Phylogeny, Resistome, and Virulome of Escherichia coli Causing Biliary Tract Infections
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Demographics, Clinical Data, and Follow Up
2.3. Antimicrobial Resistant Profile
2.4. Whole Genome Sequencing
2.5. Phylogenetic Analysis
2.6. Biofilm Formation Assay
2.7. Statistical Analysis
2.8. Nucleotide Sequence Accession Number
3. Results
3.1. Bacterial Isolates
3.2. Demographics and Clinical Data Analysis
3.3. Antimicrobial Resistance Profile
3.4. Genetic Characterization
3.4.1. Epidemiology
3.4.2. Virulome
3.4.3. Resistome
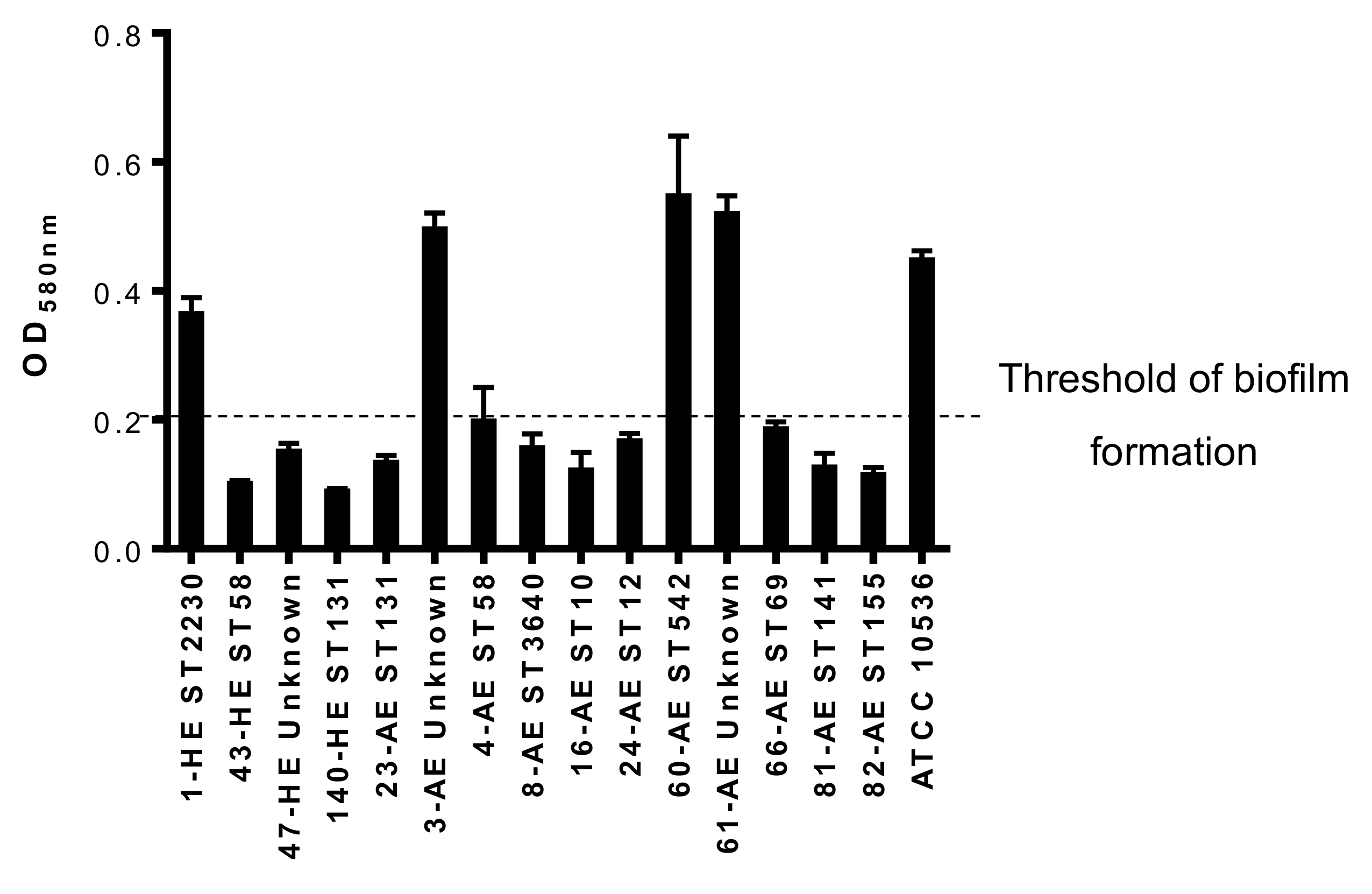
3.4.4. Biofilm Formation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rogers, B.A.; Sidjabat, H.E.; Paterson, D.L. Escherichia coli O25b-ST131: A pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 2011, 66, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lai, E.C.S.; Mok, F.P.T.; Tan, E.S.Y.; Lo, C.; Fan, S.; You, K.; Wong, J. Endoscopic biliary drainage for severe acute cholangitis. N. Engl. J. Med. 1992, 326, 1582–1586. [Google Scholar] [CrossRef] [PubMed]
- Englesbe, M.J.; Dawes, L.G. Resistant pathogens in biliary obstruction: Importance of cultures to guide antibiotic therapy. Hpb 2005, 7, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.W.; Liu, Y.L.; Lau, G.C.T.; Chan, R.C.Y.; Lai, A.C.W.; Ling, T.K.W.; Cheng, A.F. Bacteriologic analyses of bile and brown pigment stones in patients with acute cholangitis. Gastrointest. Endosc. 2001, 54, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Nicolas-Chanoine, M.H.; Bertrand, X.; Madec, J.Y. Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 2014, 27, 543–574. [Google Scholar] [CrossRef] [PubMed]
- Adams-Sapper, S.; Diep, B.A.; Perdreau-Remington, F.; Riley, L.W. Clonal composition and community clustering of drug-susceptible and -resistant Escherichia coli isolates from bloodstream infections. Antimicrob. Agents Chemother. 2013, 57, 490–497. [Google Scholar] [CrossRef]
- Manges, A.R.; Harel, J.; Masson, L.; Edens, T.J.; Portt, A.; Reid-Smith, R.J.; Zhanel, G.G.; Kropinski, A.M.; Boerlin, P. Multilocus sequence typing and virulence gene profiles associated with Escherichia coli from human and animal sources. Foodborne Pathog. Dis. 2015, 12, 302–310. [Google Scholar] [CrossRef]
- Petersen, A.M.; Nielsen, E.M.; Litrup, E.; Brynskov, J.; Mirsepasi, H.; Krogfelt, K.A. A phylogenetic group of Escherichia coli associated with active left-sided Inflammatory Bowel Disease. BMC Microbiol. 2009, 9, 171. [Google Scholar] [CrossRef]
