Next Generation Networks: Featuring the Potential Role of Emerging Applications in Translational Oncology
Abstract
1. Introduction
1.1. Background
1.2. Networks
1.3. Cancer Networks Taxonomy
2. Methodological Themes
2.1. On Entropy
2.2. Metastasis and Its Stochastic Dynamics
2.3. Therapy and Phase Transitions
2.4. On Symmetry
- (1)
- Do symmetric patterns induce localized network dynamics, i.e., those represented by modules, clusters, motifs? If this is the case, how specific these structures are?
- (2)
- Are the possibly observed patterns robust or fragile with regard to symmetry breaking? The latter would imply the presence of fluctuations leading the system to a critical point, and thus a likely change of state that in cancer might be a signature of metastasis.
- (1)
- How the influences exerted by symmetry patterns depend on cancer heterogeneity?
- (2)
- Are such patterns controllable?
2.5. On Controllability
- (i)
- The presence of interactions involved in core cell-cycle and DNA-damage repair pathways that are significantly rewired in tumors, indicating a significant impact on key genome-stabilizing mechanisms;
- (ii)
- Several flipped genes that are serine/threonine kinases which act as biological switches, reflecting cellular switching mechanisms between stages;
- (iii)
- Different sets of genes flipped during the initial and final stages, indicating a progressive pattern.
3. Results
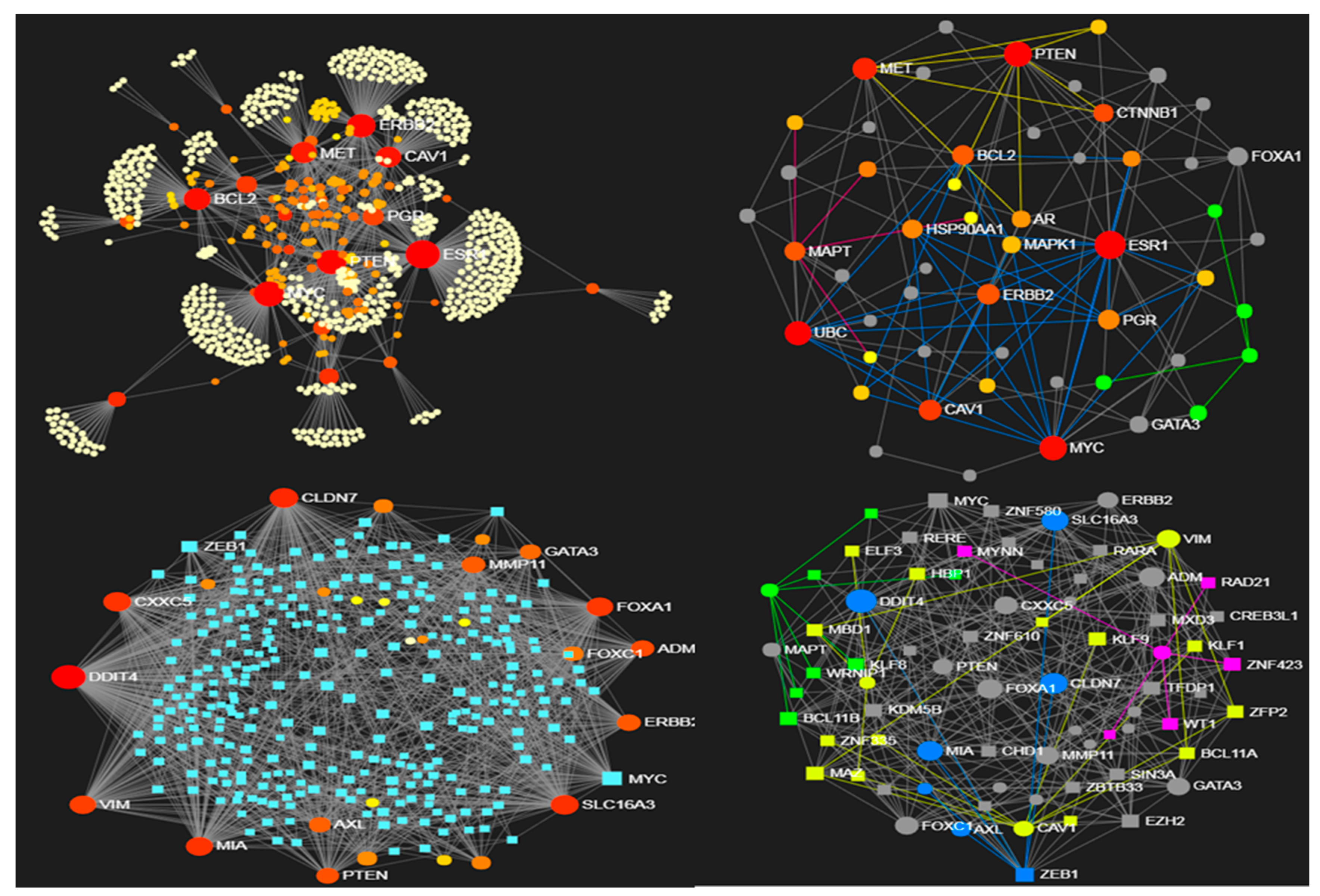
3.1. Use Case 1: Entropic Patterns in Metastatic Breast Cancer
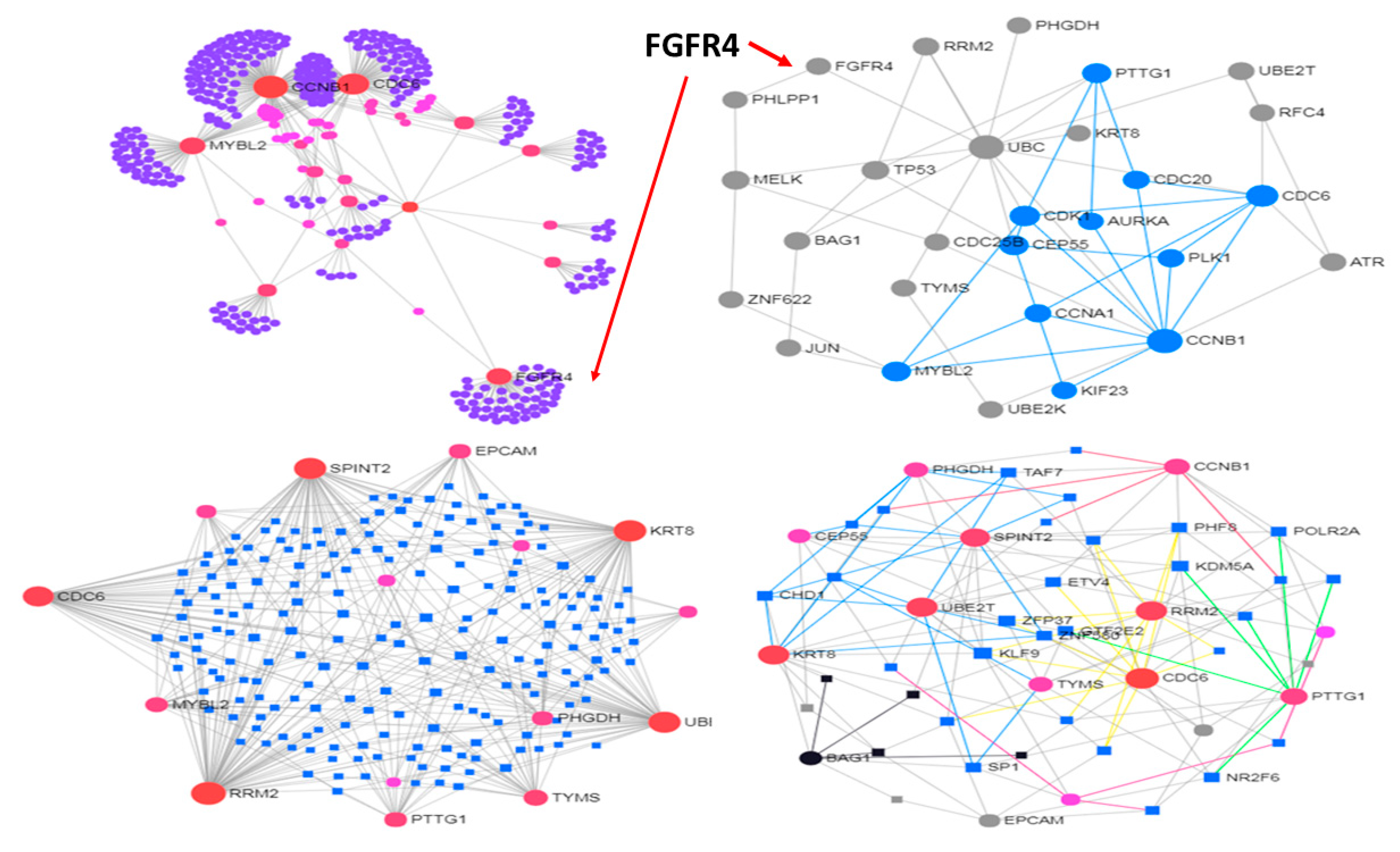
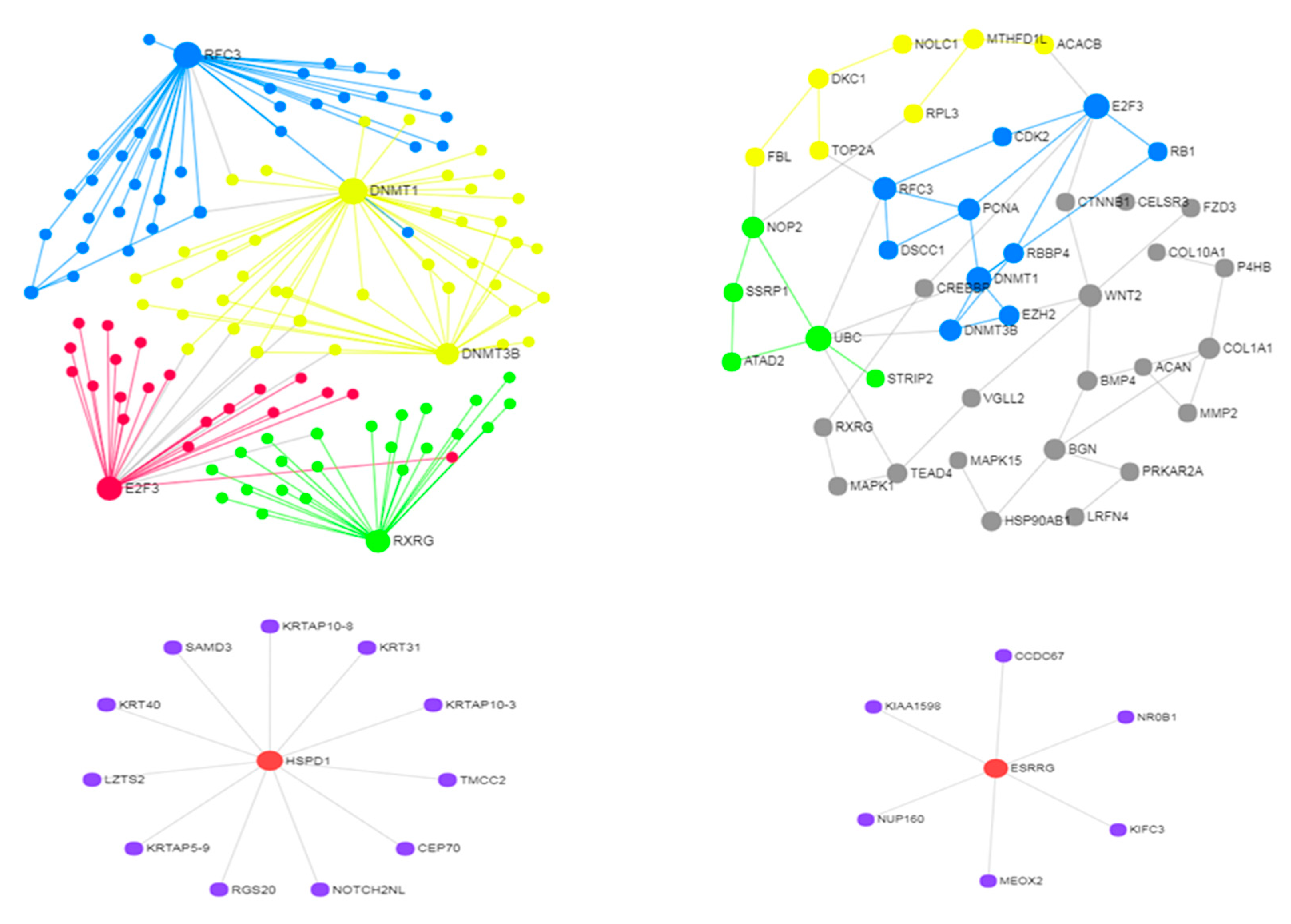
3.2. Use Case 2—Controllability in Metastatic Stomach Cancer
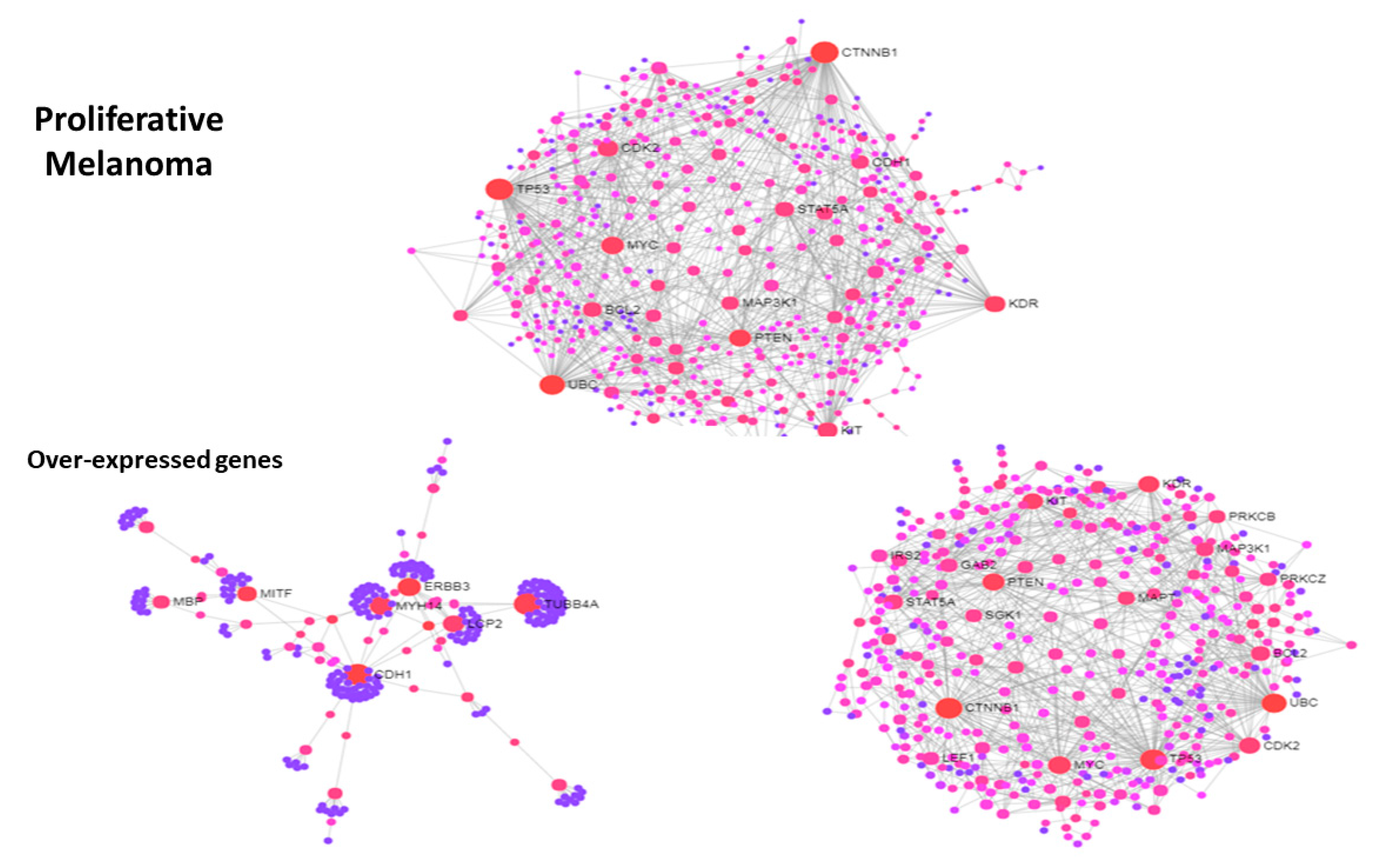
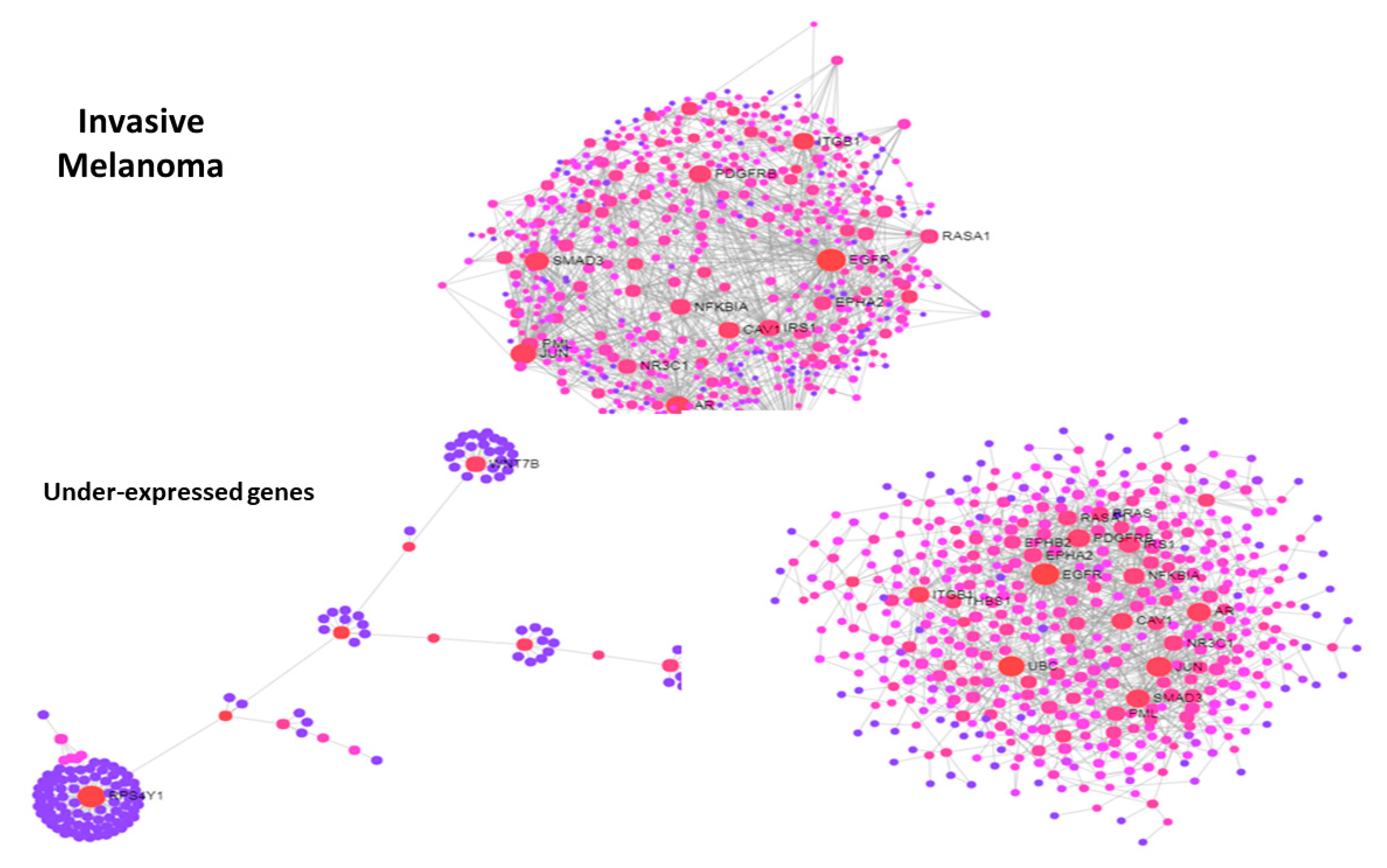
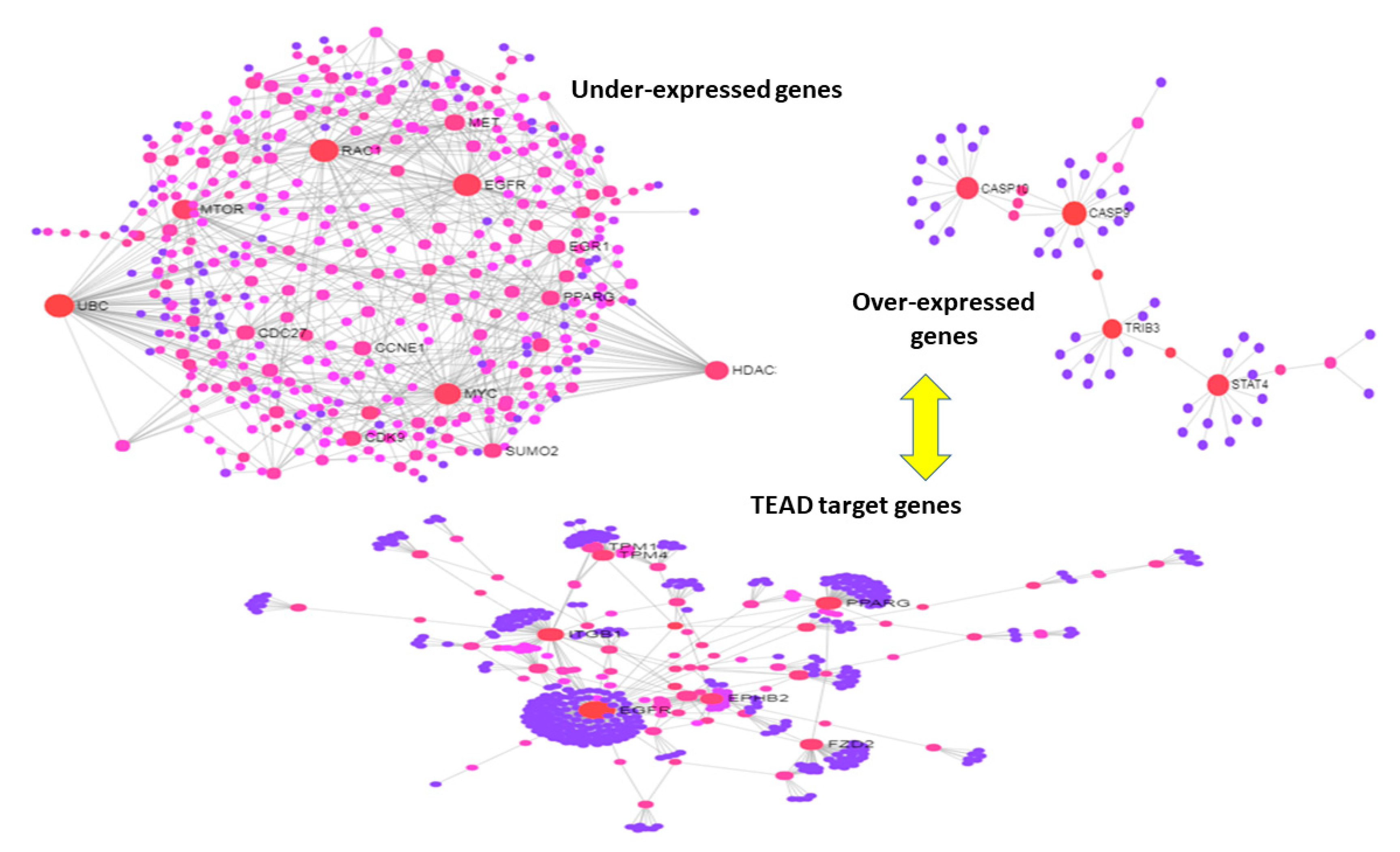
3.3. Use Case 3—Symmetry and Synchronization in Melanoma State Transitions
4. Discussion
Acknowledgments
Conflicts of Interest
Materials and Methods
References
- Lavi, O. Redundancy: A critical obstacle to improving cancer therapy. Cancer Res. 2015, 75, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Pinaire, J.; Azé, J.; Bringay, S.; Landais, P. Patient healthcare trajectory. An essential monitoring tool: A systematic review. Health Inform. Sci. Syst. 2017, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Giannoula, A.; Gutierrez-Sacristán, A.; Bravo, A.; Sanz, F.; Furlong, L. Identifying temporal patterns in patient disease trajectories using dynamic time warping: A population-based study. Sci. Rep. 2018, 8, 4216. [Google Scholar] [CrossRef] [PubMed]
