kohl, S.; Roessner, V.; Beste, C. The neuronal mechanisms underlying improvement of impulsivity in ADHD by theta/beta neurofeedback. Sci Rep. 2016, 6, 31178. [Google Scholar] [CrossRef]
- Hasler, R.; Perroud, N.; Meziane, H.B.; Herrmann, F.; Prada, P.; Giannakopoulos, P.; Deiber, M.-P. Attention-related EEG markers in adult ADHD. Neuropsychologia 2016, 87, 120–133. [Google Scholar] [CrossRef]
- Karayanidis, F.; Robaey, P.; Bourassa, M.; De Koning, D.; Geoffroy, G.; Pelletier, G. ERP differences in visual attention processing between attention-deficit hyperactivity disorder and control boys in the absence of performance differences. Psychophysiology 2000, 37, 319–333. [Google Scholar] [CrossRef]
- Strandburg, R.J.; Marsh, J.T.; Brown, W.S.; Asarnow, R.F.; Higa, J.; Harper, R.; Guthrie, D. Continuous-processing--related event-related potentials in children with attention deficit hyperactivity disorder. Biol. Psychiatry 1996, 40, 964–980. [Google Scholar] [CrossRef]
- Iannaccone, R.; Hauser, T.U.; Ball, J.; Brandeis, D.; Walitza, S.; Brem, S. Classifying adolescent attention-deficit/hyperactivity disorder (ADHD) based on functional and structural imaging. Eur. Child Adolesc. Psychiatry 2015, 24, 1279–1289. [Google Scholar] [CrossRef]
- Willcutt, E.G.; Nigg, J.T.; Pennington, B.F.; Solanto, M.V.; Rohde, L.A.; Tannock, R.; Loo, S.K.; Carlson, C.L.; McBurnett, K.; Lahey, B.B. Validity of DSM-IV attention deficit/hyperactivity disorder symptom dimensions and subtypes. J. Abnorm. Psychol. 2012, 121, 991–1010. [Google Scholar] [CrossRef]
- Saad, J.F.; Griffiths, K.R.; Kohn, M.R.; Clarke, S.; Williams, L.M.; Korgaonkar, M.S. Regional brain network organization distinguishes the combined and inattentive subtypes of Attention Deficit Hyperactivity Disorder. NeuroImage: Clinical 2017, 15, 383–390. [Google Scholar] [CrossRef]
| Layer | Layer Type | Filters | Size | Parameters | Output Dimension | Activation | Mode |
|---|---|---|---|---|---|---|---|
| 1 | Input | (C, T) | |||||
| Reshape | (1, C, T) | ||||||
| Conv2D | F | (1,64) | 64*F | (F, C, T) | Linear | same | |
| BatchNorm | 2*F | (F, 1, T) | |||||
| DepthwiseConv2D | F | (C,1) | C*F | (F, 1, T) | Linear | valid | |
| BatchNorm | (F, 1, T) | ||||||
| Activation | (F, 1, T) | ELU | |||||
| SpatialDropout2D | (F, 1, T) | ||||||
| 2 | SeparableConv2D | F | (1,8) | (F, 1, T) | Linear | same | |
| BatchNorm | 2*F | (F, 1, T) | |||||
| Activation | (F, 1, T) | ELU | |||||
| AveragePool2D | (1,4) | (F, 1, T//4) | |||||
| SpatialDropout2D | (F, 1, T//4) | ||||||
| 3 | SeparableConv2D | 2*F | (1,8) | (2*F, 1, T//4) | Linear | same | |
| BatchNorm | 2*F | (2*F, 1, T//4) | |||||
| Activation | (2*F, 1, T//4) | ELU | |||||
| AveragePool2D | (1,4) | (2*F, 1, T//16) | |||||
| SpatialDropout2D | (2*F, 1, T//16) | ||||||
| 4 | Flatten | (2*F, 1, T//16) | |||||
| Dense | N | Softmax |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vahid, A.; Bluschke, A.; Roessner, V.; Stober, S.; Beste, C. Deep Learning Based on Event-Related EEG Differentiates Children with ADHD from Healthy Controls. J. Clin. Med. 2019, 8, 1055. https://doi.org/10.3390/jcm8071055
Vahid A, Bluschke A, Roessner V, Stober S, Beste C. Deep Learning Based on Event-Related EEG Differentiates Children with ADHD from Healthy Controls. Journal of Clinical Medicine. 2019; 8(7):1055. https://doi.org/10.3390/jcm8071055
Chicago/Turabian StyleVahid, Amirali, Annet Bluschke, Veit Roessner, Sebastian Stober, and Christian Beste. 2019. "Deep Learning Based on Event-Related EEG Differentiates Children with ADHD from Healthy Controls" Journal of Clinical Medicine 8, no. 7: 1055. https://doi.org/10.3390/jcm8071055
APA StyleVahid, A., Bluschke, A., Roessner, V., Stober, S., & Beste, C. (2019). Deep Learning Based on Event-Related EEG Differentiates Children with ADHD from Healthy Controls. Journal of Clinical Medicine, 8(7), 1055. https://doi.org/10.3390/jcm8071055