- Dale, A.P.; Woodford, N. Extra-intestinal pathogenic Escherichia coli (ExPEC): Disease, carriage and clones. J. Infect. 2015, 71, 615–626. [Google Scholar] [CrossRef]
- Ron, E.Z. Distribution and evolution of virulence factors in septicemic Escherichia coli. Int. J. Med. Microbiol. 2010, 300, 367–370. [Google Scholar] [CrossRef]
- Hung, W.T.; Cheng, M.F.; Tseng, F.C.; Chen, Y.S.; Lee, S.S.J.; Chang, T.H.; Lin, H.H.; Hung, C.H.; Wang, J.L. Bloodstream infection with extended-spectrum beta-lactamase-producing Escherichia coli: The role of virulence genes. J. Microbiol. Immunol. Infect. 2019. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.C.; Tseng, C.C.; Wu, A.B.; Huang, J.J.; Sheu, B.S.; Wu, J.J. Different roles of host and bacterial factors in Escherichia coli extra-intestinal infections. Clin. Microbiol. Infect. 2009, 15, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Tarchouna, M.; Ferjani, A.; Ben-Selma, W.; Boukadida, J. Distribution of uropathogenic virulence genes in Escherichia coli isolated from patients with urinary tract infection. Int. J. Infect. Dis. 2013, 17, e450–e453. [Google Scholar] [CrossRef] [PubMed]
- Ciesielczuk, H.; Betts, J.; Phee, L.; Doumith, M.; Hope, R.; Woodford, N.; Wareham, D.W. Comparative virulence of urinary and bloodstream isolates of extra-intestinal pathogenic Escherichia coli in a Galleria mellonella model. Virulence 2015, 6, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Razaghi, M.; Tajeddin, E.; Ganji, L.; Alebouyeh, M.; Alizadeh, A.H.M.; Sadeghi, A.; Zali, M.R. Colonization, resistance to bile, and virulence properties of Escherichia coli strains: Unusual characteristics associated with biliary tract diseases. Microb. Pathog. 2017, 111, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Tseng, C.; Chen, C.; Wu, J.; Huang, J. The Role of bacterial virulence and host factors in patients with Escherichia coli bacteremia who have acute cholangitis or upper urinary tract infection. Clin. Infect. Dis. 2002, 35, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Kochar, R.; Banerjee, S. Infections of the Biliary Tract. Gastrointest. Endosc. Clin. N. Am. 2013, 23, 199–218. [Google Scholar] [CrossRef]
- Cherkaoui, A.; Hibbs, J.; Emonet, S.; Tangomo, M.; Girard, M.; Francois, P.; Schrenzel, J. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol. 2010, 48, 1169–1175. [Google Scholar] [CrossRef]
- Solomkin, J.S.; Mazuski, J.E.; Bradley, J.S.; Rodvold, K.A.; Goldstein, E.J.; Baron, E.J.; O’Neill, P.J.; Chow, A.W.; Dellinger, E.P.; Eachempati, S.R.; et al. Diagnosis and management of complicated intra-abdominal infection in adults and children: Guidelines by the Surgical Infections Society and the infectious Diseases Society of America. Clin. Infect. Dis. 2010, 50, 133–164. [Google Scholar] [CrossRef]