- Cantini, L.; Calzone, L.; Martignetti, L.; Rydenfelt, M.; Bluthgen, N.; Barillot, E.; Zinovyev, A. Classification of gene signatures for their information value and functional redundancy. NPJ Syst. Biol. Appl. 2018, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Cereda, M.; Mourikis, T.P.; Ciccarelli, F.D. Genetic Redundancy, Functional Compensation, and Cancer Vulnerability. Trends Cancer 2016, 2, 160–162. [Google Scholar] [CrossRef]
- Kitano, H. Biological robustness. Nature Rev. Genet. 2004, 5, 826–837. [Google Scholar] [CrossRef]
- Altrock, P.M.; Liu, L.L.; Michor, F. The mathematics of cancer: Integrating quantitative models. Nat. Rev. Cancer 2015, 15, 730–745. [Google Scholar] [CrossRef]
- Barabási, A.L.; Guilbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Loscalzo, J.; Barabasi, A.L.; Silverman, E.K. Network Medicine. Complex Systems in Human Disease and Therapeutics, 1st ed.; Harvard Univ Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Schadt, E.E.; Bjorkegren, J.L. NEW: Network-enabled wisdom in biology, medicine, and health care. Sci. Transl. Med. 2012, 4, 115rv1. [Google Scholar] [CrossRef] [PubMed]
- Fiscon, G.; Conte, F.; Farina, L.; Paci, P. Network-based approaches to explore complex biological systems towards network medicine. Genes 2018, 9, 437. [Google Scholar] [CrossRef] [PubMed]
- Ghiassian, S.D.; Menche, J.; Barabási, A.L. A DIseAse MOdule Detection (DIAMOnD) algorithm derived from a systematic analysis of connectivity patterns of disease proteins in the human interactome. PLoS Comput. Biol. 2015, 11, e1004120. [Google Scholar] [CrossRef] [PubMed]
- Mora, A.; Taranta, M.; Zaki, N.; Cinti, C.; Capobianco, E. Epigenetically driven network cooperativity: meta-analysis in multi-drug resistant osteosarcoma. J. Complex Netw. 2016, 4, 296–317. [Google Scholar] [CrossRef]
- Zhang, W.; Chien, J.; Yong, J.; Kuang, R. Network-based machine learning and graph theory algorithms for precision oncology. NPJ Precision Oncol. 2017, 1, 25. [Google Scholar] [CrossRef] [PubMed]
- Paci, P.; Colombo, T.; Fiscon, G.; Gurtner, A.; Pavesi, G.; Farina, L. SWIM: A computational tool to unveiling nodes in complex biological networks. Sci. Rep. 2017, 7, 44797. [Google Scholar] [CrossRef] [PubMed]
- Deming, D.A.; Leystra, A.A.; Nettekoven, L.; Sievers, C.; Miller, D.; Middlebrooks, M.; Clipson, L.; Albrecht, D.; Bacher, J.; Washington, M.K.; et al. PIK3CA and APC mutations are synergistic in the development of intestinal cancers. Oncogene 2014, 33, 2245–2254. [Google Scholar] [CrossRef]