- Charlson, M.E.; Pompei, P.; Ales, K.L.; MacKenzie, C.R. A new method of classifying prognostic comorbidity in longitudinal studies: Development and validation. J. Chronic. Dis. 1987, 40, 373–383. [Google Scholar] [CrossRef]
- McCabe, W.R.; Jackson, G.G. Gram-Negative bacteriemia. Arch. Intern. Med. 1962, 110, 847–855. [Google Scholar] [CrossRef]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, K.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing. Recommendations for MIC Determination of Colistin (Polymyxin E) As Recommended by the Joint CLSI-EUCAST Polymyxin Breakpoints Working Group; European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2016. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters; European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2019. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012, 40, W569–W572. [Google Scholar] [CrossRef]
- Rodríguez-Villodres, Á.; Gil-Marqués, M.L.; Álvarez-Marín, R.; Bonnin, R.A.; Pachón-Ibáñez, M.E.; Aguilar-Guisado, M.; Naas, T.; Aznar, J.; Pachón, J.; Lepe, J.A.; et al. Extended-spectrum resistance to β-lactams/β-lactamase inhibitors (ESRI) evolved from low-level resistant Escherichia coli. J. Antimicrob. Chemother. 2019. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Vila-Farrés, X.; Parra-Millán, R.; Sánchez-Encinales, V.; Varese, M.; Ayerbe-Algaba, R.; Bayó, N.; Guardiola, S.; Pachón-Ibáñez, M.E.; Kotev, M.; García, J.; et al. Combating virulence of Gram-negative bacilli by OmpA inhibition. Sci. Rep. 2017, 7, 14683. [Google Scholar] [CrossRef]
- Hertz, F.B.; Nielsen, J.B.; Schønning, K.; Littauer, P.; Knudsen, J.D.; Løbner-Olesen, A.; Frimodt-Møller, N. Population structure of drug-susceptible, resistant and ESBL-producing Escherichia coli from community-acquired urinary tract infections. BMC Microbiol. 2016, 16, 63. [Google Scholar]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin. Microbiol. Rev. 2019, 32, e00135-18. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Villodres, Á.; Ortiz de la Rosa, J.M.; Álvarez-Marín, R.; Pachón, J.; Aznar, J.; Lepe, J.A.; Smani, Y. Heteroresistance to piperacillin-tazobactam in clinical isolates of Escherichia coli sequence type 131. Antimicrob. Agents. Chemother. 2017, 62, e01923-17. [Google Scholar] [CrossRef] [PubMed]
- Cantón, R.; Loza, E.; Aznar, J.; Castillo, F.J.; Cercenado, E.; Fraile-Ribot, P.A.; González-Romo, F.; López-Hontangas, J.L.; Rodríguez-Lozano, J.; Suárez-Barrenechea, A.I.; et al. Monitoring the antimicrobial susceptibility of Gram-negative organisms involved in intraabdominal and urinary tract infections recovered during the SMART study (Spain, 2016 and 2017). Rev. Esp. Quimioter. 2019, 32, 145–155. [Google Scholar] [PubMed]
- Tajeddin, E.; Sherafat, S.J.; Majidi, M.R.; Alebouyeh, M.; Alizadeh, A.H.; Zali, M.R. Association of diverse bacterial communities in human bile samples with biliary tract disorders: A survey using culture and polymerase chain reaction-denaturing gradient gel electrophoresis methods. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1331–1339. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Romero, N.; Romero-Gómez, M.P.; Mora-Rillo, M.; Rodríguez-Baño, J.; López-Cerero, L.; Pascual, A.; Mingorance, J. Uncoupling between core genome and virulome in extraintestinal pathogenic Escherichia coli. Can. J. Microbiol. 2015, 61, 647–652. [Google Scholar] [CrossRef]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-Ploskonska, G.; Choroszy-Krol, I. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent reports. Gut Pathog. 2019, 11, 10. [Google Scholar] [CrossRef]
- Shin, S.; Castanie-Cornet, M.P.; Foster, J.W.; Crawford, J.A.; Brinkley, C.; Kaper, J.B. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microb. 2001, 41, 1133–1150. [Google Scholar] [CrossRef]

| Variable | BTI Patients (n = 12) | Non-BTI Patients (n = 20) | P-Value |
|---|---|---|---|
| Age, median (range) | 64 (58–72.5) | 64.5 (55.75–77.75) | 0.893 |
| Gender (female), n (%) | 7 (58.3) | 10 (50.0) | 0.647 |
| Charlson Score, median (range) | 1.5 (0–3.75) | 3 (0.25–6) | 0.146 |
| McCabe Score ultimately or rapidly fatal, n (%) | 2 (16.6) | 11 (55.0) | 0.062 |
| Acquisition, n (%) | |||
| Community | 9 (75.0) | 6 (30.0) | 0.035 |
| Healthcare | 2 (16.6) | 5 (25.0) | |
| Nosocomial | 1 (8.3) | 9 (45.0) | |
| Sepsis or septic shock, n (%) | 3 (25.0) | 10 (50.0) | 0.267 |
| Previous antibiotic exposure, n (%) | 7 (58.3) | 13 (65.0) | 0.724 |
| Days of treatment, median (range) | 12.5 (5–21) | 10 (8–13) | 0.329 |
| Death, n (%) | 3 (25.0) | 3 (15.0) | 0.647 |
| BTI E. coli (n = 15) | Non-BTI E. coli (n = 21) | P-Value | |
|---|---|---|---|
| Ampicillin, n (%) | 11 (73.3) | 19 (90.4) | 0.210 |
| Amoxicillin-clavulanic acid, n (%) | 4 (26.6) | 6 (28.6) | 1.000 |
| Piperacillin-tazobactam, n (%) | 2 (13.3) | 1 (4.7) | 0.559 |
| Cefuroxime, n (%) | 1 (6.6) | 3 (14.3) | 0.626 |
| Cefotaxime, n (%) | 0 (0.0) | 2 (9.5) | 0.500 |
| Ceftazidime, n (%) | 0 (0.0) | 2 (9.5) | 0.500 |
| Cefepime, n (%) | 0 (0.0) | 2 (9.5) | 0.500 |