- Gerlee, P.; Altrock, P.M. Complexity and stability in growing cancer cell populations. Proc. Natl. Acad. Sci. USA 2015, 112, E2742–E2743. [Google Scholar] [CrossRef]
- Chowdhury, S.A.; Nibbe, R.K.; Chance, M.R.; Koyutürk, M. Subnetwork state functions define dysregulated subnetworks in cancer. J. Comput. Biol. 2011, 18, 263–281. [Google Scholar] [CrossRef]
- Chowdhury, S.A.; Koyutürk, M. Identification of coordinately dysregulated subnetworks in complex phenotypes. Pac. Symp. Biocomput. 2010, 2010, 133–144. [Google Scholar]
- Zitnik, M.; Leskovec, J. Predicting multicellular function through multi-layer tissue networks. Bioinformatics 2017, 33, i190–i198. [Google Scholar] [CrossRef]
- Yu, L.; Yao, S.; Gao, L.; Zha, Y. Conserved Disease Modules Extracted From Multilayer Heterogeneous Disease and Gene Networks for Understanding Disease Mechanisms and Predicting Disease Treatments. Front. Genet. 2018, 9, 745. [Google Scholar] [CrossRef]
- Li, D.; Zhu, Z.; Pan, Z.; Hu, G.; He, S. Extracting active modules from multilayer PPI network: A continuous optimization approach. BioRxiv 2017, 211433. [Google Scholar] [CrossRef]
- Tarabichi, M.; Antoniou, A.; Saiselet, M.; Pita, J.M.; Andry, G.; Dumont, J.E.; Detours, V.; Maenhaut, C. Systems biology of cancer: entropy, disorder, and selection-driven evolution to independence, invasion and “swarm intelligence”. Cancer Metast. Rev. 2013, 32, 403–421. [Google Scholar] [CrossRef] [PubMed]
- Hanselmann, R.G.; Welter, C. Origin of Cancer: An Information, Energy, and Matter Disease. Front. Cell Develop. Boil. 2016, 4, 121. [Google Scholar] [CrossRef] [PubMed]
- Galas, D.J.; Nykter, M.; Carter, G.W.; Price, N.D.; Shmulevich, I. Biological information as set-based complexity. IEEE Trans. Inform. Theory 2010, 56, 667–677. [Google Scholar] [CrossRef]
- Galas, D.J.; Sakhanenko, N.A.; Skupin, A.; Ignac, T. Describing the Complexity of Systems: Multivariable Set Complexity and the Information Basis of Systems Biology. J. Comput. Biol. 2014, 21, 118–140. [Google Scholar] [CrossRef]
- Ignac, T.M.; Sakhanenko, N.A.; Galas, D.J. Relations between the set-complexity and the structure of graphs and their sub-graphs. EURASIP J. Bioinform. Syst. Biol. 2012, 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Krawitz, P.; Shmulevich, H. Basin entropy in boleean network ensembles. Phys. Rev. Lett. 2007, 98, 158701. [Google Scholar] [CrossRef]
- Daza, A.; Wagemakers, A.; Georgeot, B.; Guéry-Odelin, D.; Sanjuán, M.A. Basin entropy: A new tool to analyze uncertainty in dynamical systems. Sci. Rep. 2016, 6, 31416. [Google Scholar] [CrossRef] [PubMed]
- Ahnert, S.E.; Fink, T.M.A. Form and function in gene regulatory networks: The structure of network motifs determines fundamental properties of their dynamical state space. J. R. Soc. Interface 2016, 13, 20160179. [Google Scholar] [CrossRef]
- Sevick, E.M.; Prabhakar, R.; Williams, S.R.; Searles, D.J. Fluctuation Theorems. Annual. Rev. Phys. Chem. 2008, 59, 603–633. [Google Scholar] [CrossRef] [PubMed]
- Teschendorff, A.E.; Severini, S. Increased entropy of signal transduction in the cancer metastasis phenotype. BMC Syst. Biol. 2010, 4, 104. [Google Scholar] [CrossRef] [PubMed]
- West, J.; Bianconi, G.; Severini, S.; Teschendorff, A.E. Differential network entropy reveals cancer system hallmarks. Sci. Rep. 2012, 2, 802. [Google Scholar] [CrossRef]
- Robinson, D.R.; Wu, Y.M.; Lonigro, R.J.; Vats, P.; Cobain, E.; Everett, J.; Cao, X.; Rabban, E.; Kumar-Sinha, C.; Raymond, V.; et al. Integrative clinical genomics of metastatic cancer. Nature 2017, 548, 297–303. [Google Scholar] [CrossRef]
- Pazarentzos, E.; Bivona, T.G. Adaptive stress signaling in targeted cancer therapy resistance. Oncogene 2015, 34, 5599–5606. [Google Scholar] [CrossRef] [PubMed]
- AlQuraishi, M.; Koytiger, G.; Jenney, A.; MacBeath, G.; Sorger, P.K. A multiscale statistical mechanical framework integrates biophysical and genomic data to assemble cancer networks. Nat. Genet. 2014, 46, 1363–1371. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gupta, P.B.; Fillmore, C.M.; Jiang, G.; Shapira, S.D.; Tao, K.; Kuperwasser, C.; Lander, E.S. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 2011, 146, 633–644. [Google Scholar] [CrossRef]
- Buder, T.; Deutsch, A.; Seifert, M.; Voss-Böhme, A. CellTrans: An R Package to Quantify Stochastic Cell State Transitions. Bioinform. Biol Insights 2017, 11, 1177932217712241. [Google Scholar] [CrossRef]
- Kwang-Hyun, C.; Jae Il, J.; Dongkwan, S.; Dongsan, K.; Sang-Min, P. The reverse control of irreversible biological processes. WIREs Syst. Biol. Med. 2016, 8, 366–377. [Google Scholar] [CrossRef]
- Banerji, C.R.; Miranda-Saavedra, D.; Severini, S.; Widschwendter, M.; Enver, T.; Zhou, J.X.; Teschendorff, A.E. Cellular network entropy as the energy potential in Waddington’s differentiation landscape. Sci Rep. 2013, 3, 3039. [Google Scholar] [CrossRef] [PubMed]
- Davies, P.C.; Demetrius, L.; Tuszynski, J.A. Cancer as a dynamical phase transition. Theor. Biol. Med. Model. 2011, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Busiello, D.M.; Suweis, S.; Hidalgo, J.; Maritan, A. Explorability and the origin of network sparsity in living systems. Sci. Rep. 2017, 7, 12323. [Google Scholar] [CrossRef]