| Imipenem, n (%) | 0 (0.0) | 0 (0.0) | 1.000 |
| Meropenem, n (%) | 0 (0.0) | 0 (0.0) | 1.000 |
| Ciprofloxacin, n (%) | 2 (13.3) | 7 (33.3) | 0.252 |
| Levofloxacin, n (%) | 2 (13.3) | 7 (33.3) | 0.252 |
| Gentamicin, n (%) | 1 (6.6) | 2 (9.5) | 1.000 |
| Tobramycin, n (%) | 1 (6.6) | 2 (9.5) | 1.000 |
| Amikacin, n (%) | 0 (0.0) | 0 (0.0) | 1.000 |
| Cotrimoxazole, n (%) | 4 (26.6) | 6 (28.5) | 1.000 |
| Tigecycline, n (%) | 0 (0.0) | 0 (0.0) | 1.000 |
| Colistin, n (%) | 0 (0.0) | 0 (0.0) | 1.000 |
| ESBL, n (%) | 0 (0.0) | 2 (9.5) | 0.500 |
| MDR, n (%) | 2 (13.3) | 5 (23.8) | 0.674 |
| Strain | Source | adk | fumC | gyrB | icd | mdh | purA | recA | ST | ST Complex |
|---|---|---|---|---|---|---|---|---|---|---|
| 1-HE | Blood | 6 | 346 | 12 | 1 | 20 | 13 | 7 | 2230 | 23 |
| 43-HE ^ | Blood | 6 | 4 | 4 | 16 | 24 | 8 | 14 | 58 | 155 |
| 47-HE | Blood | 6 | 95 | 15 | 18 | 9 | 8 | 6 | Unknown | - |
| 140-HE * | Blood | 53 | 40 | 47 | 13 | 36 | 28 | 29 | 131 | 131 |
| 23-AE * | bile | 53 | 40 | 47 | 13 | 36 | 28 | 29 | 131 | 131 |
| 3-AE | Bile | 13 | 6 | 15 | 13 | 16 | 10 | 122 | Unknown | - |
| 4-AE ^ | Bile | 6 | 4 | 4 | 16 | 24 | 8 | 14 | 58 | 155 |
| 8-AE | Bile | 6 | 23 | 3 | 16 | 9 | 8 | 6 | 3640 | - |
| 16-AE | Bile | 10 | 11 | 4 | 8 | 8 | 8 | 2 | 10 | 10 |
| 24-AE | Bile | 13 | 13 | 9 | 13 | 16 | 10 | 9 | 12 | 12 |
| 60-AE ~ | Bile | 112 | 11 | 5 | 12 | 8 | 8 | 86 | 542 | - |
| 61-AE ~ | Bile | 112 | 40 | 4 | 12 | 8 | 8 | 86 | Unknown | - |
| 66-AE | Bile | 21 | 35 | 27 | 6 | 5 | 5 | 4 | 69 | 69 |
| 81-AE | Bile | 13 | 52 | 10 | 14 | 17 | 25 | 17 | 141 | - |
| 82-AE | Bile | 6 | 4 | 14 | 16 | 24 | 8 | 14 | 155 | 155 |
| Strain | 1-HE | 43-HE ^ | 47-HE | 140-HE * | 23-AE * | 3-AE | 4-AE ^ | 8-AE | 16-AE | 24-AE | 60-AE ~ | 61-AE ~ | 66-AE | 81-AE | 82-AE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source | Blood | Blood | Blood | Blood | Bile | Bile | Bile | Bile | Bile | Bile | Bile | Bile | Bile | Bile | Bile |
| iss | + | + | + | + | + | + | + | + | |||||||
| ipfA | + | + | + | + | + | + | + | + | + | ||||||
| mchC | + | + | + | ||||||||||||
| mchB | + | + | |||||||||||||
| mchF | + | + | + | + | + | + | + | ||||||||
| iroN | + | + | + | + | + | + | + | + | |||||||
| tsh | + | ||||||||||||||
| gad | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| astA | + | + | + | ||||||||||||
| senB | + | + | + | ||||||||||||
| iha | + | + | |||||||||||||
| air | + | ||||||||||||||
| ailA | + | ||||||||||||||
| mcmA | + | + | |||||||||||||
| vat | + | + | |||||||||||||
| ireA | + | ||||||||||||||
| papG | + | + | |||||||||||||