- Shahriyari, L.; Komarova, N.L. Symmetric vs. Asymmetric Stem Cell Divisions: An Adaptation against Cancer? PLoS ONE 2013, 8, e76195. [Google Scholar] [CrossRef]
- Bajaj, J.; Zimdahl, B.; Reya, T. Fearful symmetry: Subversion of asymmetric division in cancer development and progression. Cancer Res. 2015, 75, 792–797. [Google Scholar] [CrossRef]
- Mukherjee, S.; Kong, J.; Brat, D.J. Cancer Stem Cell Division: When the Rules of Asymmetry Are Broken. Stem Cells Dev. 2015, 24, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Gómez-López, S.; Lerner, R.G.; Petritsch, C. Asymmetric cell division of stem and progenitor cells during homeostasis and cancer. Cell Mol. Life Sci. 2014, 71, 575–597. [Google Scholar] [CrossRef] [PubMed]
- Morrison, S.J.; Kimble, J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 2006, 441, 1068–1074. [Google Scholar] [CrossRef]
- Meacham, C.E.; Morrison, S.J. Tumour heterogeneity and cancer cell plasticity. Nature 2013, 501, 328–337. [Google Scholar] [CrossRef]
- Izumi, H.; Kaneko, Y. Symmetry breaking in human neuroblastoma cells. Mol. Cell Oncol. 2014, 1, e968510. [Google Scholar] [CrossRef][Green Version]
- Pillai, A.S.; Jirsa, V.K. Symmetry Breaking in Space-Time Hierarchies shapes Dynamics and Behavior. Neuron 2017, 94, 1010–1026. [Google Scholar] [CrossRef] [PubMed]
- Aufderheide, H.; Rudolf, L.; Gross, T. Mesoscale symmetries explain dynamical equivalence of food webs. New J. Phys. 2012, 14, 105014. [Google Scholar] [CrossRef]
- Xiao, Y.; MacArthur, B.D.; Wang, H.; Xiong, M.; Wang, W. Network quotients: structural skeletons of complex systems. Phys. Rev. E. 2008, 78, 046102. [Google Scholar] [CrossRef]
- MacArthur, B.D.; Sanchez-Garcia, R.J. Spectral characteristics of network redundancy. Phys. Rev. E. 2009, 80, 026117. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.H.; Wu, W.T.; Wang, H.; Xiong, M.; Wang, W. Symmetry-based stricture entropy of complex networks. Phys. A 2008, 387, 2611–2619. [Google Scholar] [CrossRef]
- Garrido, A. Symmetry in Complex Networks. Symmetry 2011, 3, 1–15. [Google Scholar] [CrossRef]
- Garlaschelli, D.; Ruzzenenti, F.; Basosi, R. Complex networks and symmetry I: A review. Symmetry 2010, 2, 1683–1709. [Google Scholar] [CrossRef]
- Comellas, F.; Gago, S. Synchronizability of complex networks. J. Phys. A Math. Theor. 2007, 40, 4483–4492. [Google Scholar] [CrossRef]
- Boström, J.; Sramkova, Z.; Salašová, A.; Johard, H.; Mahdessian, D.; Fedr, R.; Marks, C.; Medalová, J.; Souček, K.; Lundberg, E.; et al. Comparative cell cycle transcriptomics reveals synchronization of developmental transcription factor networks in cancer cells. PLoS ONE 2017, 12, e0188772. [Google Scholar] [CrossRef] [PubMed]
- Motter, A.E.; Zhou, C.; Kurths, J. Network synchronization, diffusion, and the paradox of heterogeneity. Phys. Rev. E. 2005, 71, 016116. [Google Scholar] [CrossRef] [PubMed]
- Nicosia, V.; Valencia, M.; Chavez, M.; Diaz-Guilera, A.; Latora, V. Remote synchronization reveals network symmetries and functional modules. Phys. Rev. Lett. 2013, 110, 174102. [Google Scholar] [CrossRef]
- Pecora, L.M.; Sorrentino, F.; Hagerstrom, A.M.; Murphy, T.E.; Roy, R. Cluster Synchronization and isolated desynchronization in complex networks with symmetries. Nat. Comm. 2014, 5, 4079. [Google Scholar] [CrossRef] [PubMed]
- Penn, Y.; Segal, M.; Moses, E. Network synchronization in hippocampal neurons. Proc. Natl. Acad. Sci. USA 2016, 113, 3341–3346. [Google Scholar] [CrossRef]
- Malagarriga, D.; Villa, A.E.P.; Garcia-Ojalvo, J.; Pons, A.J. Consistency of heterogeneous synchronization patterns in complex weighted networks. Chaos 2017, 27, 031102. [Google Scholar] [CrossRef]
- Cho, S.H.; Park, S.M.; Lee, H.S.; Lee, H.Y.; Cho, K.H. Attractor landscape analysis of colorectal tumorigenesis and its reversion. BMC Syst. Biol. 2016, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Taherian Fard, A.; Ragan, M.A. Modeling the Attractor Landscape of Disease Progression: A Network-Based Approach. Front. Genet. 2017, 8, 48. [Google Scholar] [CrossRef]
- Choi, M.; Shi, J.; Jung, S.H.; Chen, X.; Cho, K.H. Attractor landscape analysis reveals feedback loops in the p53 network that control the cellular response to DNA damage. Sci. Signal. 2012, 5, ra83. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Song, C.; Zhang, H.; Wu, Z.; Tian, X.J.; Xing, J. Epigenetic state network approach for describing cell phenotypic transitions. Interface Focus 2014, 4, 20130068. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Z.; Su, R.Q.; Huang, Z.G.; Wang, X.; Wang, W.X.; Grebogi, C.; Lai, Y.C. A geometrical approach to control and controllability of nonlinear dynamical networks. Nat. Commun. 2016, 7, 11323. [Google Scholar] [CrossRef]
- Kim, J.; Park, S.-M.; Cho, K.-H. Discovery of a kernel for controlling biomolecular regulatory networks. Sci. Rep. 2013, 3, 2223. [Google Scholar] [CrossRef] [PubMed]
- Cornelius, S.P.; Kath, W.L.; Motter, A.E. Realistic Control of Network Dynamics. Nat. Commun. 2013, 4, 1942. [Google Scholar] [CrossRef]