| sfa | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| fimH | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| iutA | + | + | + | + | + | + | + | + | |||||||
| ipaH | + | ||||||||||||||
| cnf1 | + | ||||||||||||||
| hlyA | |||||||||||||||
| cdtB | |||||||||||||||
| stx-1 | |||||||||||||||
| stx-2 | |||||||||||||||
| eaeA | |||||||||||||||
| bfp | |||||||||||||||
| Total | 10 | 8 | 5 | 4 | 3 | 8 | 3 | 3 | 6 | 14 | 3 | 10 | 12 | 9 | 8 |
| Strain | Source | strA | blaTEM | catA | sul | tet | dfrA | aph | gyrA | parC | parE | aadA | mph |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-HE | Blood | + | 1a | + | |||||||||
| 43-HE^ | Blood | 1b | |||||||||||
| 47-HE | Blood | 1a | + | + | |||||||||
| 140-HE * | Blood | 1b | + | + | +(I529L) | + | + | ||||||
| 23-AE * | Bile | 1b | + | + | +(I529L) | + | + | ||||||
| 3-AE | Bile | + | 1b | + | + | + | + | ||||||
| 4-AE ^ | Bile | +(S83L) | |||||||||||
| 8-AE | Bile | ||||||||||||
| 16-AE | Bile | 1b | + | + | |||||||||
| 24-AE | Bile | ||||||||||||
| 60-AE ~ | Bile | 1b | +(S83L) | ||||||||||
| 61-AE ~ | Bile | + | + | + | +(S83L) | +(I529L) | + | ||||||
| 66-AE | Bile | 1b | + | + | + | ||||||||
| 81-AE | Bile | ||||||||||||
| 82-AE | Bile | 1b | + | + | + | + | + | +(S83L) (D87N) | +(S80I) | + |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Villodres, Á.; Bonnin, R.A.; Ortiz de la Rosa, J.M.; Álvarez-Marín, R.; Naas, T.; Aznar, J.; Pachón, J.; Lepe, J.A.; Smani, Y. Phylogeny, Resistome, and Virulome of Escherichia coli Causing Biliary Tract Infections. J. Clin. Med. 2019, 8, 2118. https://doi.org/10.3390/jcm8122118
Rodríguez-Villodres Á, Bonnin RA, Ortiz de la Rosa JM, Álvarez-Marín R, Naas T, Aznar J, Pachón J, Lepe JA, Smani Y. Phylogeny, Resistome, and Virulome of Escherichia coli Causing Biliary Tract Infections. Journal of Clinical Medicine. 2019; 8(12):2118. https://doi.org/10.3390/jcm8122118
Chicago/Turabian StyleRodríguez-Villodres, Ángel, Rémy A. Bonnin, José Manuel Ortiz de la Rosa, Rocío Álvarez-Marín, Thierry Naas, Javier Aznar, Jerónimo Pachón, José Antonio Lepe, and Younes Smani. 2019. "Phylogeny, Resistome, and Virulome of Escherichia coli Causing Biliary Tract Infections" Journal of Clinical Medicine 8, no. 12: 2118. https://doi.org/10.3390/jcm8122118
APA StyleRodríguez-Villodres, Á., Bonnin, R. A., Ortiz de la Rosa, J. M., Álvarez-Marín, R., Naas, T., Aznar, J., Pachón, J., Lepe, J. A., & Smani, Y. (2019). Phylogeny, Resistome, and Virulome of Escherichia coli Causing Biliary Tract Infections. Journal of Clinical Medicine, 8(12), 2118. https://doi.org/10.3390/jcm8122118