- Zhang, R.; Vinod Shah, M.; Yang, J.; Nyland, S.B.; Liu, X.; Yun, J.K.; Albert, R.; Loughran, T.P. Network model of survival signaling in large granular lymphocyte leukemia. Proc. Natl. Acad. Sci. USA 2008, 105, 16308–16313. [Google Scholar] [CrossRef] [PubMed]
- Saadatpour, A.; Wang, R.S.; Liao, A.; Liu, X.; Loughran, T.P.; Albert, I.; Albert, R. Dynamical and structural analysis of a T cell survival network identifies novel candidate therapeutic targets for large granular lymphocyte leukemia. PLoS Comput. Biol. 2011, 7, e1002267. [Google Scholar] [CrossRef]
- Maetschke, S.R.; Ragan, M.A. Characterizing cancer subtypes as attractors of Hopfield networks. Bioinformatics 2014, 30, 1273–1279. [Google Scholar] [CrossRef][Green Version]
- Kalman, R.E. Mathematical description of linear dynamical systems. J. Soc. Indus. Appl. Math. Ser. A 1963, 1, 152–192. [Google Scholar] [CrossRef]
- Luenberger, D.G. Introduction to Dynamic Systems: Theory, Models and Applications; J. Wiley & S.: New York, NY, USA, 1979. [Google Scholar]
- Liu, Y.Y.; Slotine, J.J.; Barabasi, A.L. Controllability of complex networks. Nature 2011, 473, 167–173. [Google Scholar] [CrossRef]
- Whalen, A.J.; Brennan, S.N.; Sauer, T.D.; Schift, S.J. Observability and controllability of nonlinear networks: the role of symmetries. Phys. Rev. X 2015, 5, 011005. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wang, W.X.; Liu, Y.Y.; Slotine, J.J. Intrinsic dynamics induce global symmetry in network controllability. Sci. Rep. 2015, 5, 8422. [Google Scholar] [CrossRef]
- Marras, E.; Travaglione, A.; Capobianco, E. Sub-modular resolution analysis by network mixture models. Stat. Appl. Genet. Mol. Biol. 2010, 9, 1544–6115. [Google Scholar] [CrossRef]
- Cejalvo, J.M.; Martínez de Dueñas, E.; Galván, P.; García-Recio, S.; Burgués Gasión, O.; Paré, L.; Antolín, S.; Martinello, R.; Blancas, I.; Adamo, B.; et al. Intrinsic Subtypes and Gene Expression Profiles in Primary and Metastatic Breast Cancer. Cancer Res. 2017, 77, 2213–2221. [Google Scholar] [CrossRef]
- Pons, P.; Latapy, M. Computing communities in large networks using random walks. In ISCIS’05 Proceedings of the 20th International Conference on Computer and Information Sciences, Istanbul, Turkey, 26–28 October 2005; Springer: Berlin/Heidelberg, Germany, 2005; Volume 3733, pp. 284–293. [Google Scholar]
- Raghavan, U.N.; Albert, R.; Kumara, S. Near linear time algorithm to detect community structures in large-scale networks. Phys. Rev. E 2007, 76, 036106. [Google Scholar] [CrossRef]
- Rosvall, M.; Bergstrom, C.T. Maps of random walks on complex networks reveal community structure. Proc. Natl. Acad. Sci. USA 2008, 105, 1118–1123. [Google Scholar] [CrossRef]
- Liang, L.; Ye, L.; Cui, D.; Yang, D. Gene expression signature associated with metastasis of stomach adenocarcinoma. Int. J. Clin. Exp. Med. 2017, 10, 3016–3026. [Google Scholar]
- Verfaillie, A.; Imrichova, H.; Atak, Z.K.; Dewaele, M.; Rambow, F.; Hulselmans, G.; Christiaens, V.; Svetlichnyy, D.; Luciani, F.; Van den Mooter, L.; et al. Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat. Commun. 2015, 6, 6683. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, H.; Rajurkar, M.; Li, Q.; Cotton, J.L.; Ou, J.; Zhu, L.J.; Goel, H.L.; Mercurio, A.M.; Park, J.-S.; et al. Tead and AP1 coordinate transcription and motility. Cell Rep. 2016, 14, 1169–1180. [Google Scholar] [CrossRef]
- Asgari, Y.; Salehzadeh-Yazdi, A.; Schreiber, F.; Masoudi-Nejad, A. Controllability in Cancer Metabolic Networks According to Drug Targets as Driver Nodes. PLoS ONE 2013, 8, e79397. [Google Scholar] [CrossRef]
- Srihari, S.; Raman, V.; Leong, H.W.; Ragan, M.A. Evolution and Controllability of Cancer Networks: A Boolean Perspective. IEEE/ACM Trans. Comput. Biol. Bioinform. 2014, 11, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Vinayagam, A.; Gibson, T.E.; Lee, H.J.; Yilmazel, B.; Roesel, C.; Hu, Y.; Kwon, Y.; Sharma, A.; Liu, Y.Y.; Perrimon, N.; et al. Controllability analysis of the directed human protein interaction network identifies disease genes and drug targets. Proc. Natl. Acad. Sci. USA 2016, 113, 4976–4981. [Google Scholar] [CrossRef]
- Sharma, A.; Cinti, C.; Capobianco, E. Multitype Network-Guided Target Controllability in Phenotypically Characterized Osteosarcoma: Role of Tumor Microenvironment. Front. Immunol. 2017, 8, 918. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Capobianco, E. Next Generation Networks: Featuring the Potential Role of Emerging Applications in Translational Oncology. J. Clin. Med. 2019, 8, 664. https://doi.org/10.3390/jcm8050664
Capobianco E. Next Generation Networks: Featuring the Potential Role of Emerging Applications in Translational Oncology. Journal of Clinical Medicine. 2019; 8(5):664. https://doi.org/10.3390/jcm8050664
Chicago/Turabian StyleCapobianco, Enrico. 2019. "Next Generation Networks: Featuring the Potential Role of Emerging Applications in Translational Oncology" Journal of Clinical Medicine 8, no. 5: 664. https://doi.org/10.3390/jcm8050664
APA StyleCapobianco, E. (2019). Next Generation Networks: Featuring the Potential Role of Emerging Applications in Translational Oncology. Journal of Clinical Medicine, 8(5), 664. https://doi.org/10.3390/jcm8050664



